Abstract
The eukaryotic RecA homologue Rad51 is a key factor in homologous recombination and recombinational repair. Rad51-like proteins have been identified from yeast (Rad55, Rad57 and Dmc1) to vertebrates (Rad51B, Rad51C, Rad51D, Xrcc2, Xrcc3 and Dmc1). These Rad51-like proteins are all members of the genetic recombination and DNA damage repair pathways. The sequenced genome of Arabidopsis thaliana encodes putative homologues of all six vertebrate Rad51-like proteins. We have identified and characterized an Arabidopsis mutant defective for one of these, AtXRCC3, the homologue of XRCC3. atxrcc3 plants are sterile, while they have normal vegetative development. Cytological observation shows that the atxrcc3 mutation does not affect homologous chromosome synapsis, but leads to chromosome fragmentation after pachytene, thus disrupting both male and female gametogenesis. This study shows an essential role for AtXrcc3 in meiosis in plants and possibly in other higher eukaryotes. Furthermore, atxrcc3 cells and plants are hypersensitive to DNA-damaging treatments, supporting the involvement of this Arabidopsis Rad51-like protein in recombinational repair.
Keywords: Arabidopsis thaliana, meiosis, Rad51, recombination, Xrcc3
Introduction
Meiosis is the specialized cell division used in sexually reproducing organisms to produce haploid gametes from diploid cells (reviewed by Petronczki et al, 2003). During meiosis, a single round of DNA replication is followed by two successive divisions: the first (reductional) division segregates homologous chromosomes, and the second (equational) division separates sister chromatids from each other. The first division requires pairing and synapsis of homologous chromosomes to ensure accurate segregation, while the second releases sister chromatid cohesion. Homologous chromosome recognition (pairing), synaptonemal complex assembly (synapsis) and homologous recombination are the three events that promote and ensure bivalent formation and stability before the first anaphase division (reviewed by Yamamoto and Hiraoka, 2001).
In Saccharomyces cerevisiae and most likely in all eukaryotes, meiotic recombination is initiated by the Spo11 protein, a member of the type II topoisomerase family (Bergerat et al, 1997; Keeney et al, 1997). Spo11 creates DNA double-strand breaks (DSBs) along meiotic chromosomes (reviewed by Keeney, 2001); the resulting DNA ends are then processed to produce 3′ single-stranded tails (Sun et al, 1989, 1991; reviewed by Connelly and Leach, 2002), which can invade a homologous duplex. This meiotic recombination step is promoted by Rad51 and Dmc1, a Rad51-like protein expressed only in meiotic cells (Bishop et al, 1992; Shinohara et al, 1992, 1997; reviewed by Masson and West, 2001). These two proteins are members of the Escherichia coli RecA-like recombinase family (Aboussekhra et al, 1992; Bishop et al, 1992; Shinohara et al, 1992). RecA is a key factor in bacterial homologous recombination mechanism and can promote invasion of a duplex DNA by a homologous single-stranded molecule. Consistent with the fact that they are part of the RecA-like family, in vitro experiments have shown that such strand exchange reactions are promoted by Rad51 and Dmc1 (Sung, 1994; Hong et al, 2001). In addition to Dmc1, two Rad51 paralogs, Rad55 (Lovett, 1994) and Rad57 (Kans and Mortimer, 1991), have been identified in yeast. Rad55 and Rad57 form a heterodimer (Hays et al, 1995; Johnson and Symington, 1995; Sung, 1997), which is involved in Rad51-dependent recombination events (Sung, 1997). Yeast mutants impaired in any of the Rad51-like proteins are defective in meiotic recombination and exhibit reduced spore viability (reviewed by Paques and Haber, 1999; Symington, 2002).
Homologues of Dmc1 have been identified and shown to be involved in the meiotic process of several organisms (Diener and Fink, 1996; Pittman et al, 1998; Couteau et al, 1999; Mikosch et al, 2001), but unequivocal homologues of Rad55 and Rad57 have not been characterized in higher eukaryotes. Five Rad51 paralogs (Xrcc2, Xrcc3, Rad51B/Rad51L1, Rad51C/Rad51L2 and Rad51D/Rad51L3) have been identified in mammals, with 20–30% protein sequence identity to Rad51 and to each other. Physical interactions between the five human Rad51 paralogs have been demonstrated using the yeast two-hybrid assay (Schild et al, 2000), and coexpression of Xrcc3–Rad51C and Xrcc2–Rad51D in E. coli, and Rad51B–Rad51C in S. cerevisiae has shown that they can be purified as stable complexes (Kurumizaka et al, 2001, 2002; Sigurdsson et al, 2001). In vivo the five paralogs are associated in two complexes, one containing Rad51B, Rad51C, Rad51D and Xrcc2, and the other containing Rad51C and Xrcc3 (Masson et al, 2001; Liu et al, 2002; Wiese et al, 2002). Interestingly, Rad51 was never found associated with its paralogs, although it can interact with Xrcc3 in a yeast two-hybrid assay (Schild et al, 2000). The absence of these two complexes in cells lacking Rad51C suggests that it is a key factor in mammalian homologous recombination processes (French et al, 2002; Miller et al, 2002). As is the case for the RAD51 knockout (Tsuzuki et al, 1996), targeted disruption of mouse RAD51B, RAD51D and XRCC2 leads to embryonic lethality (Shu et al, 1999; Deans et al, 2000; Pittman and Schimenti, 2000). Recently, the Drosophila genes Spindle-B (Spn-B) and Spindle-D (Spn-D) have been related to XRCC3 and RAD51C, respectively (Abdu et al, 2003). Mutants impaired for Spn-B and Spn-D are partially sterile, defective for meiotic recombination, but not hypersensitive to DSB-inducing agents (Ghabrial et al, 1998; Abdu et al, 2003). Individual knockouts of each of the five paralogs in chicken B-lymphocyte DT40 cell lines are viable, but show spontaneous chromosomal aberrations, hypersensitivity to DNA-damaging treatments and reduced levels of homologous recombination (Takata et al, 2000, 2001). In vitro studies have shown that Xrcc3–Rad51C and Xrcc2–Rad51D complexes possess homologous pairing activities similar to those of RecA or Rad51 (Kurumizaka et al, 2001, 2002). Furthermore, the Rad51B–Rad51C complex possesses a mediator role in promoting the assembly of the presynaptic Rad51 nucleofilament, in in vitro Rad51-dependent strand exchange reactions, that is similar to that of the Rad55–Rad57 heterodimer in yeast (Sung, 1997; Sigurdsson et al, 2001). These data suggest that Rad51 paralogs are important factors for homologous recombination processes and the maintenance of the integrity of the genetic material.
