Abstract
Previous studies have identified sin mutations that alleviate the requirement for the yeast SWI/SNF chromatin remodelling complex, which include point changes in the yeast genes encoding core histones. Here we characterise the biochemical properties of nucleosomes bearing these mutations. We find that sin mutant nucleosomes have a high inherent thermal mobility. As the SWI/SNF complex can alter nucleosome positioning, the higher mobility of sin mutant nucleosomes provides a means by which sin mutations may substitute for SWI/SNF function. The location of sin mutations also provides a new opportunity for insights into the mechanism for nucleosome mobilisation. We find that both mutations altering histone DNA contacts at the nucleosome dyad and mutations in the dimer–tetramer interface influence nucleosome mobility. Furthermore, incorporation of H2A.Z into nucleosomes, which also alters dimer–tetramer interactions, affects nucleosome mobility. Thus, variation of histone sequence or subtype provides a means by which eukaryotes may regulate access to chromatin through alterations to nucleosome mobility.
Keywords: ATP-dependent chromatin remodelling, nucleosome mobility, SWI/SNF, Sin mutation
Introduction
Nucleosomes are the universal molecular packaging state of DNA in nuclei. They are responsible for compacting eukaryotic genomes to allow them to fit into the limited volume of the cell nucleus. The nucleosome core particle and an additional variable length of unbound linker DNA together comprise the fundamental repeating unit of chromatin (Kornberg, 1974). The nucleosome core particle consists of a core of eight polypeptides, two copies each of the four highly conserved histone proteins H2A, H2B, H3 and H4, around which 147 bp of DNA are wrapped in 1.7 superhelical turns (Luger et al, 1997). H3 and H4 associate to form histone fold dimers as do histones H2A and H2B. Each histone fold dimer associates to form an octameric spiral with dyad symmetry. The H2A−H2B histone fold dimer units are less stably associated within the octamer than the two H3−H4 histone fold dimers (Eickbush and Moudrianakis, 1978). A number of extra protein elements decorate the regular spiral of histone fold dimers, including unstructured ‘tails' that extend outside the DNA superhelix, the additional H3 αN helix that organises the most exterior turn of bound DNA, a structured H2A C-terminal extension that passes over the top face of the histone octamer, and an H2B αC helix lying above the histone dimer (Luger and Richmond, 1998).
A consequence of the organisation of DNA into nucleosomes is that all genetic processes must contend with a chromatin substrate. In the case of gene regulation, wrapping into nucleosomes makes DNA sequences differentially accessible to transcription factors (Owen-Hughes and Workman, 1994). Accessibility of a particular site depends on the absolute ‘position' of the nucleosome (Polach and Widom, 1995), and many cases have been recognised in which nucleosome positioning affects genomic accessibility (Simpson, 1990; Lomvardas and Thanos, 2002; Miller and Widom, 2003). In this way, nucleosomes are active participants in the regulatory pathways and a range of mechanisms exist to influence their positioning.
One class of activities that can directly manipulate nucleosome positioning are the ATP-dependent chromatin remodelling complexes (Becker and Horz, 2002). These complexes comprise a central helicase-related, ATP-consuming subunit with various numbers of associated subunits that modulate and target its activity. Although their exact mechanism of action remains unknown, a common property shared by almost all such complexes is the ability to alter the positioning of nucleosomes along DNA (Hamiche et al, 1999; Langst et al, 1999; Whitehouse et al, 1999; Jaskelioff et al, 2000). ATP-dependent chromatin remodelling complexes are ubiquitous in eukaryotes, with multiple forms of the core ATP motor subunit and various ways of combining the associated subunits. For example, Saccharomyces cerevisiae encodes at least six different Snf2-related proteins that are catalytic subunits of ATP-dependent chromatin remodelling activities (Shiratori et al, 1999). Analysis of the function of these proteins has revealed involvement in processes ranging from transcription to DNA replication (Becker and Horz, 2002).
The genes that encode the S. cerevisiae SWI/SNF complex were first identified from parallel screens for defects in yeast mating type SWItching, and Sucrose Non Fermentation (Neigeborn and Carlson, 1984; Stern et al, 1984; Breeden and Nasmyth, 1987). It is now clear that the SWI/SNF complex is involved in the regulation of a significant subset of yeast genes (Holstege et al, 1998; Sudarsanam et al, 2000). Importantly, mutations that result in the loss of SWI/SNF function can be suppressed by mutations in several classes of proteins with generalised roles in gene expression. These were originally labelled SIN or SDI because they conferred Swi/Snf Independence (Nasmyth et al, 1987; Sternberg et al, 1987; Winston and Carlson, 1992). The originally identified sin mutants include mutations to the genes encoding SIN1 (Kruger and Herskowitz, 1991), an HMG1-like factor; SIN3, a component of the RPD3 histone deacetylase complex (Kadosh and Struhl, 1997); and SIN4, a component of the Pol II holoenzyme complex (Li et al, 1995; Song et al, 1996). Sin2 was found to be a point mutant of a gene encoding histone H3, and a number of other mutations in histones H3 and H4 have since been identified using genetic screens (Kruger et al, 1995). SIN-related phenotypes have also been reported for the targeted mutation of tyrosines of H4 and H2B involved in inter-histone interactions and deletion of the H2B N-terminal tail, and also from a number of H2A mutations revealed by a genetic screen (Hirschhorn et al, 1995; Santisteban et al, 1997; Recht and Osley, 1999). Taken together, these observations suggest that the structure of the histone octamer plays a central role in the mechanism by which SWI/SNF acts.
Broadly, the known core histone sin mutations fall into two groups when located on the nucleosome core particle structure (Figure 1). Mutations to H4 R45, H3 T118 and H3 R116 are very near the dyad DNA contact points at SHL0.5, whereas other sin mutation sites are more distant, mainly near tetramer–dimer packing interfaces. It is widely accepted that core histone sin mutations affect chromatin structure, yet without altering the stoichiometry of the histone octamer (Kurumizaka and Wolffe, 1997; Wechser et al, 1997; Horn et al, 2002) or the overall wrapping of DNA (Kurumizaka and Wolffe, 1997). The most significant changes observed to date involve alterations to generalised DNA accessibility. This has been observed for micrococcal nuclease (Santisteban et al, 1997; Wechser et al, 1997; Recht and Osley, 1999; Fleming and Pennings, 2001), Dam methyltransferase (Wechser et al, 1997), DNaseI (Kurumizaka and Wolffe, 1997), hydroxy radicals (Kurumizaka and Wolffe, 1997) and restriction enzymes (Horn et al, 2002). A lower chromatin-dependent superhelical density was reported in vivo for H4 R45H (Wechser et al, 1997), although this was not found for H2B mutants in vivo (Recht and Osley, 1999) or H3 mutants in vitro (Kurumizaka and Wolffe, 1997).
Figure 1.
