Abstract
Induction of p21 in senescent human fibroblasts plays a key role in the inactivation of cyclin-dependent kinases and the resulting irreversible growth arrest in the early stages of cell senescence. We found that RNA-binding proteins are critical regulators of p21 during senescence. Two RNA-binding proteins, CUGBP1 and calreticulin (CRT), interact with the same nucleotide sequences within the 5′ region of p21 mRNA, but have opposite effects on the translation of p21 mRNA. CUGBP1 increases translation of p21 mRNA, whereas CRT blocks translation of p21 via stabilization of a stem–loop structure within the 5′ region of the p21 mRNA. CUGBP1 and CRT compete for binding to p21 mRNA and thereby the regulation of p21 translation. In senescent fibroblasts, CUGBP1 displaces CRT from the p21 mRNA and releases CRT-dependent repression of p21 translation leading to growth arrest and development of a senescent phenotype. These data present evidence that competition between RNA-binding proteins for the regulation of p21 translation determines cell fate.
Keywords: aging, mRNA, p21, translation
Introduction
Normal human fibroblasts exhibit limited proliferative potential referred to as replicative senescence, and enter an irreversible growth arrest after undergoing a certain number of population doublings (Campisi, 2001). Although telomere shortening seems to play a primary role in replicative senescence (Chiu and Harley, 1997), several other aging-associated changes are required for the development of the senescent phenotype. One of the critical steps is expression of the inhibitors of cyclin-dependent kinases p21 and p16 (Stein et al, 1999; Dulic et al, 2000). A detailed analysis of the contribution of each kinase has demonstrated that the induction of p21 is required for the initial irreversible growth arrest and inhibition of cdk2–cyclin E complexes, whereas p16 is involved in the maintenance of growth arrest and in further development of the senescence phenotype (Stein et al, 1999). Elevation of p21 levels has also been observed in a variety of other differentiation/growth arrest systems, and the basis for this elevation has been intensively investigated. The results indicate that the molecular mechanisms of p21 regulation are complex and involve multiple levels of regulation of gene expression including transcription, stabilization of mRNA, changes in the rate of translation and stability of the protein (Sherr and Roberts, 1999). The regulation of mRNA stability and mRNA translation is controlled by different RNA-binding proteins that specifically interact with corresponding mRNAs. The best characterized family of RNA-binding proteins are ELAV-like proteins, the first member of which was discovered in Drosophila as a protein that causes an embryonic lethal abnormal phenotype, elav (Campos et al, 1985). Human homologs of elav, HU proteins, play a crucial role in the upregulation of mRNA encoding the cyclins (Fan and Steitz, 1998).
Another family of RNA-binding proteins, the CUGBP family, has been discovered as proteins that potentially might be involved in the development of pathology in some diseases caused by expansion of RNA triplet repeats (Timchenko et al, 1996a, 2001). One member of the family, CUGBP1, interacts with GC-rich sequences located within the 5′ region of C/EBPβ mRNA and increases translation of a dominant-negative C/EBPβ isoform, LIP (Timchenko et al, 1999). We have recently identified a new RNA-binding protein, calreticulin (CRT), which interacts with GC-rich sequences (Timchenko et al, 2002). CRT binds to GC-rich stem–loop (SL) structures (Singh et al, 1994; Pugachev and Frey, 1998; Timchenko et al, 2002) and inhibits translation of mRNA, perhaps by stabilization of the SL (Timchenko et al, 2002).
We here present evidence that competition between CUGBP1 and CRT for binding to p21 mRNA controls levels of p21 expression and determines whether cells will proliferate or undergo growth arrest and senescence. We have found that CRT inhibits translation of p21 mRNA and abolishes p21-dependent growth arrest and development of a senescent-like phenotype. CUGBP1 blocks CRT-mediated inhibition of p21 translation and promotes p21-dependent growth arrest and development of a senescent-like phenotype.
Results
Binding of CUGBP1 to p21 mRNA is increased in senescent human fibroblasts
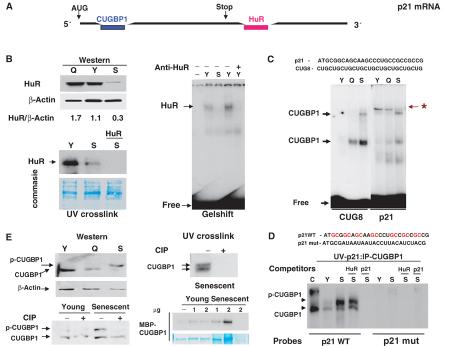
It has been previously shown that the p21 mRNA interacts with two RNA-binding proteins: CUGBP1 (Timchenko et al, 2001) and HuR (Wang et al, 2000). To determine whether these proteins might be involved in the induction of p21 in senescent fibroblasts, we examined the interactions of these proteins with p21 mRNA in young and senescent fibroblasts. We found that protein levels of HuR and binding activity of HuR are reduced in senescent fibroblasts (Figure 1B). These data are consistent with observations by others who also observed a reduction in protein levels of HuR in senescent fibroblasts (Wang et al, 2001b). Since HuR is a positive regulator of p21, these results suggested that HuR is not likely to be involved in the upregulation of p21 during cellular senescence, and we therefore focused our studies on CUGBP1 and other GC-rich RNA-binding proteins. CUG8 and GC-rich p21 probes (Figure 1C, top) were used for gel-shift studies. EMSA with the CUG8 probe demonstrated that CUGBP1 binding activity is dramatically increased in senescent compared with young fibroblast cells (Figure 1C). Gel-shift assay with the 5′ region of p21 mRNA revealed a more complex pattern of the protein–RNA interactions within this region. Whereas the binding of CUGBP1 to p21 mRNA was induced in senescent fibroblasts, the interaction of another, unknown RNA-binding protein (labeled with a star) with the 5′ region of p21 mRNA is dramatically reduced. To confirm the identity of CUGBP1, proteins were linked to either wild type (WT) or mutant p21 RNA probes by UV treatment and precipitated with antibodies to CUGBP1. Increased binding activity of CUGBP1 was observed in senescent fibroblasts (Figure 1D). The interaction of CUGBP1 with the p21 WT probe is specific since a p21 mutant probe does not interact with CUGBP1 and cold WT GC-rich p21 RNA oligomer competes for the binding, whereas the AU-rich HuR oligomer does not (Figure 1D).
Figure 1.