In the plant Arabidopsis thaliana, homologues of RAD51 (AtRAD51) and DMC1 (AtDMC1) have been identified (Klimyuk and Jones, 1997; Doutriaux et al, 1998). Unlike yeast DMC1, AtDMC1 is expressed not only in reproductive tissues but also in leaves and cultured cell suspensions. Cytological observation of a T-DNA insertion mutant has shown that the AtDmc1 protein is required for bivalent formation and proper chromosome segregation during meiosis (Couteau et al, 1999). The completion of the Arabidopsis genome sequence has revealed genes related to the five Rad51 paralogs: AT2G28560 (RAD51B), AT2G45280 (RAD51C), AT1G07745 (RAD51D), AT5G64520 (XRCC2) and AT5G57450 (XRCC3). Predicted proteins have 14.2–26.7% sequence identity to AtRad51 and to each other, and 20.1–36.4% sequence identity to their human counterpart (EMBOSS Stretcher, default parameters). AtXRCC3 and AtRAD51C cDNAs have been recently cloned and sequenced, and two-hybrid analyses have confirmed that AtXrcc3 interacts with AtRad51 and AtRad51C (Osakabe et al, 2002). Furthermore, γ-irradiation has shown that transcription of AtXRCC3 and AtRAD51C is induced in response to DNA damage (Osakabe et al, 2002). These data strongly suggest that AtXRCC3 and AtRAD51C are the functional homologues of XRCC3 and RAD51C, but no further analyses have thus far investigated the roles of these Rad51 paralogs in plants. To understand the role of AtXRCC3, most probably a key factor in homologous recombination and recombinational repair, we have characterized a T-DNA insertion line disrupting the AtXRCC3 coding sequence. Here we demonstrate that atxrcc3 mutant cells and plants are hypersensitive to DNA crosslinking agents and less so to DSB-inducing agents. This hypersensitivity is consistent with the defects previously reported for vertebrate cells mutant for Xrcc3. Furthermore, we show that atxrcc3 mutant plants undergo aberrant meiosis with extensive chromosome fragmentation seen in postpachytene stages, leading to gametophytic lethality. This finding gives a new insight concerning the meiotic role of Xrcc3 and probably the other Rad51 paralogs.
Results
Identification and molecular characterization of an AtXRCC3 T-DNA insertion mutant
To investigate the AtXRCC3 gene function in plants, we searched for mutants in public T-DNA insertion line collections. A single line, Salk_045564, carrying an insertion at the 3′ end of the AtXRCC3 coding sequence was found in the SIGnAL T-DNA express database and the corresponding allele was named atxrcc3. The AtXRCC3/T-DNA junctions were amplified and PCR products were sequenced to determine the exact position and some structural features of the insertion (Figure 1A and B). The T-DNA insertion disrupts the AtXRCC3 coding sequence at position +704. The AtXRCC3 T-DNA insertion is surrounded by two incomplete left borders designated as LB1 and LB2. These two left borders have opposite orientations, which indicate the presence of at least two T-DNAs inserted at the AtXRCC3 locus. LB1 has a 7 bp deletion and an insertion of an adenine just upstream the T-DNA, generating a TAA STOP codon in-frame with the AtXRCC3 ORF. LB2 has a 22 bp deletion and a deleted guanidine two bases after LB2. Therefore, a truncated AtXrcc3 protein, missing the last 70 amino acids, might possibly be produced from the atxrcc3 allele. Primers upstream to the T-DNA insertion were used in semiquantitative RT–PCR amplification to check whether a transcript coding for a truncated AtXrcc3 protein could be detected in atxrcc3 mutant cells (Figure 1C). We were not able to detect any AtXRCC3 transcript in atxrcc3 samples. Thus, AtXRCC3 T-DNA insertion might destabilize or produce an aberrant atxrcc3 transcript, suggesting that atxrcc3 is a null allele.
Figure 1.
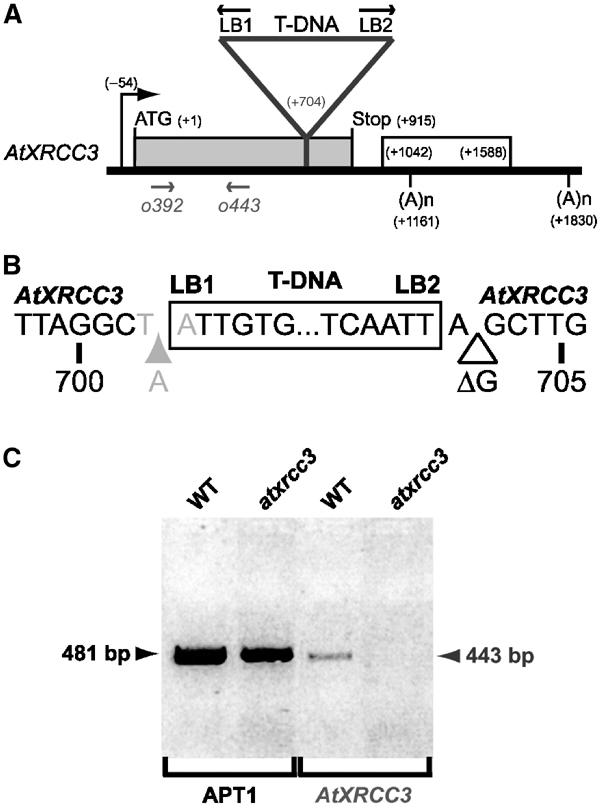
Molecular characterization of atxrcc3 T-DNA insertion. (A) Diagram showing the position of the T-DNA insertion at the AtXRCC3 locus. The black arrow indicates the AtXRCC3 transcriptional start site, the grey box represents the AtXRCC3 coding sequence and the white box the intron spliced in AtXRCC3 alpha mRNA. The triangle represents the T-DNA inserted in position +704 of the AtXRCC3 coding sequence. (B) Sequence of the atxrcc3 insertion site. The white box indicates the T-DNA insertion. LB1 and LB2 indicate the two left borders surrounding the T-DNA insertion, and their orientations are indicated in (A). The grey and white triangles represent base insertion and deletion, respectively. The in-frame TAA STOP codon resulting from LB1 integration is indicated in grey. The numbers represent positions relative to the start codon. (C) RT–PCR detection of AtXRCC3 transcript. WT and atxrcc3 indicate RT–PCR amplifications performed on wild-type and atxrcc3 total mRNAs extracted from cell suspensions. AtXRCC3 is the product amplified using primers o392 and o443 represented in (A). Amplification of the APT1 mRNA has been used as a control for reverse transcription.
atxrcc3 plants are sterile
A sterility phenotype was observed in the progeny of self-fertilized heterozygous atxrcc3+/− plants. Over 36 plants screened, eight (22.3%) were sterile and 28 (87.7%) were fertile, which correspond to the 3:1 segregation expected for a single Mendelian locus (X2, 1 d.f.=0.148). Using a PCR genotyping assay, we were able to follow the transmission of the atxrcc3 allele in these plants. Eight of the 36 plants (22.3%) were homozygous for the atxrcc3 T-DNA insertion, 19 of 36 (52.7%) were heterozygous and nine of 36 (25%) were wild type. This analysis of individual plants confirmed that sterile plants were exclusively those homozygous for the atxrcc3 allele. Compared with wild type, atxrcc3 plants produce atrophied siliques (Figure 2B and C) and almost all these siliques are devoid of any seed. atxrcc3 mutants produce an average of 2.25 seeds per plant (n=8), compared to approximately 4500 seeds per fertile plant (n=4), and this represents a residual fertility of 0.05%. Heterozygous plant fertility was similar to wild-type plants, and all plants, regardless of their AtXRCC3 genotype, had normal vegetative development (Figure 2A and B). Thus, AtXrcc3 is absolutely required for fertility, but not for vegetative development.