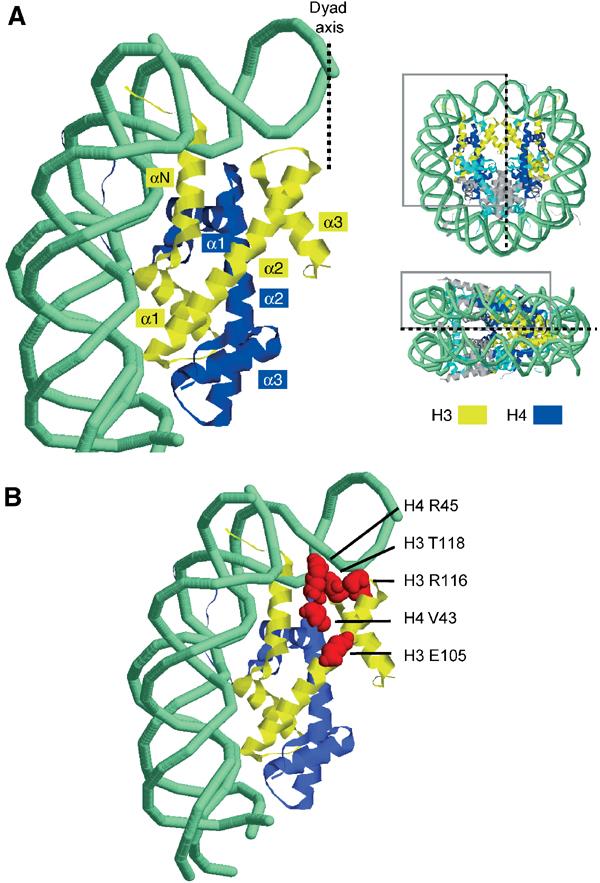
Location of core histone SIN mutants. (A) Overview showing dyad and DNA entry–exit region of nucleosome. Histone H3 and H4 ribbons are coloured yellow and blue, respectively. (B) Location of SIN mutations observed by Kruger et al. Indicated residues are red space-filled.
Although core histone sin mutations have been studied for over a decade, it has remained unclear how changes in DNA accessibility can suppress defects in SWI/SNF function. Earlier researchers had suggested that the mutant histones spontaneously generated a remodelled state of the nucleosome. However, careful analysis of the accessibility of nucleosomal DNA revealed that this was not the case. Instead it was found that there is a subtle increase in accessibility towards the outer edges of the nucleosome (Kurumizaka and Wolffe, 1997). As a potential explanation for this, it has been proposed that alterations to the nucleosome dyad might affect the association of DNA at the entry–exit point in the nucleosome. Horn et al (2002) developed this into the specific proposal that mutations on the central H3–H4 histone fold interacting directly with dyad DNA could cause changes in local DNA conformation, which would in turn disturb direct interactions of H3 αN helix with the same DNA. The H3 αN helix also controls the most terminal DNA-binding sites in the nucleosome core at SHL6.5. Since Horn et al (2002) observed that nucleosomal arrays containing H4 R45H adopt a condensed configuration less readily, they proposed that this might occur as a result of the altered DNA entry–exit trajectory, with the implication that such destabilisation of higher order chromatin structure compensates for loss of SWI/SNF activity.
Over recent years it has become clear that a shared property of many ATP-dependent chromatin remodelling complexes is the ability to redistribute nucleosomes along DNA. Nevertheless, isolated nucleosomes have long been recognised to slide along DNA in the absence of these complexes in a thermal energy-driven process (Meersseman et al, 1992). Because core histone sin mutations allow S. cerevisiae to grow in the absence of functional SWI/SNF remodelling complex, we investigated whether these mutations altered the thermal mobility of nucleosomes. We find that sin mutations reduce the temperature required for thermal nucleosome redistribution. Thus, the increased thermal mobility of sin mutant nucleosomes provides a simple means by which these mutations may substitute the requirement for the SWI/SNF complex.
Results
Quantitative assay for nucleosome mobilisation
In order to achieve quantitative measurements of mobilisation, nucleosomes were assembled onto a 255 bp fragment from the nucA region of mouse mammary tumour virus long terminal repeat (MMTV LTR). This was designed such that 54 bp extensions flank the 147 bp occupied by the nucleosome core and is referred to as 54A54. In order to confirm that nucleosomes occupy this location, we monitored nucleosome positioning by native gel electrophoresis and site-directed mapping. Site-directed mapping involves the attachment of an iron chelating compound to sites close to the dyad and enables the positions of nucleosomes to be mapped with base pair precision (Flaus et al, 1996). Following chromatin assembly by salt dialysis, site-directed mapping results in the generation of a pair of cleavage sites which indicate that the majority of nucleosomes are assembled as expected such that their dyad is located at +70 relative to the MMTV transcriptional start site (Figure 2A, lane 2). Consistent with this, nucleosomes assembled onto this DNA fragment migrate predominantly as a single species of relatively low mobility (Figure 2B, lane 1). A minor population of nucleosomes is also observed 18 bp upstream of the main position (labelled ‘p'). When nucleosomes assembled onto this DNA fragment are subject to thermal incubation for increasing times, they redistribute to positions +22 and +125, 48 bp upstream and 55 bp downstream of the original position, respectively (Figure 2A, lanes 3–5). Native gel electrophoresis of the same samples shows that the initial centrally located species is transformed into two faster migrating positions (Figure 2B, lanes 1–4). No obvious accumulation of intermediates was observed.
Figure 2.
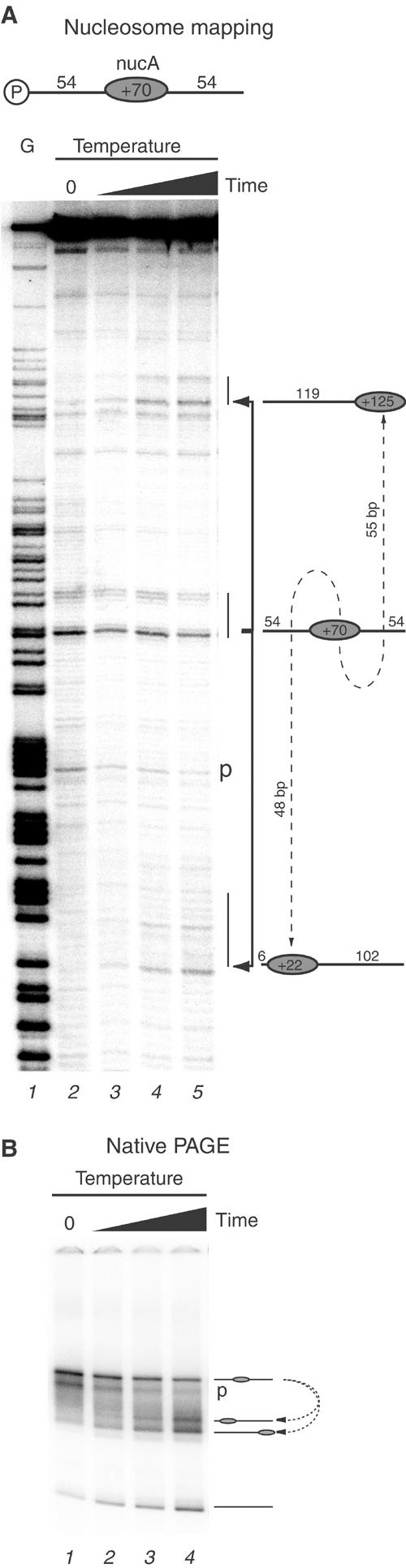
Site-directed hydroxyl radical mapping of nucleosome positions on the template 54A54, which includes 54 bp extensions on either side of a centrally located MMTV nucleosome A. (A) Initial nucleosome position at +70 (lane 2) and subsequent time course of thermal incubation for 3, 6, 12 and 24 min (lanes 3–5). Lane 1 is a G sequencing ladder. (B) Native PAGE of same samples as in (A).