Binding activity of CUGBP1 is increased in senescent human fibroblasts. (A) Locations of elements that interact with CUGBP1 (GC-rich) or with HuR (AU-rich) proteins on p21 mRNA. (B) Protein levels and binding activity of HuR are reduced in senescent fibroblasts. Upper image: Western blotting of cytoplasmic proteins from young (Y), quiescent (Q) and senescent (S) fibroblasts with antibodies to HuR. The membrane was reprobed with antibodies to β-actin. Protein levels of HuR were calculated as a ratio to β-actin (bottom). UV crosslink assay of cytoplasmic proteins from young, quiescent and senescent fibroblasts with AU-rich HuR probe from p21 mRNA. The filter was stained with Coomassie blue to verify protein loading. An RNA competitor (HuR, shown on the top) was added into the binding reaction to examine the specificity of the interaction. The right image shows a gel-shift assay with RNA HuR probe. Antibodies to HuR (shown on the top) were added to protein extracts from young cells. (C) Binding of cytoplasmic proteins to the GC-rich 5′ region of p21 mRNA. Gel-shift assay of cytoplasmic proteins from young, quiescent and senescent fibroblasts with CUG8 and p21 GC-rich probes was performed as described in Materials and methods. The upper image shows nucleotide sequences of WT p21 probe and CUG8 probe. Positions of CUGBP1-RNA complexes are shown on the left. p21 RNA–protein complex that is abundant in young cells, but is not detectable in senescent fibroblasts, is shown by a star. (D) UV crosslink: CUGBP1 precipitation. The upper image shows nucleotide sequences of WT probe and a mutant probe in which GC islands are substituted with AU or UA. Proteins were incubated with WT p21 or mut p21 probes, linked to RNA by UV, and precipitated with monoclonal Abs to CUGBP1. C: CUGBP1 purified from HeLa cells. Y and S: proteins from young and senescent fibroblasts, respectively. Cold competitors (shown on the top) were incorporated into the binding reactions. (E) The active CUGBP1 is hyper-phosphorylated in senescent fibroblasts. Western, upper: Western blotting of cytoplasmic proteins from young (Y), quiescent (Q) and senescent (S) fibroblasts with antibodies to CUGBP1. Western, bottom: Treatment of cytoplasmic proteins with alkaline phosphatase (CIP) eliminates the slow-migrating CUGBP1 isoform. UV crosslink, upper: Cytoplasmic proteins from senescent fibroblasts were treated with CIP: CUGBP1 was immunoprecipitated with specific antibodies and examined for the binding to the GC-rich p21 probe. UV crosslink, bottom: MBP-CUGBP1 was incubated with increasing amounts (1 and 2 μg, shown on the top, the last lane contains only cytoplasm from senescent cells) of cytoplasmic proteins from young or senescent cells in the presence of 1 mM ATP. Binding activity of MBP-CUGBP1 was examined with the p21 GC-rich probe after the incubations. The membrane was stained with Coomassie.
We further examined the mechanisms by which CUGBP1 is activated in senescent fibroblasts. Although the total protein levels of CUGBP1 (as ratios to β-actin) are not altered, in senescent cells there is an induction of the CUGBP1 isoform, which migrates slower than the CUGBP1 isoform in young and quiescent fibroblasts (Figure 1E, Western). The electrophoretical mobility of this isoform is similar to that of the active CUGBP1 observed in the UV crosslink assay (Figure 1D). Given the appearance of the slow-migrating CUGBP1 isoform in senescent cells, we tested the possibility that phosphorylation of CUGBP1 in senescent cells might lead to the activation of CUGBP1. Protein extracts were treated with alkaline phosphatase (CIP), and in senescent cells this resulted in a shift of the slow-migrating isoform of CUGBP1 to the position of CUGBP1 protein observed in young fibroblast cells (Figure 1E, Western). Dephosphorylation of CUGBP1 by CIP also reduces its binding to the p21 mRNA probe (Figure 1E). The increased phosphorylation of CUGBP1 in senescent fibroblasts suggests that these cells contain a kinase that activates CUGBP1. Therefore, we tested this hypothesis by incubating bacterially expressed, purified MBP-CUGBP1 with increasing amounts of cytoplasm and examined the binding of MBP-CUGBP1 to p21 mRNA. A slight (1.5- to 2-fold) induction of MBP-CUGBP1 activity was observed after incubation with cytoplasm from young cells; however, the incubation with cytoplasm from senescent cells leads to a much higher activation (8- to 10-fold) of CUGBP1 (Figure 1E). Taken together, these studies suggest that CUGBP1 is activated in senescent fibroblasts by phosphorylation.
Binding of calreticulin to the 5′ region of p21 mRNA is reduced in senescent fibroblasts
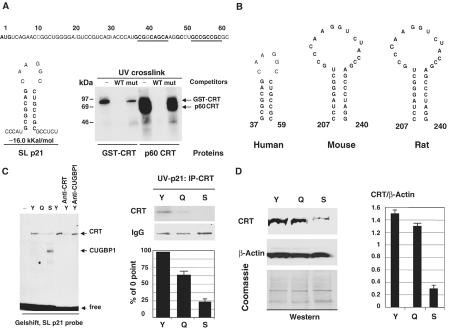
We next determined the identity of the protein that had reduced binding activity to the GC-rich p21 probe in senescent fibroblasts. We have recently found that another RNA-binding protein, CRT, interacts with SL structures within mRNAs coding for C/EBPα and C/EBPβ (Timchenko et al, 2002). Therefore, we examined the possible formation of SL structures within p21 mRNA using the RNA Secondary Structure Predictions Program, and found that a GC-rich sequence within the 5′ region of the human p21 mRNA might form a stable SL structure (Figure 2A). This GC-rich sequence has been previously identified as a binding site for CUGBP1 (Timchenko et al, 2001), and corresponds to the RNA oligomer that we used for gel-shift in experiments shown in Figure 1. To examine whether CRT binds to this SL structure, bacterially expressed, purified GST-CRT and homogenous CRT-p60 isolated from rat liver were incubated with the SL p21 probe and analyzed by the UV crosslink assay. Figure 2A (UV crosslink) shows that both GST-CRT and CRT-p60 interact with the SL p21 probe. Incorporation of WT (SL structure) and mutant RNA oligomers into the binding reactions revealed that these interactions are specific since the mutant sequence does not compete for the binding. Given the interaction of CRT with the SL structure within the 5′ region of human p21 mRNA, we searched for GC-rich SL structures within mouse and rat p21 mRNAs. Similar to human p21 mRNA, the RNA Secondary Structure Predictions Program identified GC-rich SL structures within both mouse and rat p21 mRNAs (Figure 2B). These structures contain GC-rich stems and might potentially be regulated by CRT.
Figure 2.
Binding of CRT to p21 mRNA is reduced in senescent fibroblasts. (A) Calreticulin binds to the SL structure within the 5′ region of p21 mRNA. The nucleotide sequence of the 5′ region of p21 mRNA is shown on the top. Nucleotides forming an SL structure (shown on the left) are bold and underlined. UV crosslink indicates binding of GST-CRT and CRT-p60 to the p21 SL probe. p21 WT or p21 mutant RNA oligomers (see Figure 1D) were added to the binding reactions. (B) Conserved GC-rich SL structures within human, mouse and rat p21 mRNAs. Positions of SL structures relative to the AUG start codon are shown. (C) CRT binding activity is reduced in senescent fibroblasts. The left image shows gel-shift assay of cytoplasmic proteins from young (Y), quiescent (Q) or senescent (S) cell with the p21 SL probe. Antibodies to CRT or CUGBP1 were incorporated into the binding reactions with cytoplasm from young fibroblasts. The right panel shows UV crosslink: CRT precipitation assay. Coomassie stain of the section of the membrane with heavy chains of IgG is shown. Bar graphs show a summary of three experiments. Signals in CRT IPs from quiescent and senescent cells were calculated as the percentage of signals in CRT IPs from young cells. (D) Protein levels of CRT are reduced in cytoplasmic extracts of senescent fibroblasts. Western blotting was performed with cytoplasms (shown on the top) using antibodies to CRT. The membrane was reprobed with antibodies to β-actin and stained with Coomassie blue. CRT levels were calculated as a ratio to β-actin.