Figure 2.

atxrcc3 mutant plants are sterile. (A) Wild-type (left) and atxrcc3 (right) 3-week-old plants. Flowering stems (B) and siliques (C) of the wild-type and atxrcc3 plants.
atxrcc3 plants are defective in both male and female gametophytic development
To determine the origin of the sterility phenotype observed in atxrcc3 plants, we first examined male and female gametophytic development. To assess pollen grain viability, anthers were dissected from wild-type and mutant flower buds (Figure 3A and B) and stained as described by Alexander (1969). None of the 60 observed atxrcc3 anthers contained any mature pollen grains, suggesting that male gametophytic development is arrested after meiosis. Heterozygous plants' anthers could not be differentiated from those of wild-type plants in terms of viability and number of pollen grains produced in a single anther. Male gametogenesis is thus strongly affected in the atxrcc3 mutant.
Figure 3.

Gametophytic lethality in atxrcc3 mutant plants. Anthers of wild-type (A) and atxrcc3 (B) plants have been stained according to Alexander (1969). Anthers from 2 mm buds (up) and mature flower (down) have been compared. The right of each panel presents enlargement of some pollen grains. The red-purple-stained cytoplasm indicates viability, while the pollen cell wall is counterstained in green. (C–J) Differential interference contrast (DIC) microscopy observations of embryo sac development in wild-type (C–F) and atxrcc3 (G–J) ovules, after clearing. The bottom of each panel presents an enlargement of the nuclei. MMC=megaspore mother cell, N=nucleus, DC=degenerative cell, bars=20 μm.
To check whether female gametogenesis is also affected, we monitored embryo sac development in wild-type and atxrcc3 ovules (n=200). In wild-type ovules, the megaspore mother cell (Figure 3C) undergoes meiosis and three of the four products degenerate to preserve a single functional megaspore (Figure 3D). This megaspore then undergoes three rounds of nuclear division to produce the eight nuclei embryo sac, which is the mature female gametophyte. Intermediate stages from the functional megaspore to the eight nuclei embryo sac can be observed in wild-type plants, while the simultaneous organization of integuments and growth around the megaspore can be used to determine the stage of megagametogenesis (Figure 3C–F). Observation of atxrcc3 mutants revealed that embryo sac development is disrupted after meiosis. The megaspore mother cell could not be differentiated from the wild type (Figure 3G), but the majority of atxrcc3 ovules do not preserve a functional megaspore, with only a degenerative cell being visible and persisting during embryo sac development (Figure 3H and I). In some cases one of the meiotic products survived, but precise staging of the ovules with growth of integuments indicated that these products did not undergo the nuclear divisions (Figure 3J). Thus, the atxrcc3 mutation disrupts both male and female gametophytic development, leading to a severe sterility phenotype.
Meiosis is severely disrupted in atxrcc3 plants
To further characterize the atxrcc3 sterility phenotype, we followed meiosis in pollen mother cells (PMCs). Meiotic progression in wild-type (Figure 4) and atxrcc3 (Figure 5) PMCs was examined by fluorescence microscopy after 4′,6-diamidino-2-phenylindole (DAPI) staining of chromosomes. The Arabidopsis genome consists of five pairs of chromosomes (2n=10) that can be observed as bivalents by the end of prophase I to metaphase I (Figure 4C–E). During meiotic prophase I, individual chromosomes become visible at leptotene, while the beginning of pairing is observable at zygotene (Figure 4A). The pachytene stage is characterized by fully synapsed homologous chromosomes (Figure 4B) and indicates the end of meiotic recombination events initiated in leptotene. Then, chromosomes gradually condense and separate, remaining linked by chiasmata at diplotene and diakinesis (Figure 4C and D).
Figure 4.

Meiosis in wild-type Arabidopsis: (A) zygotene, (B) pachytene, (C) diplotene, (D) diakinesis, (E) metaphase I, (F) anaphase I, (G) telophase I, (H) metaphase II, (I) anaphase II, (J) telophase II, (K) tetrad and (L) microspores. Bars=10 μm.
Figure 5.

Meiosis is severely disrupted in atxrcc3 mutant plants: (A) zygotene, (B) pachytene, (C) diplotene, (D) diakinesis, (E) metaphase I, (F) anaphase I, (G) telophase I, (H) metaphase II, (I) anaphase II, (J) telophase II, (K) polyads and (L) microspores. Yellow arrows=bivalents, red arrows=chromosome fragments, b=bridge, mn=micronucleus, bars=10 μm.
In atxrcc3 PMCs, prophase I seems to occur normally up to pachytene, as fully synapsed chromosomes can be observed (Figure 5A and B). To eliminate the possibility that the apparent synapsis results from nonhomologous interactions, fluorescent in situ hybridization (FISH) was performed on atxrcc3 pachytene chromosomes (Figure 6). Hybridization with a 5S rDNA probe yielded the expected three signals for normally paired homologous chromosomes, corresponding to the three major 5S rDNA loci in the Columbia ecotype (Murata et al, 1997; Fransz et al, 1998). The absence of univalents at metaphase I finally confirmed that homologous chromosome pairing and synapsis take place normally in atxrcc3 mutants (Figure 5E). At diplotene, the presence of chromosome fragments (red arrows) in addition to bivalents (yellow arrows) (Figure 5C) underlines the severe meiotic deficiency. At diakinesis, bivalents are difficult to differentiate from chromosome fragments (Figure 5D).
Figure 6.

Pachytene chromosomes of atxrcc3 hybridized with 5S rDNA (red) and DAPI counterstained: (A) DAPI, (B) 5S rDNA signals and (C) merged images. Arrows=5S rDNA signals.
At metaphase I in the wild type, chromosomes reach their maximum condensation state, and can be seen as five bivalents orientated on the spindle (Figure 4E). Homologous chromosomes separate from each other and migrate to the opposite poles of the cell in anaphase I (Figure 4F). The first meiotic division ends with partial decondensation of chromosomes at telophase I (Figure 4G). In atxrcc3 PMCs, in addition to the five bivalents, a variable number of chromosome fragments are seen (Figure 5E). Chromosome fragmentation continues during anaphase I, and bridges between separating chromosomes are frequently observed (Figure 5F). First division finally produces two pools of ‘chromosomes' containing more than the five chromosomes expected for Arabidopsis (Figure 5G), presumably due to random assortment of acentric chromosome fragments at anaphase I.