Introduction of sin mutations alters the inherent thermal mobility of nucleosomes
The well-separated and relatively homogenous initial position to which nucleosomes are deposited on this fragment, together with the defined products of thermal shifting, make it possible to accurately quantitate nucleosome movement using native gel electrophoresis. Under standard conditions with 50 mM Tris–HCl (pH 7.5) and 150 mM NaCl buffer, we measured the rate of change in the proportion of initial and final positions for wild-type (WT) nucleosomes and those containing histone H4 substitution Arg45 → His (H4 R45H). Figure 3A shows that at 46°C H4 R45H nucleosomes migrated from their initial location considerably faster than the control nucleosomes. As over half of the H4 R45H nucleosomes had vacated their initial location within 3 min at 46°C, a series of thermal redistribution time-course reactions were carried out at a range of different temperatures. These show that there is a 4- to 6-fold difference in the rate of nucleosome movement between WT and H4 R45H across the temperature range (Figure 3B).
Figure 3.
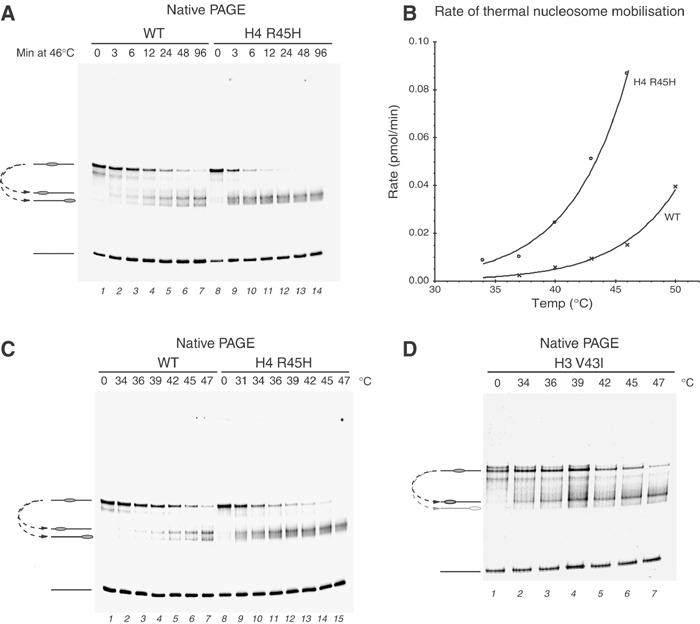
Comparison of rates of thermal mobilisation of WT and H4 R45H-containing nucleosomes. (A) Time course of nucleosome sliding at 46°C for the times shown. (B) Plot of initial rates of nucleosome sliding at different temperatures for WT (crosses) and H4 R45H (open circles). (C) Temperature comparison of nucleosome sliding rates in 60 min at the temperatures indicated. (D) Thermal shifting of nucleosomes bearing the H3 V43I mutation illustrating the preferential accumulation of one product.
Because the observed rates differed by such a significant amount at any particular temperature, we chose instead to determine the temperature sufficient for mobilising 50% of the nucleosomes in 60 min (i.e. a rate of 0.08 pmol/min under our experimental conditions). This characteristic temperature for each nucleosome variant was determined by incubating identical reaction aliquots in parallel across a range of temperatures followed by gel electrophoresis, as shown for H4 R45H in Figure 3C. A plot of the proportion of nucleosomes shifted at each temperature was then used to calculate the temperature required to shift 50% of the nucleosomes under these conditions. In the case of R45H mutation, the temperature required for thermal shifting is 33°C, which is a substantial 10°C lower than that for the WT octamer (43°C; Table I). Seven replicates of the thermal shifting reaction with WT nucleosomes assembled from three independently refolded histone octamer preparations showed the variation in measurements to be less than 1°C (data not shown). Thus, the thermal mobilisation assay provides a means of measuring differences in the inherent thermal mobility of nucleosomes with an accuracy of approximately 1°C.
Table 1.
Observed thermal mobilisation of original sin mutant containing nucleosomes on 54A54, and temperature for 50% sliding in 60 min (column 3) and ratio of +22 to +125 products (column 4)
| Histone |
Thermal mobilisation |
Relative in vivo expression |
|||
|---|---|---|---|---|---|
| Mutation | Allele | Temp. for 50% shifting (°C) | Ratio upstream to downstream product | HO-LacZ, sole source of histone (Wechser et al, 1997) | Pho5 (Wechser et al, 1997) |
| WT | 43 | 1.2 | |||
| H4 V43I | hhf2-8 | 42 | 2.5 | 84 | 8 |
| H3 E105K | hht2-2 | 40 | 1.2 | 2 | |
| H3 R116H | hht1-1/hht2-1 | 39 | 3.3 | ||
| H3 T118I | hht2-3 | 34 | 5.0 | 3 | |
| H4 R45H | hhf2-13 | 33 | 1.2 | 66 | 31 |
| H4 R45C |
hhf2-7 |
33 |
1.3 |
100 |
23 |
| Corresponding allele names used by Kruger et al, together with in vivo HO-LacZ and Pho5 reporter assay results (Wechser et al, 1997) are listed in columns 2, 5 and 6, respectively. | |||||
The H4 R45H mutation was characterised initially as it was identified in the initial screen for suppressors of defects in the SWI/SNF complex (Kruger et al, 1995) and has a substantial effect on HO expression when present as the sole source of histone H4 in yeast (Wechser et al, 1997). Additional mutations were identified in the initial genetic screens and the effect of these mutations on nucleosome mobility was investigated next. All products formed unique bands similar to those of WT and the effects on thermal mobility are summarised in Table I. With the exception of H4 V43I, all of the sin mutants tested significantly reduced the temperature required for thermal nucleosome movement. The magnitude of the effect ranged from a substantial change of approximately 10°C for the H3 T118I, H4 R45C and H4 R45H mutations, to less severe, but still significant changes of 3 and 4°C for H3 E105K and H3 R116H, respectively.
We also noted that the products of the thermal shifting reaction differed for some of the nucleosomes containing sin mutations. For example, the V43I mutation alters the relative proportions of the two products obtained from the thermal shifting reaction (Figure 3C and D). As a subset of sin mutations were found to alter the directionality of nucleosome mobilisation, these data are included in Tables I and II. In order to confirm that these products represent nucleosomes located at the +22 and +125 locations as observed with WT octamer, one of the sin mutations that caused asymmetric accumulation of products, H3 R116H, was combined with H4 to which the site-directed mapping reagent had been attached. Site-directed mapping confirmed that these nucleosomes moved to the same locations as WT nucleosomes, although a higher proportion accumulated at the +22 location (Supplementary Figure 1). The preferential accumulation of nucleosomes at one end of the fragment could occur as a result of the preferential movement of sin mutant nucleosomes in the upstream direction, or be caused by a difference in the stability of nucleosomes to dissociation. In order to test this latter possibility, we monitored the accumulation of each species during incubation at successively higher temperatures. The results (shown in Supplementary Figure 2) indicate that preferential dissociation of one species is unlikely to explain the asymmetric redistribution.