We next examined the activity of CRT in young and senescent fibroblasts. Figure 2C (left image) shows the results of a gel-shift assay. The incorporation of antibodies to CRT into the binding reactions with cytoplasmic proteins from young fibroblasts leads to the neutralization of the CRT–RNA complex, while antibodies to CUGBP1 do not affect the formation of the CRT–RNA complex. UV-IP assay confirmed that binding of CRT to the SL p21 probe is reduced in senescent fibroblasts (Figure 2C). Measurements of radioactivity in CRT IPs showed a four- to five-fold reduction of the activity in senescent cells (Figure 2C, bar graphs). Western blotting indicates that protein levels of CRT (as a ratio to β-actin) are three- to four-fold reduced in the cytoplasm of senescent fibroblasts (Figure 2D). Thus, these data suggest that the reduction of CRT binding activity in senescent fibroblasts is due to the decrease of its protein levels.
CRT inhibits translation of p21 mRNA
We next determined whether CRT regulates p21 translation. WT p21 and a p21 deletion construct that lacks the 5′ region of p21 (Δ1–41, Figure 3A; Nakanishi et al, 1997) were translated in reticulocyte lysate in the presence of increasing amounts of p60-CRT purified from rat liver. As can be seen, p60-CRT inhibits translation of WT p21 mRNA and translation of p21-Δ36–40 mutant containing an unrelated internal deletion. Both these mRNAs have an intact CRT-binding site. However, the construct p21-Δ1—41, which lacks the CRT-binding site, SL structure, is not inhibited by p60-CRT. Examination of two additional p21 deletion/mutation constructs (that are not able to form the SL structure, SL-mut p21 and p21-Δ17–22) in reticulocyte lysate showed that GST-CRT does not inhibit translation of these mRNAs (Figure 3B). The inhibition of WT p21 translation by bacterially expressed GST-CRT is slightly weaker than the inhibition by CRT-p60 isolated from rat liver (Figure 3A). This difference is consistent with the observations that bacterially expressed GST-CRT binds to p21 mRNA weaker than CRT-p60 (Figure 2A).
Figure 3.
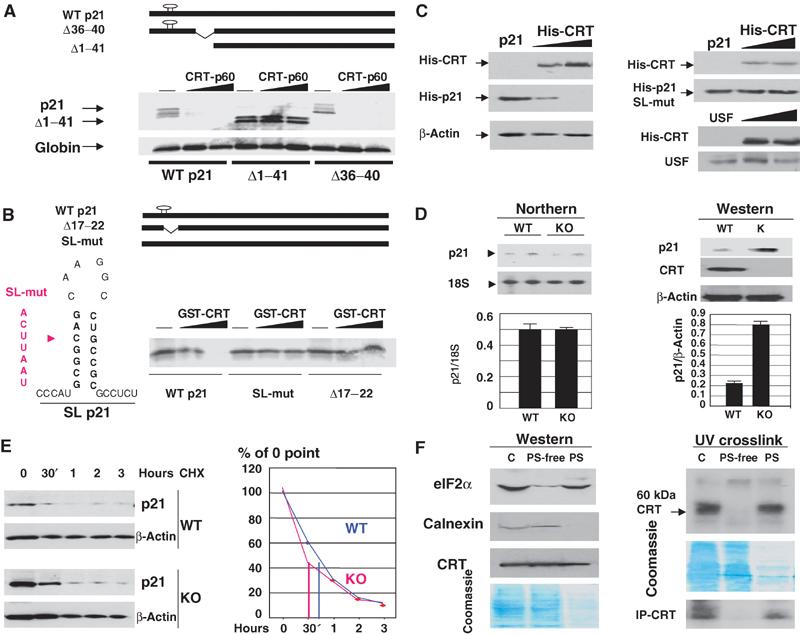
CRT inhibits translation of p21 mRNA. (A) CRT-dependent inhibition of p21 translation in a cell-free translation system. The upper panel shows p21 constructs used in these experiments. Bottom: The p21 constructs (shown below) were translated in RL in the presence of increasing amounts of CRT-p60. The bottom panel shows translation of an endogenous globin mRNA in the same translation mixtures. (B) The SL structure is required for CRT-mediated inhibition of p21 translation. A diagram of p21 constructs is shown on the top. The left section shows mutations (red) incorporated into the left side of the stem within the SL-mut-p21 construct. The right image shows translation of WT, SL-mut-p21 and Δ17–22 p21 mRNAs in the presence of purified GST-CRT. (C) CRT inhibits translation of p21 in cultured cells. Left: WT His-p21 and increasing amounts of His-CRT plasmid were cotransfected into HT1080 cells. Expression of His-p21 and His-CRT was examined by Western blotting with His-tag antibodies. Right: A similar experiment with the SL-mut-p21 and cotransfection of CRT with a vector coding for USF, the translation of which is not affected by CRT in a cell-free translation system. (D) CRT −/− MEF express high levels of p21 protein. Left image: Northern blotting was performed with RNA isolated from CRT+/+ and CRT −/− MEF. The levels of p21 mRNA were calculated as ratios to18S RNA (bar graphs). Right image: Western blotting with antibodies to p21 and CRT. The same membrane was reprobed with β-actin. Bar graphs show a summary of three experiments. (E) The half-life of p21 protein is identical in CRT+/+ and CRT −/− cells. Cells were treated with cyclohexamide and protein extracts were isolated at different time points (shown on the top) and analyzed by Western blotting with antibodies to p21. The membrane was reprobed with β-actin, and p21 levels were calculated as a ratio to β-actin. Bar graphs show p21 levels relative to the 0 time point. (F) CRT capable of binding to RNA is associated with polysomes. Western. Total cytoplasm (C), polysome-free (PS-free) and polysomal (PS) fractions (1/50, 1/50 and 1 ratios, see text) were loaded on the gel and examined by Western blotting with antibodies to CRT, calnexin and eIF2α. The bottom part shows Coomassie stain of the membrane. UV-crosslink: Proteins from subcellular fractions (shown on the top) were incubated with the SL p21 probe and linked by UV treatment. The position of CRT is shown on the right. The membrane was stained with Coomassie. The bottom image (IP-CRT) shows immunoprecipitation of CRT from the binding reactions treated with UV.
To examine whether CRT inhibits p21 translation in cultured cells, we cotransfected p21 mRNA with His-CRT into HT1080 cells. The expression of WT p21 mRNA is dramatically reduced in cells overexpressing CRT. This inhibition is mediated through SL structure, since translation of the SL-mut p21 mRNA is not affected by CRT (Figure 3C). We have previously shown that CRT does not affect translation of upstream stimulatory factor (USF) in a cell-free translation system (Timchenko et al, 2002). As can be seen in Figure 3C, translation of USF also is not affected by overexpression of CRT in cultured cells. These data indicate that CRT specifically inhibits translation of p21 mRNA and does not affect the entire translational machine in transfected cells.