The second meiotic division starts with rapid condensation and alignment of chromosomes (metaphase II, Figure 4H). In anaphase II, sister chromatids separate and migrate to the opposite poles of the cell (Figure 4I), resulting in four groups of five chromosomes. Telophase II ensues, chromosomes decondense (Figure 4J) and cytoplasm is partitioned to produce a tetrad containing four haploid microspores (Figure 4K and L). In atxrcc3 metaphase II, most visible fragments are aligned on the spindle, with some fragments being isolated (Figure 5H). During anaphase II, four groups of ‘chromosomes' are separated, with fragments being scattered throughout the cytoplasm (Figure 5I). At telophase II, more than the four normal nuclei are observed (Figure 5J) and polyads containing three or four major products, associated with several micronuclei, are generated (Figure 5K). These meiotic products finally give rise to microspores with varying size and DNA content (Figure 5L). This cytological analysis of meiosis in atxrcc3 PMCs demonstrates that AtXrcc3 is required to ensure chromosome integrity during meiosis.
atxrcc3 cells and plants are hypersensitive to mitomycin C and less so to bleomycin
Previous studies with CHO (Chinese hamster ovary) and DT40 (chicken B-lymphocyte) mutant cells have shown that Xrcc3 is required for efficient repair of DNA damage and especially DNA crosslinks (Tebbs et al, 1995; Liu et al, 1998; Cui et al, 1999; Pierce et al, 1999; Brenneman et al, 2000; Takata et al, 2000, 2001). To investigate the role of AtXrcc3 in DNA damage repair, we first determined whether the atxrcc3 mutation confers hypersensitivity to mitomycin C (MMC), a DNA crosslinking agent (Warren et al, 1998). Cultured cells derived from wild-type and atxrcc3 plants were transferred to plates containing from 0 to 40 μM of MMC and growth was scored visually after 3 weeks. As shown in Figure 7A, atxrcc3 cells are highly sensitive to MMC, as a dose of 5 μM is sufficient to reduce callus growth. In all, 10 μM MMC severely affected the growth of mutant cells, while wild-type cells grow normally at this MMC concentration. Doses of 20 μM MMC and above strongly reduce the growth of both mutant and wild-type cells. Thus, AtXrcc3 is involved in the repair of DNA crosslinks.
Figure 7.
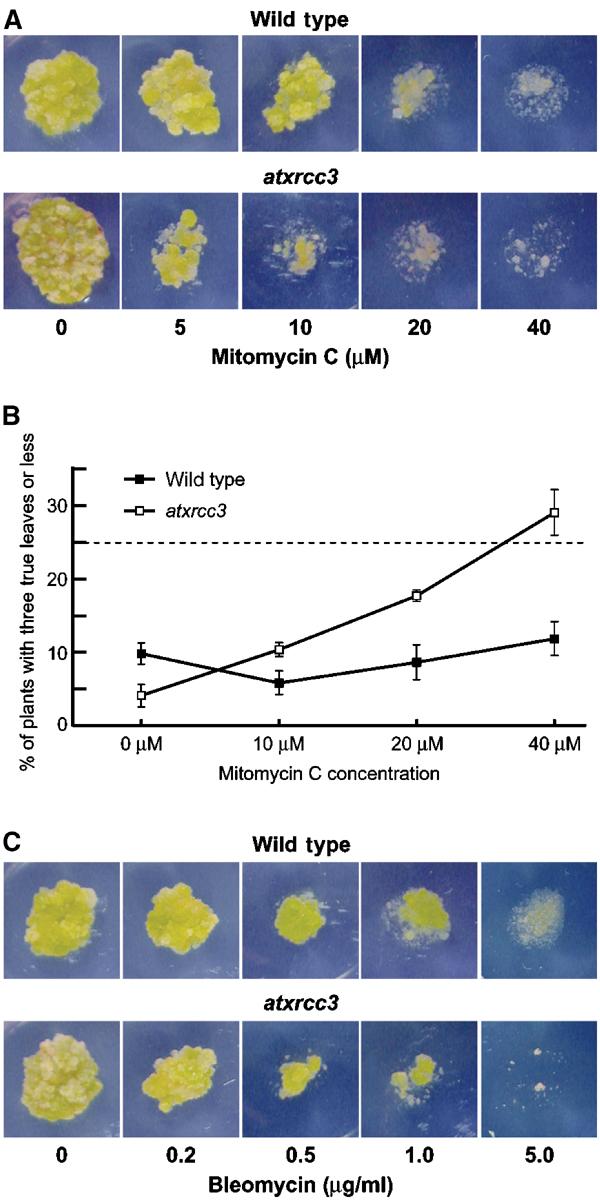
atxrcc3 cells and plants are hypersensitive to DNA crosslinking agents, but not to DSB-inducing agents. (A, C) Five-day-old wild-type and atxrcc3 cell cultures were transferred and grown on plates containing increasing doses of mitomycin C (A) or bleomycin (C). After 3 weeks, callus growth was used to score sensitivity to DNA-damaging agent. (B) Seeds of wild-type and atxrcc3+/− heterozygote plants were sown on plates containing increasing doses of mitomycin C. After 14 days, the percentage of hypersensitive plants (plants with three leaves or less) was used to produce a mitomycin C dose–response curve. Values represent three replicates, each replicate containing an average of 100 plants per dose. Error bars=standard deviation.
To confirm the MMC hypersensitivity phenotype observed with atxrcc3 cells, seeds from wild-type and self-fertilized heterozygous atxrcc3+/− plants were sown on plates containing germination medium and increasing doses of MMC. After 14 days the plants were scored for MMC hypersensitivity. Without treatment, most of the plants developed at least four true leaves (excluding the cotyledons); thus, plants with three true leaves or less, in the presence of MMC, were considered as hypersensitive. A dose–response curve was produced for the percentage of plants with three leaves or less at 14 days (Figure 7B), showing a clear MMC hypersensitivity of the atxrcc3 plants. In the wild type, the percentage of hypersensitive plants rises from 5.8% at 10 μM MMC to 11.9% at 40 μM MMC, with most of them having two or three leaves. The MMC hypersensitivity of atxrcc3 plants was assayed on the progeny of an atxrcc3+/− plant (atxrcc3 mutants are sterile), and the atxrcc3 mutant plants thus represent only one quarter of the plants sown. In the presence of MMC, the percentage of hypersensitive plants in the progeny of an atxrcc3+/− plant rises from 10.4% at 10 μM MMC to 29% at 40 μM MMC, with half of them having 0 or 1 leaves. That these hypersensitives correspond to the atxrcc3 mutants was verified by PCR genotyping in one experiment: of 103 plants scored, 28 (27.2%) were hypersensitive to MMC and 26 of these were atxrcc3 mutant plants. These results confirm that AtXrcc3 is involved in the repair of DNA crosslinks and show that AtXrcc3 is required for plant development in the presence of such lesions.