Table 2.
Observed thermal mobilisation of selected histone variants, and temperature for 50% sliding in 60 min (column 2) and ratio of +22 to +125 products (column 3)
| Histone mutation | Temp. for 50% shifting (°C) | Ratio upstream to downstream product |
|---|---|---|
| xWT | 43 | 1.2 |
| xH2A L115S | 37 | 1.1 |
| xH2A L116C | 39 | 1.4 |
| xH2A.Z | 39 | 1.1 |
| hWT | 43 | 1.6 |
| hH3.3 | 42 | 2.1 |
| xH4 S47C | 42 | 1.1 |
In order to compare the changes in mobility we have detected to the severity of sin mutations in vivo, we have compiled data from the previously published functional characterisation of sin mutations. These are also summarised in Table I. One problem in assembling these data is that the effects of significant numbers of sin mutations have not been monitored quantitatively in the absence of the corresponding WT protein under otherwise controlled conditions for many different reporter genes. Despite the fact that not all the desirable information is available, it is clear that sin mutations differ in their severity depending upon the assay used to monitor function. For example, the H4 V43I mutation is effective in overcoming the requirement for SWI/SNF at the HO promoter, but does not have much effect on expression of the PHO5 promoter in the presence of phosphate (Table I; Wechser et al, 1997). Thus, the effect of sin mutations varies depending on the context, suggesting that the sensitivity of specific genes to different attributes of chromatin structure varies. The effect of sin mutations on thermal mobility also varies, but overall there is a reasonable correlation between the severity of sin mutations and the degree to which they can suppress the requirement for the SWI/SNF complex.
Sin mutations alter nucleosome mobility under different ionic conditions
The original observations of thermal nucleosome mobilisation in vitro by Bradbury and colleagues showed that sliding could be blocked by 2 mM MgCl2 (Pennings et al, 1991). As ionic conditions alter nucleosome mobility, we wanted to establish whether the alteration to nucleosome mobility we detected was unique to the experimental conditions we had selected, or whether the effect was generally applicable over a range of conditions. Firstly, we confirmed that moderate concentrations of MgCl2 have an inhibitory effect on nucleosome mobility, although even 2 mM MgCl2 did not totally prevent the nucleosome sliding (Figure 4A). The mobility of H4 R45H sin mutant nucleosomes was also reduced by MgCl2 (Figure 4B). However, as the R45H octamers have generally higher mobility, the majority were still redistributed within 48 min even in the presence of 2 mM MgCl2. Figure 4C shows the proportion of nucleosomes redistributed over time for WT and sin mutant nucleosomes in the presence of a range of MgCl2 and CaCl2 concentrations. MgCl2 and CaCl2 were found to affect nucleosome mobility similarly whether WT or R45H octamer was used (Figure 4C and D). Importantly, over a range of divalent or monovalent salt concentrations, the H4 R45H mutation still exhibited dramatically enhanced nucleosome mobility (Figure 4B).
Figure 4.
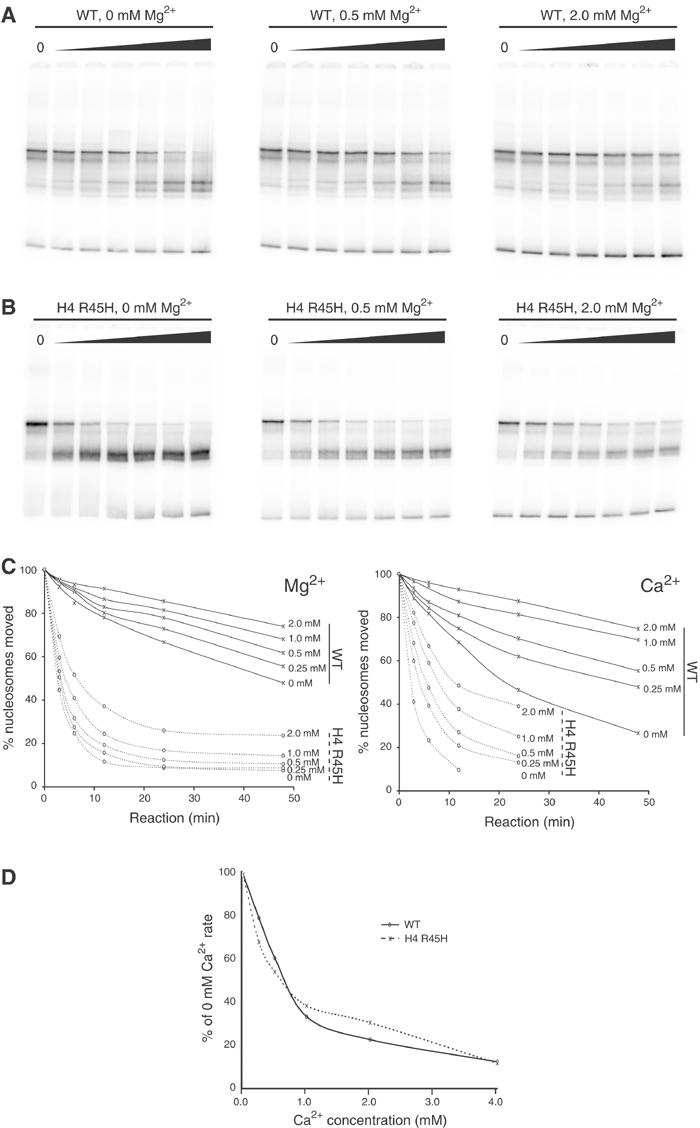
Effect of ionic strength on nucleosome sliding. (A) Time course of nucleosome sliding at 47°C in 50 mM Tris–HCl (pH 7.5) with indicated amounts of MgCl2 using WT octamer. (B) Time course of nucleosome sliding at 47°C in 50 mM Tris–HCl (pH 7.5) with indicated amounts of MgCl2 using H4 R45H octamer. (C) Plots indicating the proportion of nucleosomes redistributed against time for different concentrations of divalent cation using WT and R45H octamers as indicated. (D) Nucleosome sliding indicated as a proportion of the rate observed in the absence of divalent cations for WT and R45H octamers. The similarity of the two curves indicates that divalent cations affect WT and sin mutant octamers similarly.
Sin mutations do not affect the rate of remodelling by the SWI/SNF and RSC complexes
To investigate the relationship between thermal- and ATP-dependent nucleosome mobilisation, we next determined the effect of nucleosomes containing H4 R45H on sliding by the yeast RSC and SWI/SNF remodelling complexes. In order to do this, preparations of WT and sin mutant nucleosomes were prepared that were carefully matched for concentration and in the proportion of free DNA. Incubation of WT nucleosomes with yeast RSC complex over a time course results in the accumulation of faster migrating products (Figure 5A, lanes 2–7). Previous studies suggest that the faster migrating species that are generated represent nucleosomes relocated to a series of positions at and beyond the ends of this DNA fragment (Fan et al, 2003; Flaus and Owen-Hughes, 2003a; Kassabov et al, 2003). Incubation of a well-matched preparation of H4 R45H sin mutant nucleosomes displays an identical RSC-dependent accumulation of faster migrating products (Figure 5A, lanes 9–14). We next determined whether there was any effect of sin mutations on the action of the yeast SWI/SNF complex. Figure 5B shows that this complex causes the mobility of nucleosomes assembled onto this fragment to increase. When H4 R45H-containing nucleosomes are incubated with SWI/SNF in the presence of ATP, the amount of nucleosomes at the original location decreases at a rate similar to the WT octamer (Figure 5B). Thus, the rates at which WT and R45H nucleosomes are remodelled are comparable.