Translation of p21 is increased in CRT knockout mouse embryonic fibroblasts
We next examined whether CRT is an important regulator of p21 under physiological conditions using CRT −/− mouse embryonic fibroblasts (MEF) (Mesaeli et al, 1999). Although the levels of p21 mRNA are identical in WT and in CRT knockout (KO) cells, CRT −/− MEF express three- to four-fold higher levels of p21 protein (Figure 3D). Examination of p21 stability in cells lacking CRT showed that the half-life of p21 is identical in WT and CRT KO cells (Figure 3E). These results suggest that the lack of CRT leads to the increase of p21 translation. We next examine subcellular compartments where CRT might regulate translation of p21. Cytoplasm from WT MEF was fractionated on two subfractions: polysomal fraction (PS) and PS-free fraction as described in Materials and methods. These fractions were analyzed by Western blotting with antibodies to CRT and by UV crosslink-IP assay with the p21 SL probe. Control experiments showed that another ER-specific protein calnexin is not detectable on PS, while eIF2α, which initiates translation of mRNAs, is located on polysomes. Contrary to calnexin, CRT is observed in polysomal fraction (Figure 3F). Western blotting with dilution curves for cytoplasm and the PS fraction showed that approximately equal signals of CRT are observed with the whole PS fraction and 1/50 part of the original cytoplasm from which PS fractions were isolated. Figure 3F shows the typical picture of Western blotting with these loadings. These experiments suggest that the PS-associated CRT represents a minor (less than 2%) portion of the total protein. We next examined RNA-binding activity of CRT in PS-free and PS fractions. UV crosslink identified in PS fraction a protein (p60) that binds to the SL p21 probe, while the RNA-binding activity of this protein is not detectable in PS-free fraction (Figure 3F, UV crosslink). Immunoprecipitation of CRT from reaction mixtures revealed that the p60 protein is CRT. These studies demonstrate that the PS-associated CRT binds to the SL p21 RNA probe, while PS-free CRT does not interact with the RNA probe. Thus, a small portion (less than 2%) of CRT is associated with polysomes and this portion is able to interact with the SL structure of p21 mRNA.
CRT −/− cells proliferate slowly and undergo senescence at early passages
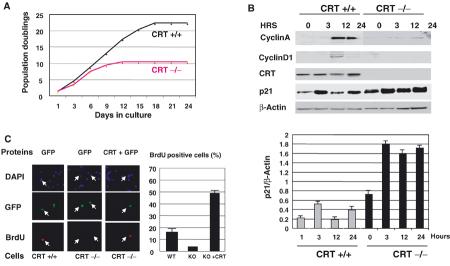
To examine whether the release of CRT-mediated repression of p21 causes growth arrest and premature senescence, we calculated population doublings (PDs) for WT and CRT KO MEF. Figure 4A shows that WT MEF slow down proliferation after 22–23 PDs, while CRT KO cells stop proliferation after 10–12 PDs. We next analyzed cell cycle progression in CRT KO cells. Cells were synchronized by serum starvation, and then stimulated by the addition of 10% FBS. Although serum induces expression of p21 in both WT and CRT KO cells, p21 levels (as ratios to β-actin) are three- to four-fold higher in CRT KO MEF than those in WT cells within 24 h after serum addition (Figure 4B, bar graphs). Expression of cyclins A and D1 is increased in WT cells at 12 h after initiation of proliferation, while CRT-deficient cells fail to increase the expression of these proteins (Figure 4B). Thus, these data demonstrate that the CRT-deficient cells contain high levels of p21 and fail to induce cell cycle proteins after stimulation of proliferation.
Figure 4.
CRT −/− cells have a short proliferative lifespan. (A) Lifespan of CRT −/− MEF. Primary MEF with indicated genotypes were propagated in culture as described in Materials and methods. Accumulated numbers of population doublings are shown. The data represent experiments using two independent MEF preparations for each genotype. (B) Expression of cyclins A and D1 in WT and CRT-deficient cells during cell cycle progression. Cells were synchronized by serum starvation, stimulated by 10% FBS and harvested at the times shown on the top. In all, 50 μg of proteins were analyzed by SDS–PAGE, followed by immunoblotting with antibodies to CRT, cyclin D1, cyclin A, p21 and β-actin as a loading control. Bar graphs show levels of p21 as a summary of three independent experiments. (C) Reloading of CRT-deficient MEF with CRT increases cell proliferation. Incorporation of BrdU (red) was examined in WT or CRT −/− MEF after nine PDs transfected with His-CRT. DAPI staining shows all cells in the field, and GFP indicates transfected cells. Bar graphs show a summary of three independent experiments.
Next, we tested whether the delivery of CRT into CRT-deficient cells restores proliferation. CRT −/− cells (9–10 PDs) were cotransfected with green fluorescent protein (GFP)+CRT or GFP alone to visualize transfected cells. Cells were labeled with BrdU and immunostained with antibodies to BrdU. Calculations of BrdU-positive cells indicated that 15–20% of WT cells synthesize DNA, while only 5–7% of CRT −/− cells incorporate BrdU (Figure 4C). The delivery of CRT into CRT −/− MEF leads to a significant increase of BrdU-positive cells. Under experimental conditions (partially synchronized cells), we observed up to 50% of BrdU-positive CRT −/− cells after transfection with CRT. Thus, CRT KO cells express higher levels of p21, proliferate slowly and undergo senescence at early passages. The delivery of CRT into CRT −/− cells stimulates proliferation.
Overexpression of p21 in HT1080 cells causes growth arrest and induces a senescent-like phenotype
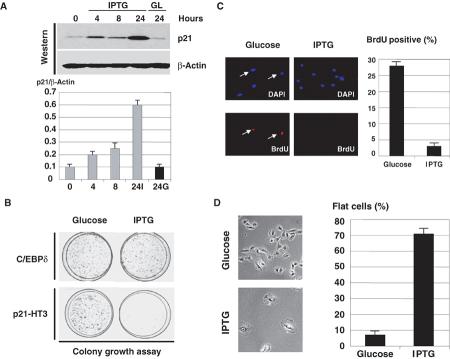
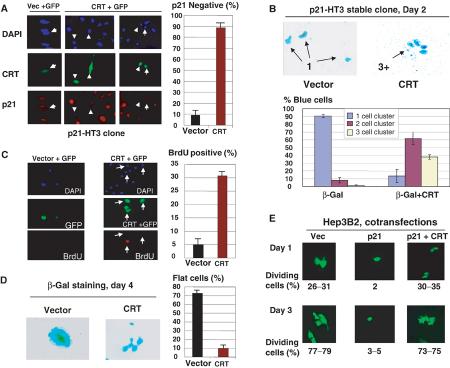
We next examined whether CRT and CUGBP1 compete for the regulation of p21 and for the regulation of cell growth. For these studies, we generated a stable clone of HT1080 cells (clone p21-HT3) in which p21 was placed under Lac-Repressor control as previously described (Timchenko et al, 1996a, 1996b). Figure 5 presents a characterization of the clonal cell line, p21-HT3. Addition of IPTG induces p21 expression with maximum levels (8- to 10-fold induction) observed at 24 h (Figure 5A). Colony growth assay showed that these levels of p21 are sufficient to cause growth arrest (Figure 5B). The growth arrest is specific to p21 since the IPTG-mediated induction of a noninhibitory protein C/EBPδ in the control clone does not cause growth arrest. Examination of BrdU uptake showed that, in control glucose-treated cells, 25–30% cells incorporate BrdU, while only 2–3% of p21-HT3 cells induced by IPTG are positive for BrdU (Figure 5C). In the course of these experiments, we found that IPTG-treated p21-HT3 cells develop a flattened shape, which has been previously described as an indication of a senescent phenotype. The development of the flattened shape is specific for p21 induction, since the ITPG treatment of the control C/EBPδ cells did not cause the formation of flat cells. A typical picture of p21 growth-arrested, flat cells (IPTG) and control dividing cells (glucose) is shown in Figure 5D. Approximately 70% cells are flat on IPTG-treated plates, whereas only 2–3% flat cells are observed on plates treated with glucose (Figure 5D). These alterations suggested that the induction of p21 causes a senescent-like phenotype.