To further investigate the role of AtXrcc3 in DNA damage repair and especially in DSB repair, hypersensitivity to bleomycin was scored on cell cultures and plants. Bleomycin is a γ-ray mimetic agent known to induce DNA DSBs (Favaudon, 1982). Cells and plants were grown on plates containing from 0 to 5 μg/ml of bleomycin. As shown in Figure 7C, a dose of 0.2 μg/ml bleomycin does not affect callus growth in either wild-type or mutant cell cultures. For doses of 0.5 μg/ml bleomycin and above, the growth of atxrcc3 cells was significantly more affected than wild type. The highest dose of 5.0 μg/ml bleomycin was fully lethal for atxrcc3 cells and considerably affects wild-type callus growth. However, in contrast to the MMC data, no significant difference was found between wild-type and atxrcc3 plants germinated on medium containing the same range of doses of bleomycin (data not shown). We have also observed the sensitivity of atxrcc3 cells for doses of 100 ppm methylmethane sulphonate (MMS) and above, while the sensitivity of the wild type is only seen at 166 ppm MMS and above (data not shown). These findings suggest that, like its vertebrate counterpart, AtXrcc3 is involved in repair of DNA DSBs, but is not required for plant survival in the presence of DSB-inducing agent.
Discussion
The sterility of atxrcc3 mutant is due to a meiotic defect
The most visible defect of the atxrcc3 mutant is the almost complete absence of seed following flowering (Figure 2). Cytological observations of male and female gametogenesis show a very high lethality of gametophytes (Figure 3), which is consistent with the strong sterility phenotype. Although we did not observe any mature gametophytes, the presence of arrested female gametophytes (Figure 3J) suggests that a few viable gametophytes can be produced, thus explaining the very low residual fertility of mutant plants.
Arabidopsis mutants defective for genes coding proteins involved in recombination (AtDMC1 and AtSPO11-1) and DNA damage repair (AtATM and MEI1) during meiosis have reduced gametophytic viability and fertility (Couteau et al, 1999; Grelon et al, 2001, 2003; Garcia et al, 2003). In these mutants, the gametophytic lethality has been correlated with a strong meiotic defect, without meiotic arrest. We show here that the atxrcc3 mutation leads to aberrant meiosis, with extensive chromosome fragmentation (Figure 5). The production of such (acentric) chromosome fragments would lead to aneuploid gametes and so explain the high gametophytic lethality.
AtXrcc3 is not required for synapsis
Rad51 paralogs play significant roles in homologous recombination and recombinational repair (reviewed by Thompson and Schild, 2001), but the embryonic lethality of mice defective in the Rad51 paralogs has precluded the investigation of their role in meiotic recombination and meiosis in a more general way. Our results demonstrate the requirement for AtXrcc3 to ensure the integrity of the genetic material during meiosis in Arabidopsis, suggesting a role for the AtXrcc3 protein in meiotic recombination. Bishop et al (1998) have shown that Xrcc3 is required for assembly of damage-induced Rad51 foci in CHO cells. Rad51 foci are also formed along meiotic chromosomes, presumably to promote recombinational repair of Spo11-induced DSBs (Barlow et al, 1997); however, the inviability of mouse xrcc3 mutants has precluded determining whether Xrcc3 plays a role in this. Thus, AtXrcc3 could be required for early steps of meiotic recombination. Previous studies have reported the meiotic phenotypes of Arabidopsis mutants defective for DMC1 and SPO11 homologues, two proteins involved in early steps of meiotic recombination (Couteau et al, 1999; Grelon et al, 2001). AtSpo11-1 is presumably required to introduce DSBs along meiotic chromosomes, while AtDmc1 might promote strand exchange between homologous chromosomes (Couteau et al, 1999; Grelon et al, 2001). These two mutants are both deficient for meiotic homologous chromosome synapsis, attested to by the 10 univalents, rather than five bivalents, observed at metaphase I (Couteau et al, 1999; Grelon et al, 2001). These results are consistent with the phenotypes reported for yeast and mice defective for either SPO11 or DMC1 (Giroux et al, 1989; Bishop et al, 1992; Rockmill et al, 1995; Bergerat et al, 1997; Keeney et al, 1997; Schwacha and Kleckner, 1997; Pittman et al, 1998; Yoshida et al, 1998; Romanienko and Camerini-Otero, 2000). The atxrcc3 mutant phenotype differs markedly from those observed for atdmc1 and atspo11-1. The presence of normal pachytene figures (Figures 5B and 6A) and the observation of bivalents at metaphase I (Figure 5E) show that synapsis takes place normally in atxrcc3 mutants and was confirmed by FISH analysis of the 5S rDNA loci (Figure 6). AtXrcc3 is thus not required for the establishment of inter-homologue synapsis and presumably not for early steps of meiotic recombination.
AtXrcc3 is essential for postsynaptic events in meiosis
Diplotene and later stages of atxrcc3 meiosis show the presence of chromosome fragments (Figure 5), while atdmc1 and atspo11-1 do not accumulate chromosomal breaks (Couteau et al, 1999; Grelon et al, 2001). The chromosome fragmentation observed in the atxrcc3 mutant is more similar to the meiotic phenotypes reported for Arabidopsis atatm and mei1 mutants (Garcia et al, 2003; Grelon et al, 2003). These two proteins are involved in DNA damage repair, AtAtm being required for the repair of DSBs (Garcia et al, 2003) and Mei1 presumably for premeiotic repair of damage associated with replication (Grelon et al, 2003). As we show here for atxrcc3, atatm and mei1 mutations lead to extensive chromosome fragmentation associated with bridges in both anaphase I and II (Garcia et al, 2003; Grelon et al, 2003). Such phenotypes are also seen in the Arabidopsis dif1/syn1 cohesin mutant (Bai et al, 1999; Bhatt et al, 1999; Peirson et al, 1997) and an atrad50 mutant (J-Y Bleuyard and C White unpublished). As reported for mei1 (Ross et al, 1997; Grelon et al, 2003) and in contrast to atatm (Garcia et al, 2003), atxrcc3 chromosome fragmentation is observed prior to anaphase I, as early as the diplotene stage (Figure 5C). The chromosome fragmentation seen in atxrcc3 thus cannot be solely explained by the breakage of anaphase bridges (as for atatm), but must derive from the nonrepair of DNA breaks. Liu et al (2003) have recently shown that in vitro, Rad51C and Xrcc3 are involved in the resolution of Holliday junctions, supporting a late role in recombination for Rad51 paralogs. A lack of resolvase activity in atxrcc3 mutants would explain the presence of bridges at anaphase I and II, as well as the early fragmentation of chromosomes, when synapsed homologues start to separate at diplotene. In the case of mei1, Grelon et al (2003) showed that the chromosome fragmentation phenotype is not Atspo11 dependent and thus the DNA breaks presumably are of premeiotic origin. We are performing the cross between atxrcc3+/− and atspo11-1+/− heterozygous plants to produce and characterize an atxrcc3/atspo11-1 double mutant.