Figure 5.
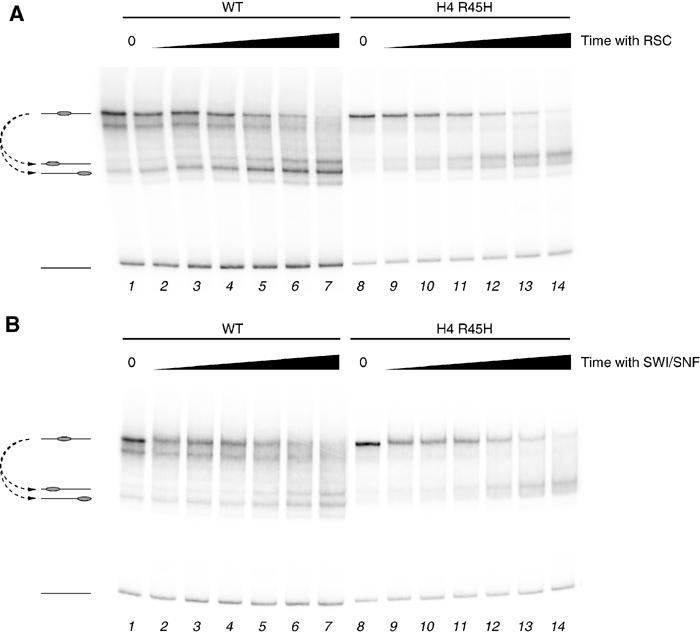
Comparison of rates of sliding in WT and H4 R45H-containing nucleosomes by ATP-dependent remodelling activities. (A) Remodelling for 0, 2, 4, 8, 16, 32 and 64 min (lanes 1–7 and 8–14) in the presence of 0.021 pmol RSC of WT (lanes 1–7) or H4 R45H (lanes 8–14) octamers. (B) Remodelling for 0, 2, 4, 8, 16, 32 and 64 min (lanes 1–7 and 8–14) in the presence of 0.060 pmol SWI/SNF of WT (lanes 1–7) or H4 R45H (lanes 8–14) octamers.
A role for alterations to histone DNA contacts at the nucleosome dyad and reduced association of H2A with the H3–H4 histone fold in nucleosome mobility
The mutations at H4 R45 and H3 T118 occur at locations in the nucleosome structure in close proximity to DNA at the nucleosome dyad. This raises the possibility that the means by which they alter nucleosome mobility involves destabilisation of histone DNA contacts in the region of the nucleosome dyad. In support of this, the crystal structures of nucleosomes bearing these mutations show a loss of histone DNA contacts in this region (Muthurajan et al, 2004). As DNA is bound to the surface of the histone octamer by more interactions in this central region, loosening histone DNA contacts here provides an attractive basis for affecting a rate-limiting step in nucleosome mobilisation. Nevertheless, the actual reason for this awaits clarification of the actual mechanistic steps in nucleosome sliding.
However, the mutation at H3 E105K is distant from DNA and appears unlikely to affect histone DNA contacts directly. As we detect a significant change in the mobility of nucleosomes bearing this mutation (Table I), there is likely to be another route by which nucleosome structure can be altered so as to increase nucleosome mobility. One possibility is that this mutation destabilises the interaction of the H2A C-terminal extension with histone H3. In support of this, a significant number of mutations that might affect association of the C-terminal extension with the H3–H4 histone fold have been previously found to confer sin phenotypes in vivo. These include H4 Y98 and H2B H2A L115 (Hirschhorn et al, 1995; Santisteban et al, 1997), the locations of which are indicated in Figure 6A. To investigate this further, a surface interaction map for the H2A C-terminal region was generated (Figure 6B). Remarkably, the H3 E105, H4 Y98 and H2A L115 mutations map to regions of close contact in this region. In order to test whether the H4 Y98H, H3 L48A or H3 Q55A alterations cause an alteration to nucleosome mobility, we attempted to make nucleosmes containing these mutations. Surprisingly, these mutations destabilised dimer–tetramer association to the extent that we were unable to prepare these histone octamers in vitro (data not shown). We were able to generate H2A L115S and its neighbour H2A L116C, and find that these increase thermal mobility by 4 and 6°C, respectively (Table II).
Figure 6.
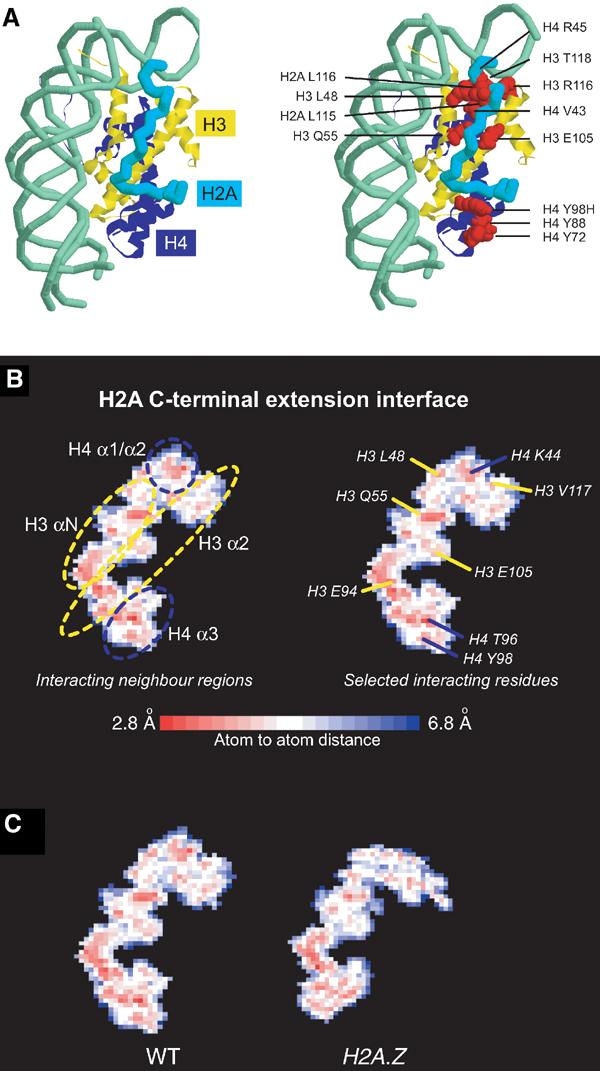
Interaction of H2A C-terminal extension with surrounding structural elements. (A) Path of H2A C-terminal extension shown as a cyan backbone over histone H3 and H4 in yellow and blue, respectively, as in Figure 1. In the right panel, mutated residues of interest are indicated in red. (B) Molsurfer plot (Gabdoulline et al, 2003) of H2A C-terminal extension interaction surface by interatomic distance, coloured according to the scale below. Secondary structure regions of the interface are indicated in the left panel, and selected residues making close approach are indicated in the right panel. (C) Comparison of interaction surfaces for X. laevis WT (PDB 1AOI) and X. laevis WT with Mus musculus H2A.Z (PDB 1F66) generated identically as for (B).