Figure 5.
p21 inhibits cell proliferation and causes a senescent-like phenotype in the stable p21-HT3 clonal line. (A) Levels of p21 induction after the addition of IPTG. Upper: Protein extracts were isolated from p21-HT3 cells at different time points after IPTG addition, and analyzed by Western blotting with antibodies to p21and β-actin. p21 levels are calculated as a ratio to β-actin (bar graphs). (B) IPTG-mediated induction of p21 is sufficient to cause growth arrest in p21-HT3 cells. p21-HT3 cells were plated at low density, treated with glucose (control) or with IPTG and stained with hematoxylin at day 7 after IPTG addition. A stable clone with C/EBPδ under Lac repressor control was used as the negative control. (C) p21 inhibits DNA synthesis in p21-HT3 clonal cells. p21 was induced by IPTG in p21-HT3 cells, BrdU was added for 20 min at 24 h after IPTG addition, and cells were fixed and stained with monoclonal Abs to BrdU. Bar graphs show a summary of three independent experiments. (D) IPTG-mediated induction of p21 causes the development of senescent-like morphology: flat cells. Left: Glucose and IPTG-treated cells at 5 days after the addition of IPTG. Right: Bar graphs show a summary of three experiments.
Overexpression of CRT blocks p21-mediated growth arrest and development of the senescent-like phenotype
To determine if CRT inhibitory activity is sufficient to block p21-mediated growth arrest, p21-HT3 cells were transfected with a CRT plasmid and examined for the expression of p21, growth arrest and cell morphology. A summary of five experiments indicates that p21 translation is inhibited in approximately 96% of CRT-expressing cells, while GFP alone has minor effects on p21 translation in 3–5% of cells (Figure 6A). We next examined the effects of CRT on the growth-inhibitory activity of p21. His-CRT was cotransfected with a plasmid coding for β-gal into p21-HT3 cells, and the cells were stained for β-gal activity. The majority of the control IPTG-induced p21-HT3 cells are arrested and present as single cells. However, more than 85% of CRT-expressing cells divide and form colonies containing two and three cell clusters (Figure 6B, bar graphs). We next asked whether overexpression of CRT abolishes p21-mediated inhibition of DNA synthesis and alterations in cell morphology. The DNA synthesis in p21-HT3 clone was examined by measuring BrdU uptake at 48 h after transfection with CRT+GFP or GFP alone. Figure 6C shows that cells transfected with control GFP do not incorporate BrdU, while more than 30% cells overexpressing CRT synthesize DNA. These data show that CRT can abolish p21-mediated inhibition of DNA synthesis. We next tested whether CRT could eliminate the p21-mediated formation of flat cells through the inhibition of p21. Figure 6D shows the morphology of cells at day 4 after induction of p21. IPTG-mediated induction of p21 in the majority of control cells results in the formation of flat cells. However, cells overexpressing CRT do not form flat cells and appear as multiple cell colonies (Figure 6D). These studies in stable p21-HT3 clone revealed that CRT blocks the growth-inhibitory activity of p21 in human fibrosarcoma cells. We next examined whether CRT blocks p21 activity in other types of cells. AdTrack-p21 plasmid (coding for both p21 and GFP) was cotransfected with His-CRT into Hep3B2 cells. Figure 6E shows that p21 inhibits the proliferation of Hep3B2 cells; however, cells transfected with p21 and CRT proliferate.
Figure 6.
Overexpression of CRT blocks the biological functions of p21. (A) CRT inhibits the translation of p21 in a stable clonal line p21-HT3. Vector +GFP (control) or CRT+GFP (ratio 10:1) were cotransfected into p21-HT3 cells, p21 was induced by IPTG and stained with specific antibodies at 16 h after IPTG addition. DAPI staining shows all cells on the field. Green shows transfected cells. Red represents p21 staining. Bar graphs show a summary of five independent experiments. (B) CRT blocks p21-mediated growth arrest. His-CRT or empty vectors were cotransfected with a β-gal plasmid, and transfected cells were visualized at day 2 by β-gal staining. Bar graphs show a summary of five independent experiments. (C) Overexpression of CRT abolishes p21-mediated inhibition of DNA synthesis. BrdU uptake was examined in p21-HT3 cells induced by IPTG, and transfected with plasmid coding for CRT or with empty vector. GFP was cotransfected to visualize transfected cells. Bar graphs show a summary of three independent experiments. (D) CRT blocks the p21-mediated formation of flat cells. CRT or empty vectors were cotransfected with plasmid coding for β-gal into p21-HT3 cells. p21 was induced by IPTG, and cells were stained for β-gal activity 4 days after p21 induction. The right shows a summary of three experiments. (E) Overexpression of CRT blocks p21 inhibitory activity in hepatoma 3B2 cells. AdTrack-p21 plasmid (coding for p21 and GFP) was cotransfected with empty vector or with His-CRT into Hep3B2 cells. Transfected cells were visualized at days 1 and 3 after transfection. The percentage of dividing green cells (two and more cells per colony) is shown below as a summary of two experiments.
CUGBP1 competes with CRT for binding to the 5′ region of p21 mRNA and relieves CRT-mediated repression of p21 translation
Given the reverse pattern of CRT and CUGBP1 expression in senescent cells (Figure 1), we examined the hypothesis that these proteins compete for the interaction with p21 mRNA and thereby for the regulation of p21 mRNA translation. Bacterially expressed, purified full-length CUGBP1 and GST-CRT were added into the binding reactions in different molar ratios to each other. This UV crosslink assay was performed under conditions of two- to four-fold molar excess of proteins to the SL-p21 RNA probe. When CRT and CUGBP1 are present in the same binding reactions, the resulting interaction depends on the molar ratio of these proteins (Figure 7A). These experiments showed that CUGBP1 had to be present in the binding reactions in four- to eight-fold molar excess to CRT in order to replace CRT from the p21 RNA (Figure 7A).
Figure 7.