AtXrcc3 is also important for somatic DNA repair
The fragmentation of meiotic chromosome in atxrcc3 mutant thus derives from the breakage of anaphase bridges and the nonrepair of (previously existing?) DNA breaks. Vertebrate Rad51 paralogs are required for efficient DNA damage repair in somatic cells (reviewed by Thompson and Schild, 2001). The hypersensitivity of atxrcc3 cells and/or plants to MMC and bleomycin clearly demonstrates that AtXrcc3 is involved in the repair of DNA crosslinks and DSBs in Arabidopsis (Figure 7), supporting the hypothesis that AtXrcc3 is the Arabidopsis functional homologue of vertebrate Xrcc3.
In conclusion, Arabidopsis AtXrcc3 plays essential roles in DNA damage repair in both somatic and meiotic cells. As is the case with Drosophila and in contrast to the situation in mice, AtXrcc3 is not required for normal development in Arabidopsis. Further studies will be necessary to understand fully at a mechanistic level the roles of AtXrcc3 in Arabidopsis, in particular in meiotic and somatic recombination and recombinational repair.
Materials and methods
Plant material, growth conditions and mutant screening
A. thaliana seeds (Columbia ecotype) were sown directly into damp compost, and plants were grown in a greenhouse under standard conditions.
The atxrcc3 T-DNA insertion line (Salk_045564) was found in the public searchable database established by the Salk Institute Genomic Analysis Laboratory (Alonso et al, 2003). The T-DNA Express database is accessible from the SIGnAL website at http://signal.salk.edu.
As the kanamycin resistance gene present in atxrcc3 T-DNA insertion is silenced, plants heterozygous and homozygous for the atxrcc3 mutation were identified by a PCR genotyping assay. For each individual plant, the following primer combinations were used to amplify the wild-type ATXRCC3 locus, o438 (5′-ATGCAAAATGGGAAAATTAAGCCG-3′) and o439 (5′-CTACGCTTGAACCGCACAAATC-3′), and the mutant locus, o447 (5′-GGATTTGGTTGAAACTTCTGATGG-3′) and o405 (5′-TGGTTCACGTAGTGGGCCATCG-3′). Mutant identification was confirmed based on their sterility phenotype.
Sequencing of T-DNA insertion sites
The following primer combinations were used to amplify DNA flanking the T-DNA: at the LB1 left border, o392 (5′-CGAATCGTAAACTAACCACAGGC-3′) and o406 (5′-GCGTGGACCGCTTGCTGCAACT-3′), and at the LB2 left border, o439 (5′-CTACGCTTGAACCGCACAAATC-3′) and o405 (5′-TGGTTCACGTAGTGGGCCATCG-3′).
The PCR products were then purified on a QIAquick column (Qiagen) and directly sequenced. Sequence reactions were performed using one of the primers used for amplification and the CEQ DTCS Quick Start Kit (Beckman Coulter), and analysed on a CEQ 2000 DNA Analysis System (Beckman Coulter).
Semiquantitative RT–PCR
For semiquantitative RT–PCR, total RNAs extracted from 7-day-old wild-type and atxrcc3 cell suspensions were treated with RNase-free DNase I (Roche). In all, 1 μg of DNA-free total RNA was reverse transcribed in 20 μl of reaction mixture containing 50 U of Expand Reverse Transcriptase (Roche), 1 × random hexanucleotide mix (Roche), 1 mM of each deoxyribonucleotide triphosphate and 20 Us of RNasin ribonuclease inhibitor (Promega). PCR was carried out in 25 μl of reaction mixture containing 2 μl of RT reaction mixture, 1 U of HotStarTaq DNA polymerase (Qiagen), 2.5 mM MgCl2, 100 μM of each deoxyribonucleotide triphosphate and 0.4 μM of gene-specific primers. The gene-specific primers were o392 (5′-CGAATCGTAAACTAACCACAGGC-3′) and o443 (5′-CAAACTCCGATCTAAACAATGC-3′) for AtXRCC3, and apt1 (5′-TCCCAGAATCGCTAAGATTGC-3′) and apt2 (5′-CCTTTCCCTTAAGCTCTG-3′) for APT1 (adenine phosphoribosyl transferase) (Moffatt et al, 1994). The initial denaturation was carried out at 94°C for 15 min, and then amplification was performed for 35 cycles with a denaturation time of 30 s at 94°C, followed by annealing for 30 s at 52°C and extension for 1 min at 72°C.
Light and fluorescence microscopy
Mature pollen grain viability was assayed according to Alexander (1969). Anthers from fixed flowers were isolated, stained and observed using a Leica MZFLIII stereomicroscope, and photographs were taken using a JVC digital camera.
Embryo sac development was monitored according to Motamayor et al (2000). Full inflorescences were collected, fixed and cleared in lactic acid/phenol. Ovules from different sized pistils were then dissected on a slide in a drop of the lactic acid/phenol solution and mounted in a drop of the same mix. Slides were observed with differential interference contrast (DIC) using a Zeiss Axioplan 2 Imaging microscope.
DAPI staining of meiotic chromosomes was performed as originally described by Maluszynska and Heslop-Harrison (1991), and modified as follows. Whole inflorescences were collected and fixed in fresh ethanol:acetic acid (3:1) for 1 h on ice. Fixative solution was renewed several times, until it remained clear. Fixed inflorescences were washed for 5 min at room temperature in 10 mM citrate buffer (pH 4.8) and digested in citrate buffer with 0.2% (w/v) cellulase (Sigma #C-1794) and 2% (v/v) pectinase (Sigma 1#P-4716) for 30 min at 37°C, in a moist chamber. Enzymes solution was carefully removed and inflorescences were washed for 5 min and then kept in citrate buffer. Buds of 0.1–0.5 mm were selected and anthers were dissected on a slide, in 10 μl of 45% acetic acid. Anthers were slightly squashed under a coverslip by gently tapping them with the base of a needle (release of the meiocytes was monitored under a stereomicroscope). Slides were frozen for 30 s in liquid nitrogen, and the coverslip was quickly removed with a razor blade. The slides were air dried and mounted in VECTASHIELD mounting medium with 1.5 μg/ml DAPI. UV fluorescence microscopy observations were performed using a Zeiss Axioplan 2 Imaging microscope.
Fluorescent in situ hybridization
FISH experiments were performed according to Schubert et al (2001). 5S rDNA was amplified with primers 5Suniv1 (5′-CTTTTCGGGCNTTTTNGTG-3′) and 5Suniv2 (5′-CGAAAAGGTATCACATGCC-3′), and labelled using the DIG-Nick Translation Mix (Roche). Mouse antidigoxigenin (1:250, Roche) followed by rabbit anti-mouse conjugated with fluorescein isothiocyanate (FITC) (1:1000, Sigma) and goat anti-rabbit conjugated with Alexa 488 (1:200, Molecular Probes) antibodies were used for the detection of the digoxigenin-labelled probe.