Point mutation of amino acids is not the only means by which the association of H2A with nucleosomes can be manipulated. Most eukaryotes encode multiple variants of histone H2A, one of the best characterised of which is H2A.Z. We also noted that many of the changes between WT and histone variant H2A.Z are clustered in this H2A C-terminal extension, and that the essential regions of its sequence for biological activity lay in this region between residues 97 and 117 (Clarkson et al, 1999). Figure 6C shows interaction surface maps for the WT and H2A.Z nucleosomes in this region. This illustrates that the number of close contacts between the H2A C-terminal region and the histone octamer is qualitatively reduced in this region. Because sin mutations that reduce these contacts affect nucleosome mobility, we expect that the incorporation of H2A.Z into nucleosomes might also increase nucleosome mobility. H2A.Z-containing nucleosomes were found to relocate at 39°C, 4°C lower than was observed for WT nucleosomes (Table II).
For completeness, we also include data for the thermal mobilisation of other histone variants and mutations we had available. Nucleosomes assembled using the human histone genes have mobility identical to that of Xenopus nucleosomes, despite the fact that human octamers contain over 40 amino-acid changes. Incorporation of the histone H3.3 variant into the human histone octamer had little effect on its mobility. Histone H3.3 differs from H3 at four amino acids. Finally, we also monitored the mobility of the mutant H4 S47C that we use for site-directed nucleosome mapping. This mutation causes a strong sin phenotype at the SUC2 and FLO1 promoters (Fleming and Pennings, 2001), but we find it has little effect on nucleosome mobility.
Discussion
The observation that the requirement for the SWI/SNF complex can be compensated for by histone mutations suggests that the means by which this complex functions involves manipulation of chromatin structure. However, the basis for this has not been clear. Although it is known that the SWI/SNF complex can alter nucleosome positioning in vitro, there is limited evidence to support a general role for nucleosomes redistribution in the function of SWI/SNF complexes in vivo. Here we report that increased thermal mobility is a shared property of most of the core histone sin mutations. Furthermore, there is a qualitative correlation between the ability of the H3 sin mutations to rescue SWI/SNF deletions and the magnitude of the effect on sliding. This is consistent with the function of SWI/SNF involving the manipulation of nucleosome positioning. The increased inherent thermal mobility of nucleosomes bearing sin mutations provides a simple means of enabling nucleosomes to redistribute independently of SWI/SNF complex.
The possibility that sin mutant histones affected nucleosome mobility has previously been considered by Kurumizaka and Wolffe (1997), who cited unpublished data comparing WT and H3 R116H-containing nucleosomes using the two-dimensional gel approach of Bradbury and co-workers. In these experiments, it was reported that both WT and sin mutant nucleosomes exhibited significant mobility at 37°C. Unfortunately, this is not very informative as the temperature selected does not allow an accurate assessment of any differences in thermal mobility to be made. It is likely that the low ionic strength electrophoresis buffer and the relatively high mobility of nucleosomes on 5S DNA contributed to the high levels of sliding observed.
Of the mutants identified in the original genetic screens of Kruger et al, only H4 V43I does not significantly increase thermal mobility in our assay. Although H4 V43I did not reduce the temperature required for thermal shifting, it altered the directionality of nucleosome sliding, a feature shared with a subset of the sin mutations we have characterised (Tables I and II). As the histone octamer is symmetrical, this asymmetry must be conferred by the DNA sequence. Consistent with this, we find that when asymmetric derivatives of the MMTV nuc A sequence are used in sliding assays, H3 V43I caused a difference in the rate of sliding in the upstream but not the downstream direction (Supplementary Figure 3). This suggests that some feature of the DNA sequence favours movement of sin mutant octamers in the +22 direction. If a similar effect occurred in vivo, it could profoundly alter the function of a nucleosome in gene regulation. Thus, it is possible that this DNA sequence-dependent effect could contribute to the gene-specific effects of some sin mutations; for example, only a subset of sin mutations are able to substitute the requirement for SWI/SNF at SUC2 (Hirschhorn et al, 1995; Wechser et al, 1997; Recht and Osley, 1999). H4 V43I had significantly lower effects than H4 R45 mutants on Dam methylation and expression from the PHO5 promoter in vivo (Wechser et al, 1997) (Table I).
Our observation that sin mutations alter nucleosome mobility does not preclude them from acting in other ways as well. In this respect, an obvious candidate would be H4 S47C mutation, which was found to have little effect on nucleosome mobility in our study (Table II), but does affect expression of the FLO1 gene in vivo (Fleming and Pennings, 2001). One alternative means by which it is possible that sin mutations alter chromatin structure involves altering the ability of nucleosome arrays to adopt higher order structures. Previously, it has been observed that the H4 R45H and R45C mutations reduce the ability of nucleosomes to form more compact structures (Horn et al, 2002). However, this has not been tested for the V43I or S47C mutations. The finding that the H4 R45H and R45C mutations both alter nucleosome mobility and chromatin compaction raises the possibility that the defective compaction into nucleosomal arrays could be a secondary effect of increased nucleosome mobility. This is attractive as the spacing between nucleosomes will dramatically affect the way in which neighbouring nucleosomes are aligned within a nucleosomal array. The spacing of more mobile nucleosomes might be expected to be less ordered and to impede the formation of a chromatin fibre. Our results show that nucleosomes remain mobile in the 1–2 mM MgCl2 conditions used to generate compacted arrays (Figure 4A–C), and we have previously observed that nucleosomes assembled on the 5S-derived DNA used in the compaction studies are significantly more thermally mobile than on the MMTV nucA sequence (Flaus et al, 1996). It will be interesting to learn whether the full spectrum of sin mutations affects the ability of nucleosomal arrays to adopt condensed structures and whether there is any correlation with the effects on thermal mobility we have observed.
With respect to H2A.Z, the situation is more complicated because two contradictory reports exist as to whether this histone variant affects higher order chromatin compaction (Abbott et al, 2001; Fan et al, 2002). However, recent functional studies suggest that H2A.Z functions to prevent the spread of heterochromatin (Meneghini et al, 2003). Importantly, yeast H2A.Z is unable to act like a conventional sin mutation in suppressing the effect of SWI/SNF deletion, but instead cells are highly dependent on SWI/SNF when H2A.Z is deleted. Synergy between H2A.Z and SWI/SNF has been characterised at the PHO5 and GAL1 genes, suggesting that incorporation of H2A.Z primes genes for activation (Santisteban et al, 2000). Our observation that H2A.Z-containing nucleosomes have high inherent mobility provides an alternative mechanism by which this could occur. If enrichment for H2A.Z increases nucleosome mobility, then the requirement for SWI/SNF would be reduced. Conversely, depletion of H2A.Z would be expected to increase the requirement for a nucleosome mobilising activity such as SWI/SNF. It is tempting to speculate that H2A.Z acts as a ‘plug-in' cassette to alter the biophysical properties of targeted nucleosomes.