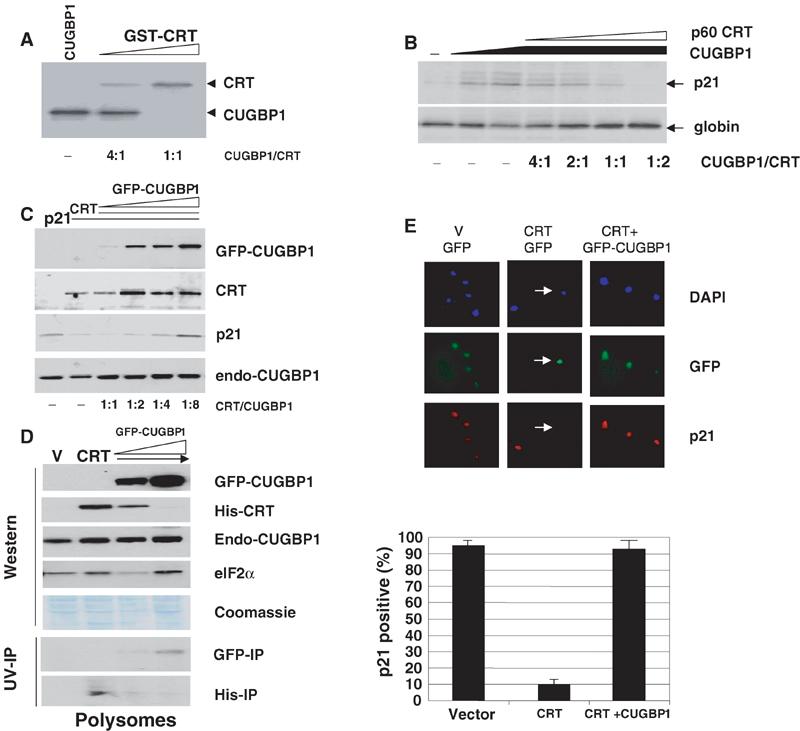
CUGBP1 and CRT compete for the interaction with p21 mRNA and for the regulation of its translation. (A) CRT and CUGBP1 compete for the binding to p21 mRNA. SL p21 probe was incubated with an excess of CRT or CUGBP1 at different molar ratios (shown below), linked by UV treatment and separated by gel electrophoresis. (B) CUGBP1 and CRT compete for the regulation of p21 translation in a cell-free translation system. WT p21 mRNA was translated in reticulocyte lysate containing CRT and CUGBP1 at different molar ratios. Translation of an unrelated globin mRNA in the same reactions is shown below. (C) CUGBP1 releases CRT-mediated repression of p21 translation in cultured cells. p21 was cotransfected with His-CRT and GFP-CUGBP1 at different molar ratios. The same membranes were probed with antibodies to p21, His-tag and CUGBP1. (D) Overexpression of CUGBP1 reduces the association of CRT with polysomes. Polysomal fractions were isolated from cells transfected with empty vector (V), with His-CRT and with CRT:CUGBP1 (1:4 and 1:8 ratios). A construct expressing p21 mRNA was included in each transfection. Western blotting was performed with antibodies to CUGBP1 and to His. The membrane was reprobed with antibodies to eIF2α to verify loading of polysomal fractions. The bottom part shows UV crosslink assay with His and GFP immunoprecipitates from polysomal fractions. (E) CUGBP1 blocks CRT-mediated repression of p21 in p21-HT3 clone. p21-HT3 cells were cotransfected with CRT+GFP or CRT+CFP-CUGBP1 (1:4 ratios). Expression of p21 was examined by immunostaining (red). The percentage of p21-positive transfected cells was calculated. Bar graphs show a summary of three independent experiments.
We next determined if the competition of CUGBP1 and CRT for binding to p21 mRNA affected the translation of p21. Figure 7B shows data obtained in a cell-free translation system. The addition of CUGBP1 increases translation of p21 (Figure 7B, lanes 2 and 3), while the addition of increasing amounts of p60-CRT to the translation mixtures abolishes the CUGBP1-dependent increase in p21 translation (Figure 7B). To examine whether CUGBP1 and CRT compete for the regulation of p21 in vivo, two approaches were used: triple cotransfection studies and analysis of p21 in the cells of a stable p21-HT3 clone transfected with CUGBP1 and CRT in different molar ratios. For triple cotransfection studies, p21 mRNA was cotransfected with His-CRT and GFP-CUGBP1 at different molar ratios. Antibodies to CUGBP1 recognize both GFP-CUGBP1 (80 kDa) and endogenous CUGBP1 (endo-CUGBP1, 51 kDa), which serves as a loading control. Figure 7C shows that CRT inhibits translation of p21 (lanes 2 and 3) and increasing expression of CUGBP1 relieves CRT-mediated inhibition of p21 translation.
To examine if the regulation of p21 expression by CRT and CUGBP1 takes place at the translational level, polysomal fractions were isolated from cells cotransfected with WT p21 mRNA, His-CRT and GFP-CUGBP1 at different ratios. Preliminary studies showed that the transfections of CUGBP1 and CRT together with WT p21 mRNA significantly increase the concentrations of each protein on polysomes (data not shown). This p21 mRNA-mediated enhancement of association of CRT and CUGBP1 with polysomes increases the sensitivity of subsequent Western and UV crosslink assays, and reduces a possible contribution of endogenous mRNAs. Western blotting of polysomal fractions shows that His-CRT localizes on polysomes in cells that translate very low levels of p21 (Figure 7D). Although total levels of CRT in the cytoplasm are not changed in cells cotransfected with GFP-CUGBP1 (Figure 7C, lanes 5 and 6), amounts of CRT are reduced in polysomal fractions of these cells. The polysomal fractions of these cells contain high concentrations of GFP-CUGBP1, which correlate with the induction of p21 translation (Figure 7C, lane 6). Examination of RNA-binding activities of His-CRT and GFP-CUGBP1 in polysomal fractions shows similar patterns of His-CRT and GFP-CUGBP1 associations with polysomes (Figure 7D, bottom). Taken together, these data demonstrate that the competitive regulation of p21 translation by CRT and CUGBP1 takes place on polysomes. To confirm these observations, we examined whether CUGBP1 can abolish CRT-mediated inhibition of p21 translation in the stable p21-HT3 clone. Figure 7E demonstrates that expression of CUGBP1 over CRT levels abolishes the inhibition of p21 translation in more than 90% transfected cells.
CUGBP1 enhances the biological functions of p21 by abolishing the inhibitory effect of CRT
To test whether the competition of CUGBP1 and CRT for the regulation of p21 might be involved in the irreversible growth arrest, we expressed CUGBP1 and CRT in p21-HT3 clone at the ratio under which CRT blocks CUGBP1 binding and at the ratio under which CUGBP1 relieves the inhibitory effect of CRT, and determined the biological consequences of the competition between these RNA-binding proteins. The results of colony formation assay are shown in Figure 8A. Expression of CRT blocks p21-mediated growth arrest; however, when CUGBP1 is expressed in the same cells at four- to eight-fold excess to CRT, it abolishes CRT-dependent inhibition of p21 growth arrest. We also examined DNA synthesis in p21-HT3 cells expressing CRT and CUGBP1 at different ratios. The measurements of BrdU uptake revealed that CUGBP1 relieves the CRT-dependent block of p21-mediated inhibition of DNA synthesis (Figure 8B). The expression of CUGBP1 over CRT levels also rescues p21-dependent development of flat cell formation (Figure 8C). These studies demonstrate that the competition between CRT and CUGBP1 for the regulation of p21 translation plays a critical role in the regulation of biological processes that are dependent on p21, such as growth arrest and development of the senescent-like phenotype.
Figure 8.