MMC, bleomycin and MMS sensitivity tests
Callus cultures were derived from leaves and maintained in SIM medium as described previously by Gallego and White (2001). A droplet of a 5-day-old liquid culture was transferred onto the surface of agar plates containing fresh solid SIM medium and different concentrations of bleomycin (Sigma #B-5507), MMC (Sigma #M-0503) or MMS (Sigma #M-4016). The plates were then incubated (23°C, 16 h light), and resistance or sensitivity was scored visually 3 weeks later.
Seeds were surface-sterilized with a 7% calcium hypochlorite solution (w/v). After sterilization, seeds were sown on plates containing fresh solid germination medium with different concentrations of bleomycin (Sigma #B-5507) or MMC (Sigma #M-0503). The plates were then incubated for 14 days (23°C, 16 h light), and resistance or sensitivity was scored by the number of true leaves (excluding the cotyledons) per plant.
Images processing
All images were further processed with Adobe Photoshop 6.0 to enhance their quality.
Acknowledgments
We thank members of BIOMOVE for their help and discussions. Olivier Mathieu is thanked for his help with Arabidopsis cytology and FISH. We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis TDNA insertion mutants. This work was partly financed by a European Union research grant (QLG2-CT-2001-01397).
References
- Abdu U, Gonzalez-Reyes A, Ghabrial A, Schupbach T (2003) The Drosophila spn-D gene encodes a RAD51C-like protein that is required exclusively during meiosis. Genetics 165: 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboussekhra A, Chanet R, Adjiri A, Fabre F (1992) Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol Cell Biol 12: 3224–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander MP (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bai X, Peirson BN, Dong F, Xue C, Makaroff CA (1999) Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell 11: 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow AL, Benson FE, West SC, Hulten MA (1997) Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J 16: 5207–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P (1997) An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414–417 [DOI] [PubMed] [Google Scholar]
- Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C (1999) The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J 19: 463–472 [DOI] [PubMed] [Google Scholar]
- Bishop DK, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum RR, Shinohara A (1998) Xrcc3 is required for assembly of Rad51 complexes in vivo. J Biol Chem 273: 21482–21488 [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N (1992) DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456 [DOI] [PubMed] [Google Scholar]
- Brenneman MA, Weiss AE, Nickoloff JA, Chen DJ (2000) XRCC3 is required for efficient repair of chromosome breaks by homologous recombination. Mutat Res 459: 89–97 [DOI] [PubMed] [Google Scholar]
- Connelly JC, Leach DR (2002) Tethering on the brink: the evolutionarily conserved Mre11–Rad50 complex. Trends Biochem Sci 27: 410–418 [DOI] [PubMed] [Google Scholar]
- Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, Doutriaux MP (1999) Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell 11: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Brenneman M, Meyne J, Oshimura M, Goodwin EH, Chen DJ (1999) The XRCC2 and XRCC3 repair genes are required for chromosome stability in mammalian cells. Mutat Res 434: 75–88 [DOI] [PubMed] [Google Scholar]
- Deans B, Griffin CS, Maconochie M, Thacker J (2000) Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J 19: 6675–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener AC, Fink GR (1996) DLH1 is a functional Candida albicans homologue of the meiosis-specific gene DMC1. Genetics 143: 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doutriaux MP, Couteau F, Bergounioux C, White C (1998) Isolation and characterisation of the RAD51 and DMC1 homologs from Arabidopsis thaliana. Mol Gen Genet 257: 283–291 [DOI] [PubMed] [Google Scholar]
- Favaudon V (1982) On the mechanism of reductive activation in the mode of action of some anticancer drugs. Biochimie 64: 457–475 [DOI] [PubMed] [Google Scholar]
- Fransz P, Armstrong S, Alonso-Blanco C, Fischer TC, Torres-Ruiz RA, Jones G (1998) Cytogenetics for the model system Arabidopsis thaliana. Plant J 13: 867–876 [DOI] [PubMed] [Google Scholar]
- French CA, Masson JY, Griffin CS, O'Regan P, West SC, Thacker J (2002) Role of mammalian RAD51L2 (RAD51C) in recombination and genetic stability. J Biol Chem 277: 19322–19330 [DOI] [PubMed] [Google Scholar]
- Gallego ME, White CI (2001) RAD50 function is essential for telomere maintenance in Arabidopsis. Proc Natl Acad Sci USA 98: 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Bruchet H, Camescasse D, Granier F, Bouchez D, Tissier A (2003) AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial A, Ray RP, Schupbach T (1998) okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev 12: 2711–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux CN, Dresser ME, Tiano HF (1989) Genetic control of chromosome synapsis in yeast meiosis. Genome 31: 88–94 [DOI] [PubMed] [Google Scholar]
- Grelon M, Gendrot G, Vezon D, Pelletier G (2003) The Arabidopsis MEI1 gene encodes a protein with five BRCT domains that is involved in meiosis-specific DNA repair events independent of SPO11-induced DSBs. Plant J 35: 465–475 [DOI] [PubMed] [Google Scholar]
- Grelon M, Vezon D, Gendrot G, Pelletier G (2001) AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J 20: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays SL, Firmenich AA, Berg P (1995) Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc Natl Acad Sci USA 92: 6925–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EL, Shinohara A, Bishop DK (2001) Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J Biol Chem 276: 41906–41912 [DOI] [PubMed] [Google Scholar]
- Johnson RD, Symington LS (1995) Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol Cell Biol 15: 4843–4850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kans JA, Mortimer RK (1991) Nucleotide sequence of the RAD57 gene of Saccharomyces cerevisiae. Gene 105: 139–140 [DOI] [PubMed] [Google Scholar]
- Keeney S (2001) Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol 52: 1–53 [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384 [DOI] [PubMed] [Google Scholar]
- Klimyuk VI, Jones JD (1997) AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J 11: 1–14 [DOI] [PubMed] [Google Scholar]
- Kurumizaka H, Ikawa S, Nakada M, Eda K, Kagawa W, Takata M, Takeda S, Yokoyama S, Shibata T (2001) Homologous-pairing activity of the human DNA-repair proteins Xrcc3.Rad51C. Proc Natl Acad Sci USA 98: 5538–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurumizaka H, Ikawa S, Nakada M, Enomoto R, Kagawa W, Kinebuchi T, Yamazoe M, Yokoyama S, Shibata T (2002) Homologous pairing and ring and filament structure formation activities of the human Xrcc2*Rad51D complex. J Biol Chem 277: 14315–14320 [DOI] [PubMed] [Google Scholar]
- Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, Brookman KW, Siciliano MJ, Walter CA, Fan W, Narayana LS, Zhou ZQ, Adamson AW, Sorensen KJ, Chen DJ, Jones NJ, Thompson LH (1998) XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol Cell 1: 783–793 [DOI] [PubMed] [Google Scholar]
- Liu N, Schild D, Thelen MP, Thompson LH (2002) Involvement of Rad51C in two distinct protein complexes of Rad51 paralogs in human cells. Nucleic Acids Res 30: 1009–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Masson JY, Shah R, O'Regan P, West SC (2003) Rad51C is required for Holliday junction processing in mammalian cells. Science, In Press [DOI] [PubMed] [Google Scholar]
- Lovett ST (1994) Sequence of the RAD55 gene of Saccharomyces cerevisiae: similarity of RAD55 to prokaryotic RecA and other RecA-like proteins. Gene 142: 103–106 [DOI] [PubMed] [Google Scholar]
- Maluszynska J, Heslop-Harrison JS (1991) Localization of tandemly repeated DNA sequences in Arabidopsis thaliana. Plant J 1: 159–166 [Google Scholar]
- Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, Benson FE, West SC (2001) Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev 15: 3296–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson JY, West SC (2001) The Rad51 and Dmc1 recombinases: a non-identical twin relationship. Trends Biochem Sci 26: 131–136 [DOI] [PubMed] [Google Scholar]
- Mikosch TS, Sonnenberg AS, Van Griensven LJ (2001) Isolation, characterization, and expression patterns of a DMC1 homolog from the basidiomycete Pleurotus ostreatus. Fungal Genet Biol 33: 59–66 [DOI] [PubMed] [Google Scholar]
- Miller KA, Yoshikawa DM, McConnell IR, Clark R, Schild D, Albala JS (2002) RAD51C interacts with RAD51B and is central to a larger protein complex in vivo exclusive of RAD51. J Biol Chem 277: 8406–8411 [DOI] [PubMed] [Google Scholar]
- Moffatt BA, McWhinnie EA, Agarwal SK, Schaff DA (1994) The adenine phosphoribosyltransferase-encoding gene of Arabidopsis thaliana. Gene 143: 211–216 [DOI] [PubMed] [Google Scholar]
- Motamayor JC, Vezon D, Bajon C, Sauvanet A, Grandjean O, Marchand M, Bechtold N, Pelletier G, Horlow C (2000) Switch (swi1) an Arabidopsis thaliana mutant affected in the female meiotic switch. Sex Plant Reprod 12: 209–218 [Google Scholar]
- Murata M, Heslop-Harrison JS, Motoyoshi F (1997) Physical mapping of the 5S ribosomal RNA genes in Arabidopsis thaliana by multi-color fluorescence in situ hybridization with cosmid clones. Plant J 12: 31–37 [DOI] [PubMed] [Google Scholar]
- Osakabe K, Yoshioka T, Ichikawa H, Toki S (2002) Molecular cloning and characterization of RAD51-like genes from Arabidopsis thaliana. Plant Mol Biol 50: 71–81 [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirson BN, Bowling SE, Makaroff CA (1997) A defect in synapsis causes male sterility in a T-DNA-tagged Arabidopsis thaliana mutant. Plant J 11: 659–669 [DOI] [PubMed] [Google Scholar]
- Petronczki M, Siomos MF, Nasmyth K (2003) Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112: 423–440 [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13: 2633–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, Handel MA, Schimenti JC (1998) Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell 1: 697–705 [DOI] [PubMed] [Google Scholar]
- Pittman DL, Schimenti JC (2000) Midgestation lethality in mice deficient for the RecA-related gene, Rad51d/Rad51l3. Genesis 26: 167–173 [DOI] [PubMed] [Google Scholar]
- Rockmill B, Sym M, Scherthan H, Roeder GS (1995) Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev 9: 2684–2695 [DOI] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD (2000) The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6: 975–987 [DOI] [PubMed] [Google Scholar]
- Ross KJ, Fransz P, Armstrong SJ, Vizir I, Mulligan B, Franklin FC, Jones GH (1997) Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA-transformed lines. Chromosome Res 5: 551–559 [DOI] [PubMed] [Google Scholar]
- Schild D, Lio YC, Collins DW, Tsomondo T, Chen DJ (2000) Evidence for simultaneous protein interactions between human Rad51 paralogs. J Biol Chem 275: 16443–16449 [DOI] [PubMed] [Google Scholar]
- Schubert I, Fransz PF, Fuchs J, de Jong JH (2001) Chromosome painting in plants. Methods Cell Sci 23: 57–69 [PubMed] [Google Scholar]
- Schwacha A, Kleckner N (1997) Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90: 1123–1135 [DOI] [PubMed] [Google Scholar]
- Shinohara A, Gasior S, Ogawa T, Kleckner N, Bishop DK (1997) Saccharomyces cerevisiae recA homologues RAD51 and DMC1 have both distinct and overlapping roles in meiotic recombination. Genes Cells 2: 615–629 [DOI] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T (1992) Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69: 457–470 [DOI] [PubMed] [Google Scholar]
- Shu Z, Smith S, Wang L, Rice MC, Kmiec EB (1999) Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can be partially rescued in a p53(−/−) background. Mol Cell Biol 19: 8686–8693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S, Van Komen S, Bussen W, Schild D, Albala JS, Sung P (2001) Mediator function of the human Rad51B–Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev 15: 3308–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Treco D, Schultes NP, Szostak JW (1989) Double-strand breaks at an initiation site for meiotic gene conversion. Nature 338: 87–90 [DOI] [PubMed] [Google Scholar]
- Sun H, Treco D, Szostak JW (1991) Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64: 1155–1161 [DOI] [PubMed] [Google Scholar]
- Sung P (1994) Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265: 1241–1243 [DOI] [PubMed] [Google Scholar]
- Sung P (1997) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev 11: 1111–1121 [DOI] [PubMed] [Google Scholar]
- Symington LS (2002) Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev 66: 630–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Fukushima T, Morrison C, Albala JS, Swagemakers SM, Kanaar R, Thompson LH, Takeda S (2000) The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol Cell Biol 20: 6476–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S (2001) Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol 21: 2858–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbs RS, Zhao Y, Tucker JD, Scheerer JB, Siciliano MJ, Hwang M, Liu N, Legerski RJ, Thompson LH (1995) Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc Natl Acad Sci USA 92: 6354–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LH, Schild D (2001) Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res 477: 131–153 [DOI] [PubMed] [Google Scholar]
- Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci USA 93: 6236–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren AJ, Maccubbin AE, Hamilton JW (1998) Detection of mitomycin C-DNA adducts in vivo by 32P-postlabeling: time course for formation and removal of adducts and biochemical modulation. Cancer Res 58: 453–461 [PubMed] [Google Scholar]
- Wiese C, Collins DW, Albala JS, Thompson LH, Kronenberg A, Schild D (2002) Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic Acids Res 30: 1001–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Hiraoka Y (2001) How do meiotic chromosomes meet their homologous partners? Lessons from fission yeast. Bioessays 23: 526–533 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Kondoh G, Matsuda Y, Habu T, Nishimune Y, Morita T (1998) The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol Cell 1: 707–718 [DOI] [PubMed] [Google Scholar]