Although the sliding assay is highly sensitive and quantitative, the histones and DNA used in the nucleosome mobilisation assay reported here are not of yeast origin, and the recombinant histones are not post-translationally modified. Despite extensive efforts, we have so far been unable to assemble S. cerevisiae histone-containing nucleosomes of sufficient quality to carry out measurements of comparable accuracy. Comparison of the Xenopus laevis and S. cerevisiae nucleosome core particle structures shows 85% sequence identity and only 1.1 Å RMS deviation for the structured core of the histones, suggesting that all general properties of nucleosomes are maintained between species. The only major identifiable structural difference is the absence of an inter-H2A/H2B dimer interaction in the S. cerevisiae nucleosome (White et al, 2001). This may contribute to the difficulty in assembling yeast histone octamers in vitro, but the high degree of similarity between yeast and Xenopus histones means that it is possible to introduce yeast mutations in the Xenopus background.
With respect to DNA sequence, we have observed qualitatively similar effects on sliding using the MMTV LTR nucB positioning sequence (data not shown), and an accompanying manuscript shows consistent results for 5S and alpha satellite-derived sequences (Muthurajan et al, 2004). However, it is likely that DNA sequence may influence the magnitude of the temperature-dependent effects observed and may contribute to the gene-specific effects of some sin mutations.
Our observation that sin mutations affect nucleosome sliding provides an opportunity for new insights into the mechanisms by which nucleosomes move along DNA. The finding that sin mutations which alter histone DNA contacts at the nucleosome dyad (Muthurajan et al, 2004) affect nucleosome sliding suggests that the breakage of these contacts could be a rate-limiting step for nucleosome movement. We have also found that alterations to nucleosome structure that do not directly alter DNA contacts also affect nucleosome mobility. In particular, we note that alterations that would be predicted to loosen the association of histone H2A/H2B dimers with the H3/H4 tetramer affect nucleosome mobility. The dimer–tetramer interface represents a weak link in the structure of the nucleosomes and is the first major rearrangement to occur upon incubation of nucleosomes in increasing salt (Germond et al, 1976). Dimer loss has also been reported to occur as a result of transcription and the action of ATP-dependent remodelling enzymes (Bruno et al, 2003). The finding that mutations that are expected to alter the stability of association between the H2A/H2B dimer and the nucleosome also alter nucleosome mobility raises the possibility that a rearrangement of the histone dimers occurs during nucleosome mobilisation. This would require a substantial refinement to the currently popular ‘twist defect diffusion' and ‘bulge diffusion' models for nucleosome sliding (Flaus and Owen-Hughes, 2003b). It is also possible that rearrangement of the dimer–tetramer interface could cause alterations to nuclease sensitivity as has been detected at the DNA entry/exit for sin-containing mononucleosomes in vitro (Kurumizaka and Wolffe, 1997) and observed for sin mutant chromatin structure in vivo (Santisteban et al, 1997; Wechser et al, 1997; Recht and Osley, 1999; Fleming and Pennings, 2001) or affect chromatin fibre formation (Horn et al, 2002).
Although the H4 R45H mutation has a very significant effect on thermally driven nucleosome sliding, it has no effect on the rate of sliding catalysed by the SWI/SNF and RSC ATP-dependent remodelling complexes (Figure 5). This is consistent with the observations of Horn et al (2002), who found that H4 R45H nucleosomes enabled SWI/SNF-dependent restriction enzyme access at qualitatively similar rates to WT nucleosomes. Although this suggests that there are different rate-limiting steps for ATP-dependent nucleosome redistribution when compared to thermal redistribution, it does not mean that the same interactions do not need to be broken during each process.
Although core histone proteins are among the most conserved in nature, it remains unclear exactly why such extreme conservation is required. Nucleosomes must be sufficiently stable to effectively package DNA, but also retain the fluidity for rearrangement during gene regulation. Our finding that relatively subtle alterations to the histone proteins can have dramatic effects on nucleosome mobility suggests that nucleosomes are finely tuned to meet these opposing criteria. Consistent with this, nucleosomes made from Xenopus and human histones were found to have identical mobilities despite a difference of 40 amino acids between them.
Materials and methods
WT histone sequences were as described by Luger et al. Histone mutagenesis was carried out using the Quikchange method (Stratagene), and histones were prepared and refolded into octamers as previously described (Luger et al, 1999). 54A54 DNA spanning from bases −58 to +197 of the MMTV 3′ LTR promoter and flanking mouse genomic sequences (Donehower et al, 1983; Flaus and Richmond, 1998) was prepared by PCR and purified by ion exchange chromatography. DNA was labelled using 5′ Cy5 dyes on PCR primers or with 5′ 32P ATP using standard methods. Nucleosomes were assembled as previously described (Luger et al, 1999) and maintained at 0–4°C at all times. Site-directed mapping and ATP-dependent remodelling activities were carried out as described previously (Flaus and Owen-Hughes, 2003a). Thermal nucleosome mobilisation was carried out by mixing 1 pmol nucleosomes in a total volume of 10 μl with final conditions of 50 mM Tris–HCl (pH 7.5) and 150 mM NaCl except in Figure 4, where ionic conditions were as indicated with 50 mM Tris–HCl (pH 7.5) buffer. All assays were carried out by incubating samples in an Eppendorf Mastercycler PCR machine whose temperature accuracy had been confirmed to ±0.5°C according to the manufacturer's protocols. For temperature gradient experiments, samples were incubated for 60 min at six appropriate temperatures in <3°C intervals, followed by cooling to 4°C. Gel electrophoresis was carried out at 4°C as previously described (Flaus and Richmond, 1998), and results were quantitated using a Fuji FLA2000 imaging system with fluorescent detection or phosphor plates that had been tested for linearity. Temperatures for 50% sliding were determined by graphical interpolation.
Interaction plots were prepared using Molsurfer software with default settings (Gabdoulline et al, 2003; http://projects.villa-bosch.de/dbase/molsurfer/) based on PDB structures 1AOI (WT) and 1F66 (H2A.Z). All structure images were prepared using RasMol (http://www.openrasmol.org/).
Supplementary Material
Supplementary Figures
Acknowledgments
We thank Uma Muthurajan and Karolin Luger for communication on results prior to publication, exchange of reagents and advice. The expression clone for Xenopus laevis H2A.Z was generously provided by Michael Studer and Tim Richmond. Andrew Travers, Chris Stockdale, Michael Bruno, and members of the division of gene regulation and expression also provided helpful discussion and advice. This work was supported by a Wellcome Trust Senior Fellowship (AF, CR, NW, TOH) and a Wellcome Studentship (HF).