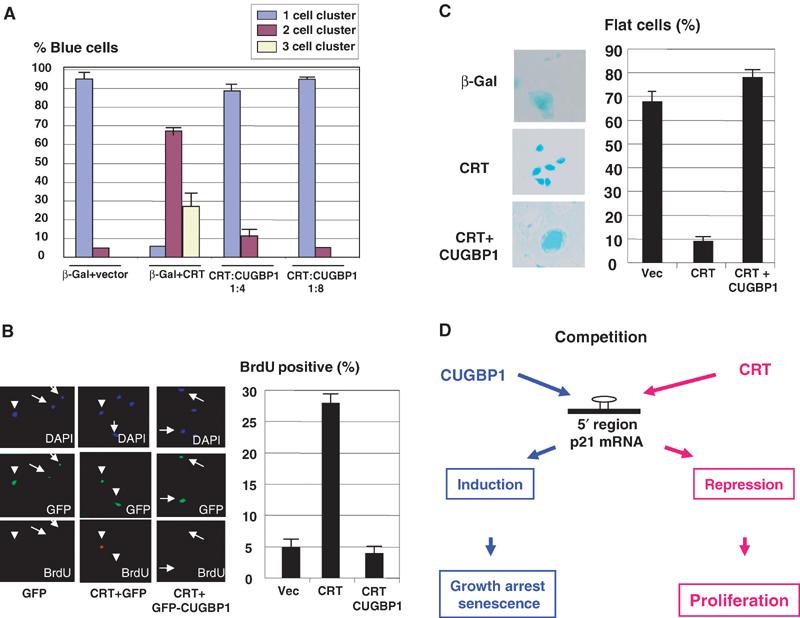
Competition of CRT and CUGBP1 determines whether cells are arrested by p21 or proliferate. (A) Colony formation assay in p21-HT3 cells expressing CRT and CUGBP1 at different ratios. p21-HT3 cells were transfected with β-gal and CRT+CUGBP1 at different molar ratios. p21 was induced at 4 h after transfections. The number of single cell clusters (arrested cells), two and three cell clusters (divisions) was calculated and presented as a summary of three repeats. (B) Overexpression of CUGBP1 abolishes CRT-mediated block of the inhibition of DNA synthesis by p21. p21-HT3 cells were transfected with GFP, CRT+GFP or CRT+GFP-CUGBP1. p21 was induced by IPTG, and BrdU was added 24 h after IPTG addition. Bar graphs show a summary of four independent experiments. (C). CUGBP1 enhances p21-mediated formation of flat cells. p21-HT3 cells were transfected with (β-gal+CRT) and with (β-gal+CRT+CUGBP1). Cells were stained for β-gal activity 4 days after p21 induction. A summary of three independent experiments is shown on the right. (D) Hypothetical model for the role of CUGBP1 and CRT in cell proliferation and senescence.
Discussion
The first step in the senescence, irreversible growth arrest, requires the biological function of an inhibitor of cyclin-dependent kinases, p21 (Stein et al, 1999; Dulic et al, 2000). Here, we present evidence that competition of two RNA-binding proteins for the regulation of p21 translation determines whether a cell will proliferate or undergo growth arrest and senescence. The analysis of protein–RNA interactions with a GC-rich sequence of the 5′ region of p21 mRNA demonstrated that binding of CUGBP1 to the p21 mRNA is dramatically induced in senescent fibroblasts. The aging-dependent activation of CUGBP1 is mediated by increased phosphorylation of CUGBP1, since treatment of CUGBP1 isolated from senescent cells with alkaline phosphatase dramatically reduces its binding to p21 mRNA. Human fibroblasts contain a kinase that is more active in senescent cells and is able to activate CUGBP1 (Figure 1E). A number of observations had demonstrated that the induction of p21 mRNA does not necessarily lead to the translation of p21 protein, suggesting an additional level of p21 control: inhibition of p21 mRNA translation. We found that the translation of p21 mRNA might be blocked by the RNA-binding protein, CRT, which interacts with the 5′ region of human p21 mRNA capable of forming the stable SL structure.
CRT is a multifunctional protein that is located on the endoplasmic reticulum (Michalak et al, 1999). Given the observed inhibitory activity of CRT on translation of C/EBP (Timchenko et al, 2002) and p21 mRNAs (this paper), we examined subcellular compartments where CRT might mediate this kind of activity. A relatively small portion of CRT (less than 2%) is associated with polysomes and this portion of CRT is able to interact with p21 mRNA. Interestingly, the PS-associated CRT represents the only portion of CRT capable of interacting with RNA. These findings suggest that the ‘active' CRT is operating on polysomes and seems to be involved in the regulation of translation of mRNAs. Although the precise mechanisms of translocation of CRT to polysomes are not known, one could suggest that the interaction of CRT with mRNAs might mediate its localization on polysomes. We have previously found that CUGBP1 is also associated with polysomes (Timchenko et al, 1999). Data in this paper suggest that the competition of CRT and CUGBP1 for p21 regulation on polysomes determines whether cells proliferate (when CRT binds to p21 mRNA) or undergo growth arrest (if CUGBP1 displaces CRT from p21 mRNA), and that this competition is also involved in the regulation of senescence (Figure 8D).
Here, we present data showing the control of cell growth and senescence by RNA-binding proteins through competitive regulation of p21 translation. This pathway of the regulation perhaps is not limited to growth arrest/proliferation by p21 mRNA alone, but may operate in other systems via control of translation of other mRNAs. It is interesting to note that CRT and CUGBP1 also interact with the 5′ region of C/EBPβ mRNA (Timchenko et al, 2002), and potentially might compete for the regulation of C/EBPβ translation. Given a key role of p21 and C/EBPβ in a variety of biological processes (Poli, 1998; Sherr and Roberts, 1999), one can suggest that RNA-binding proteins might be critical players of many biological systems via regulation of translation of these mRNAs and perhaps other as yet unknown mRNAs.
Materials and methods
RNA probes
Gel-shift and UV crosslink assays were performed with CUG8, p21-AU-rich (HuR binding site) and p21-GC-rich (CUGBP1 binding site) RNA oligomers. The locations of these oligomers on p21 mRNA and nucleotide sequences are shown in Figure 1.
Plasmids
Expression constructs for wild-type p21 and mutant p21 constructs were described in our earlier papers (Nakanishi et al, 1997; Timchenko et al, 2001). AU substitutions were incorporated into a left site of the SL of full-length p21 mRNA as shown in Figure 3B. The mutant SL-mut p21 was linked to the His-tag to distinguish its expression from endogenous p21. GST-CRT protein was isolated from bacteria as described (Timchenko et al, 2002). For expression in mammalian cells, human CRT cDNA was obtained in RT–PCR reaction and cloned into pcDNA-6-His vector. The nucleotide sequences of the cloned SL-mut-p21 and His-CRT were verified by sequencing. GFP-CUGBP1 was generated by fusing CUGBP1 cDNA to GFP.
UV-crosslink assay
Equal amounts of radioactive RNA probes (50–200 000 cpm) were incubated with proteins from young, quiescent and senescent fibroblasts for 30 min at room temperature and subjected to UV treatment for 5 min at 125 mJ. After electrophoresis, proteins were transferred onto the membrane and autoradiographed. The membranes were stained with Coomassie blue to verify protein loading.
UV crosslink-immunoprecipitation assay (UV-IP)
To identify CUGBP1 or CRT in RNA–protein complexes, proteins linked to the SL p21 probe were precipitated using antibodies to CUGBP1 or CRT. After washing, immunoprecipitates were loaded on a denaturing gel, transferred onto a membrane and autoradiographed.
Western analysis
Proteins were isolated from young, quiescent and senescent fibroblasts and analyzed by Western blotting as described (Wang et al, 2001a). Proteins were detected with antibodies against CUGBP1 (3B1), p21 (H164) and His-tag (Santa Cruz). After detection of proteins, the membranes were stripped and reprobed with antibodies against β-actin to verify protein loading.
Gel-shift analysis
The Conditions for electrophoretic mobility shift assay were described in our previous publication (Timchenko et al, 2002). Proteins (5–25 μg) were incubated with the SL p21 probe labeled with 32P-γ-ATP and T4 kinase. Where indicated, RNA competitors (100 ng) were added prior to probe addition.
Effect of CRT on translation of p21 mRNA in reticulocyte lysate
To examine the result of interaction of CRT with p21 mRNA, a cell-free coupled transcription/translation system in rabbit reticulocyte lysate (RL) was used. p60-CRT or GST-CRT was added into translation mixtures programmed with WT or mutant p21 constructs in the presence of 35S–methionine. After translation, p21 was immunoprecipitated from the reaction mixtures with specific antibodies and analyzed by electrophoresis and autoradiography.
Effect of CRT on translation of p21 mRNA in cultured cells
HT1080 cells were cotransfected with His-p21+His-CRT or His-p21+His-CRT:GFP-CUGBP1 constructs using the Fugene 6 transfection reagent (Boehringer, Mannheim) according to the manufacturer's protocol. To visualize transfected cells, His-CRT vector was cotransfected with a plasmid coding for a GFP at a ratio of 10:1. The efficiency of transfection was monitored by GFP green fluorescence. A summary of three to four independent experiments with two to three independent protein isolates is presented in this paper.
Generation of stable clonal line p21-HT3 and colony formation assay
A stable HT1080 cell line, which contains p21 under Lac-Repressor control, was generated as described in our paper (Timchenko et al, 1996b). p21 was induced by the addition of 10 mM IPTG. The formation of single- and multiple-cell colonies was induced in p21-HT3 cells as described in our earlier paper (Wang et al, 2001a). In each experiment, 150–200 transfected cells were used for the calculations. Bar graphs data in the manuscript represent summaries of three to four independent transfections.
Analysis of CRT−/− and CRT+/+ MEF
MEF were grown in DMEM media as described (Mesaeli et al, 1999). The experiments presented in this paper were performed with low-density cell plating (30%). For measurements of BrdU incorporation, CRT −/− MEF and WT MEF were partially synchronized by high density, plated and incubated with BrdU 24 h after plating for 1 h. Cells were fixed and stained with antibodies to BrdU as described (Wang et al, 2001a). For examination of cell cycle progression, cells were starved for 72 h in medium supplemented with 0.1% FBS and then stimulated by the addition of 10% FBS. Proteins were isolated at 3, 12 and 24 h after the stimulation and analyzed by Western blotting with antibodies to cyclins D1 and A (Santa Cruz Biotechnology).
Isolation of polysomes and examination of CRT in subcellular fractions
WT MEF were homogenized with polysomal buffer (PB) containing 25 mM Tris–HCl (pH 7.5), 100 mM NaCl, 5 mM MgCl2 and 25% sucrose. Nuclei and mitochondria were spun down by centrifugation at 15 000 rpm for 45 min. The supernatant (cytoplasm) was treated with 1% Triton X-100 and 0.5% DOX for 2 h and divided into two portions. Two methods were used for further isolation of PS: centrifugation through 1 M sucrose and precipitation of PS with high concentrations of MgCl2. One half of the cytoplasm was centrifuged through 1.5 M sucrose prepared on PB. PS pellet was resuspended in PB buffer and used for Western blotting and for UV crosslink. MgCl2 (80 mM) was added to the second half of the cytoplasm. After overnight incubation, polysomes were collected by centrifugation at 12 000 rpm for 15 min and resuspended in PB. The quality of ER and PS separation was examined by Western analysis with ER-specific protein, calnexin and PS-specific protein eIF2α (Figure 3).
Acknowledgments
This work was supported by NIH grants GM55188, CA100070 and AG20752 (NAT), AR49222 and AR44387 (LTT), and by grants from Muscular Dystrophy Association (LTT).
References
- Campisi J (2001) From cells to organisms: can we learn about aging from cells in culture? Exp Gerontol 36: 607–618 [DOI] [PubMed] [Google Scholar]
- Campos AR, Grossman D, White K (1985) Mutant alleles at the locus elav in Drosophila melanogaster lead to nervous system defects: a developmental genetic analysis. J Neurogenet 2: 197–218 [DOI] [PubMed] [Google Scholar]
- Chiu CP, Harley CB (1997) Replicative senescence and cell immortality: the role of telomeres and telomerase. Proc Soc Exp Biol Med 214: 99–106 [DOI] [PubMed] [Google Scholar]
- Dulic V, Beney G-E, Freburg G, Drullinger LF, Stein GH (2000) Uncoupling between phenotypic senescence and cell cycle arrest in aging p21-deficient fibroblasts. Mol Cell Biol 20: 6741–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan XC, Steitz JA (1998) Overexpression of HuR, a nuclear-cytoplasmic shutting protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J 17: 448–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesaeli N, Nakamura K, Zvaritch E, Dickie P, Dziak E, Krause K-H, Opas M, MacLennan DH, Michalak M (1999) Calreticulin is essential for cardiac development. J Cell Biol 144: 857–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak M, Corbett EF, Mesaeli NK, Nakamura K, Opas M (1999) Calreticulin: one protein, one gene, many functions. Biochem J 344: 281–292 [PMC free article] [PubMed] [Google Scholar]
- Nakanishi M, Robetorye RS, Pereira-Smith OM, Smith JR (1997) The C-terminal region of p21 is involved in proliferating cell nuclear antigen binding but does not appear to be required for growth inhibition. J Biol Chem 270: 17060–17063 [DOI] [PubMed] [Google Scholar]
- Poli V (1998) The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem 273: 29279–29282 [DOI] [PubMed] [Google Scholar]
- Pugachev K, Frey T (1998) Effects of defined mutations in the 5′ nontranslated region of rubella virus genomic RNA on virus viability and macromolecule synthesis. J Virol 72: 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr ChJ, Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 13: 1501–1512 [DOI] [PubMed] [Google Scholar]
- Singh NK, Atreya ChD, Nakashi HL (1994) Identification of calreticulin as a rubella virus RNA-binding protein. Proc Natl Acad Sci USA 91: 12770–12774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein GH, Drullinger LF, Soulard A, Dulic V (1999) Differential roles of cyclin dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Mol Cell Biol 19: 2109–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Iakova P, Cai Z-J, Smith JR, Timchenko LT (2001) Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol Cell Biol 21: 6927–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS (1996a) Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res 24: 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Wilde M, Nakanishi M, Smith JR, Darlington GJ (1996b) CCAAT/enhancer binding protein (C/EBPα) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev 10: 804–815 [DOI] [PubMed] [Google Scholar]
- Timchenko LT, Iakova P, Welm LA, Cai Z-J, Timchenko NA (2002) Calreticulin interacts with C/EBPα and C/EBPβ mRNAs and represses translation of C/EBP proteins. Mol Cell Biol 22: 7242–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko NA, Welm AL, Lu X, Timchenko LT (1999) CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPβ mRNA and regulates translation of C/EBPβ isoforms. Nucleic Acids Res 27: 4517–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Furnaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holdbrook N, Gorospe M (2000) HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol 20: 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Iakova P, Wilde M, Goode T, Welm AL, Roesler W, Timchenko NA (2001a) C/EBPα arrests cell proliferation through direct inhibition of cdk2 and cdk4. Mol Cell 8: 817–828 [DOI] [PubMed] [Google Scholar]
- Wang W, Yang X, Cristofalo VJ, Holbrook NJ, Gorospe M (2001b) Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol Cell Biol 21: 5880–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]