References
- Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausio J (2001) Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J Biol Chem 276: 41945–41949 [DOI] [PubMed] [Google Scholar]
- Becker PB, Horz W (2002) ATP-dependent nucleosome remodeling. Annu Rev Biochem 71: 247–273 [DOI] [PubMed] [Google Scholar]
- Breeden L, Nasmyth K (1987) Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell 48: 389–397 [DOI] [PubMed] [Google Scholar]
- Bruno M, Flaus A, Stockdale C, Rencurel C, Ferreira H, Owen-Hughes T (2003) Histone H2A/H2B dimer exchange driven by ATP-dependent chromatin remodelling activities Bruno et al. Mol Cell 12, (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson MJ, Wells JR, Gibson F, Saint R, Tremethick DJ (1999) Regions of variant histone His2AvD required for Drosophila development. Nature 399: 694–697 [DOI] [PubMed] [Google Scholar]
- Donehower LA, Fleurdelys B, Hager GL (1983) Further evidence for the protein coding potential of the mouse mammary tumor virus long terminal repeat: nucleotide sequence of an endogenous proviral long terminal repeat. J Virol 45: 941–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH, Moudrianakis EN (1978) The histone core complex: an octamer assembled by two sets of protein–protein interactions. Biochemistry 17: 4955–4964 [DOI] [PubMed] [Google Scholar]
- Fan HY, He X, Kingston RE, Narlikar GJ (2003) Distinct strategies to make nucleosomal DNA accessible. Mol Cell 11: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ (2002) The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Biol 9: 172–176 [DOI] [PubMed] [Google Scholar]
- Flaus A, Luger K, Tan S, Richmond TJ (1996) Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc Natl Acad Sci USA 93: 1370–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T (2003a) Dynamic properties of nucleosomes during thermal and ATP-driven mobilization. Mol Cell Biol 23: 7767–7779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T (2003b) Mechanisms for nucleosome mobilization. Biopolymers 68: 563–578 [DOI] [PubMed] [Google Scholar]
- Flaus A, Richmond TJ (1998) Positioning and stability of nucleosomes on MMTV 3′LTR sequences. J Mol Biol 275: 427–441 [DOI] [PubMed] [Google Scholar]
- Fleming AB, Pennings S (2001) Antagonistic remodelling by Swi–Snf and Tup1–Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation. EMBO J 20: 5219–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabdoulline RR, Wade RC, Walther D (2003) MolSurfer: a macromolecular interface navigator. Nucleic Acids Res 31: 3349–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond JE, Bellard M, Oudet P, Chambon P (1976) Stability of nucleosomes in native and reconstituted chromatins. Nucleic Acids Res 3: 3173–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C (1999) ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97: 833–842 [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Bortvin AL, Ricupero-Hovasse SL, Winston F (1995) A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol Cell Biol 15: 1999–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Horn PJ, Crowley KA, Carruthers LM, Hansen JC, Peterson CL (2002) The SIN domain of the histone octamer is essential for intramolecular folding of nucleosomal arrays. Nat Struct Biol 9: 167–171 [DOI] [PubMed] [Google Scholar]
- Jaskelioff M, Gavin IM, Peterson CL, Logie C (2000) SWI–SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol Cell Biol 20: 3058–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Struhl K (1997) Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89: 365–371 [DOI] [PubMed] [Google Scholar]
- Kassabov SR, Zhang B, Persinger J, Bartholomew B (2003) SWI/SNF unwraps, slides and rewraps the nucleosome. Mol Cell 11: 391–403 [DOI] [PubMed] [Google Scholar]
- Kornberg R (1974) Chromatin structure: a repeating unit of histones and DNA. Science 184: 868–871 [DOI] [PubMed] [Google Scholar]
- Kruger W, Herskowitz I (1991) A negative regulator of HO transcription, SIN1 (SPT2), is a nonspecific DNA-binding protein related to HMG1. Mol Cell Biol 11: 4135–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger W, Peterson CL, Sil A, Coburn C, Arents G, Moudrianakis EN, Herskowitz I (1995) Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev 9: 2770–2779 [DOI] [PubMed] [Google Scholar]
- Kurumizaka H, Wolffe AP (1997) Sin mutations of histone H3: influence on nucleosome core structure and function. Mol Cell Biol 17: 6953–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langst G, Bonte EJ, Corona DF, Becker PB (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97: 843–852 [DOI] [PubMed] [Google Scholar]
- Li Y, Bjorklund S, Jiang YW, Kim Y-J, Lane WS, Stillman DJ, Kornberg RD (1995) Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc Natl Acad Sci USA 92: 10864–10868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Thanos D (2002) Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110: 261–271 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol 304: 3–19 [DOI] [PubMed] [Google Scholar]
- Luger K, Richmond TJ (1998) The histone tails of the nucleosome. Curr Opin Genet Dev 8: 140–146 [DOI] [PubMed] [Google Scholar]
- Meersseman G, Pennings S, Bradbury EM (1992) Mobile nucleosomes—a general behavior. EMBO J 11: 2951–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD (2003) Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112: 725–736 [DOI] [PubMed] [Google Scholar]
- Miller JA, Widom J (2003) Collaborative competition mechanism for gene activation in vivo. Mol Cell Biol 23: 1623–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Stillman D, Kipling D (1987) Both positive and negative regulators of HO transcription are required for mother-cell-specific mating-type switching in yeast. Cell 48: 579–587 [DOI] [PubMed] [Google Scholar]
- Neigeborn L, Carlson M (1984) Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen-Hughes T, Workman JL (1994) Experimental analysis of chromatin function in transcription control. Crit Rev Eukaryot Gene Expr 4: 403–441 [PubMed] [Google Scholar]
- Pennings S, Meersseman G, Bradbury EM (1991) Mobility of positioned nucleosomes on 5S rDNA. J Mol Biol 220: 101–110 [DOI] [PubMed] [Google Scholar]
- Polach KJ, Widom J (1995) Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol 254: 130–149 [DOI] [PubMed] [Google Scholar]
- Recht J, Osley MA (1999) Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi–Snf in yeast. EMBO J 18: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban MS, Arents G, Moudrianakis EN, Smith MM (1997) Histone octamer function in vivo: mutations in the dimer–tetramer interfaces disrupt both gene activation and repression. EMBO J 16: 2493–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban MS, Kalashnikova T, Smith MM (2000) Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell 103: 411–422 [DOI] [PubMed] [Google Scholar]
- Shiratori A, Shibata T, Arisawa M, Hanaoka F, Murakami Y, Eki T (1999) Systematic identification, classification, and characterization of the open reading frames which encode novel helicase-related proteins in Saccharomyces cerevisiae by gene disruption and Northern analysis. Yeast 15: 219–253 [DOI] [PubMed] [Google Scholar]
- Simpson RT (1990) Nucleosome positioning can affect the function of a cis-acting DNA element in vitro. Nature 343: 387–389 [DOI] [PubMed] [Google Scholar]
- Song W, Treich I, Qian N, Kuchin S, Carlson M (1996) SSN genes that affect transcriptional repression in Saccharomyces cerevisiae encode SIN4, ROX3, and SRB proteins associated with RNA polymerase II. Mol Cell Biol 16: 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M, Jensen R, Herskowitz I (1984) Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol 178: 853–868 [DOI] [PubMed] [Google Scholar]
- Sternberg PW, Stern MJ, Clark I, Herskowitz I (1987) Activation of the yeast HO gene by release from multiple negative controls. Cell 48: 567–577 [DOI] [PubMed] [Google Scholar]
- Sudarsanam P, Iyer VR, Brown PO, Winston F (2000) Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 97: 3364–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechser MA, Kladde MP, Alfieri JA, Peterson CL (1997) Effects of Sin- versions of histone H4 on yeast chromatin structure and function. EMBO J 16: 2086–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Suto RK, Luger K (2001) Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J 20: 5207–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T (1999) Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400: 784–787 [DOI] [PubMed] [Google Scholar]
- Winston F, Carlson M (1992) Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet 8: 387–391 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures


