Abstract
YidC is a recently discovered bacterial membrane protein that is related to the mitochondrial Oxa1p and the Alb3 protein of chloroplasts. These proteins are required in the membrane integration process of newly synthesized proteins that do not require the classical Sec machinery. Here we demonstrate that YidC is sufficient for the membrane integration of a Sec-independent protein. Microgram amounts of the purified single-spanning Pf3 coat protein were efficiently inserted into proteoliposomes containing the purified YidC. A mutant Pf3 coat protein with an extended hydrophobic region was inserted independently of YidC into the membrane both in vivo and in vitro, but its insertion was accelerated by YidC. These results show that YidC can function separately from the Sec translocase to integrate membrane proteins into the lipid bilayer.
Keywords: membrane insertion, reconstitution, Sec-independence, translocation, YidC
Introduction
Most membrane proteins and exported proteins require protein catalysts for their membrane insertion and transport across membranes. In eukaryotes, the Sec61 translocon is involved in transporting both secreted and membrane proteins from the cytosol into the endoplasmic reticulum (ER) (Goerlich and Rapoport, 1993; Panzer et al, 1995). In the mitochondrion and chloroplast, the TOM/TIM and TOC/TIC translocases, respectively, function in importing proteins from the cytosol into the organelle (Keegstra and Froehlich, 1999; Jarvis and Soll, 2001; Pfanner and Geissler, 2001).
In prokaryotes, the SecAYEG translocase is required for the export of proteins into the periplasm (Driessen et al, 2001). Similarly, the membrane insertion of a number of inner membrane proteins has been shown to depend on the Sec translocase, although it seems that the majority of them do not need the SecA and SecG components that are required for the exported proteins (Beck et al, 2000; Koch and Müller, 2000). In addition, most membrane proteins use the bacterial SRP for targeting to the membrane, whereas secreted proteins use the SecB–SecA pathway (reviewed in Driessen et al, 2001).
One membrane component that has been recently identified in Escherichia coli and shown to play an important role in the insertion process of newly synthesized membrane proteins is the 61 kDa YidC protein. YidC is specifically used for the insertion of membrane proteins and not for the translocation of exported proteins (Dalbey and Kuhn, 2000; Samuelson et al, 2000). Possibly, the function of YidC is to recognize hydrophobic regions of a membrane protein and to catalyse the integration of these regions in a transmembrane orientation into the membrane bilayer. As a consequence, YidC works in conjunction with the Sec translocase to transfer the transmembrane regions of Sec-dependent substrate proteins into the hydrophobic bilayer. Indeed, YidC was found to copurify with SecYEGDF (Scotti et al, 2000). Moreover, it was shown that nascent chains of FtsQ, a membrane protein involved in E. coli cell division (Carson et al, 1991), first contact SecY and then YidC (Urbanus et al, 2001). The functional relationship of YidC and Sec was also inferred by the observed inhibition of the export of proteins when Sec-dependent membrane proteins were stalled within the Sec translocase under conditions where YidC is limiting and the membrane protein is overproduced (Samuelson et al, 2001).
Homologues to YidC are present in mitochondria (Oxa1p) and chloroplasts (Alb3). They are involved in the membrane insertion of a subset of proteins (Herrmann et al, 1997; Moore et al, 2000). The Oxa1p protein plays a central role in the insertion of proteins from the matrix into the mitochondrial inner membrane as mitochondria lack the Sec translocase components (Glick and von Heijne, 1996; Nargang et al, 2002).
Some bacterial membrane proteins have been shown to insert independently of the Sec translocase (Kuhn, 1995; de Gier et al, 1998; Roos et al, 2001). These are the single- or double-spanning coat proteins of the Pf3 and M13 filamentous phage, which have small periplasmic regions. Whereas the single-spanning Pf3 coat protein comprises only 44 amino acids, the 73-amino-acid-long M13 procoat protein is synthesized with a leader peptide and is inserted as a double-spanning protein. Both these proteins require YidC for their insertion and the translocation of their periplasmic region across the membrane (Samuelson et al, 2001; Chen et al, 2002). Since these proteins apparently do not require the Sec translocase, YidC might operate on its own.
To understand the membrane insertion process of newly synthesized proteins at the molecular level, one important step is to reconstitute this reaction in vitro with purified components. Previously, we have shown that the Sec-independent Pf3 coat protein is inserted into inverted inner membrane vesicles (INV) with the help of the membrane potential ΔΨ (Kiefer and Kuhn, 1999). Protease-treated INVs that were blocked for Sec-dependent transport allowed normal Pf3 coat insertion, suggesting that there are two separate membrane insertion pathways. We show here that YidC is sufficient in promoting the membrane insertion of the Pf3 coat protein in vitro, demonstrating that YidC can function distinct of the Sec components.
Results
Insertion of purified Pf3 and 3L-Pf3 coat protein into liposomes
To test whether the Pf3 coat protein can insert into large unilamellar vesicles (LUVs), the protein was purified from E. coli membranes. This was achieved by first extracting the Pf3 coat protein from the membranes with 8 M urea, followed by reversed phase and size exclusion chromatography. Microgram amounts of the purified coat protein were incubated for 60 min with freshly prepared LUVs made from E. coli lipids. The protein bound to the liposomes as indicated from its presence in the pellet fraction (Figure 1A, lane 1). The proportion of the coat proteins that was inserted into the membrane was estimated by the protease-protected fraction of the protein (lane 2). Proteinase K was added to the outside of the liposomes and the digestion was carried out for 30 min. We observed that most of the Pf3 coat protein was digested by the proteinase and was therefore not inserted into the LUVs, suggesting that the insertion of the protein requires additional factors.
Figure 1.

Insertion of Pf3 coat protein into liposomes. Purified Pf3 coat protein (A, B) and purified 3L-Pf3 coat protein (C, D) were added to E. coli liposomes with a 0.25 μm mean diameter, generated with an extruder. The reactions were incubated at 37°C for 1 h and pelleted at 130 000 g. The Pf3 coat protein was found binding to the liposomes (lane 1). An aliquot was digested with 0.5 mg/ml proteinase K at 0°C for 30 min in the absence (lane 2) or presence of detergent (lane 3). The proteins were applied to liposomes (A, C) and energized liposomes (B, D). The samples were acid precipitated, analysed by PAGE and visualized by silver stain.
A membrane potential was generated across the LUVs by the addition of valinomycin after the Pf3 coat protein had been added. When the liposomes contained 100 mM Na2SO4 inside and 100 mM K2SO4 outside, the addition of valinomycin generated a positive inside potential of about 200 mV visualized by oxonol fluorescence (compare Figure 2B with Figure 2A). The membrane potential had only a small effect on the insertion of the Pf3 coat protein into the liposomes (Figure 1B). Less than 10% was found protected to the outside added protease (lane 2).
Figure 2.
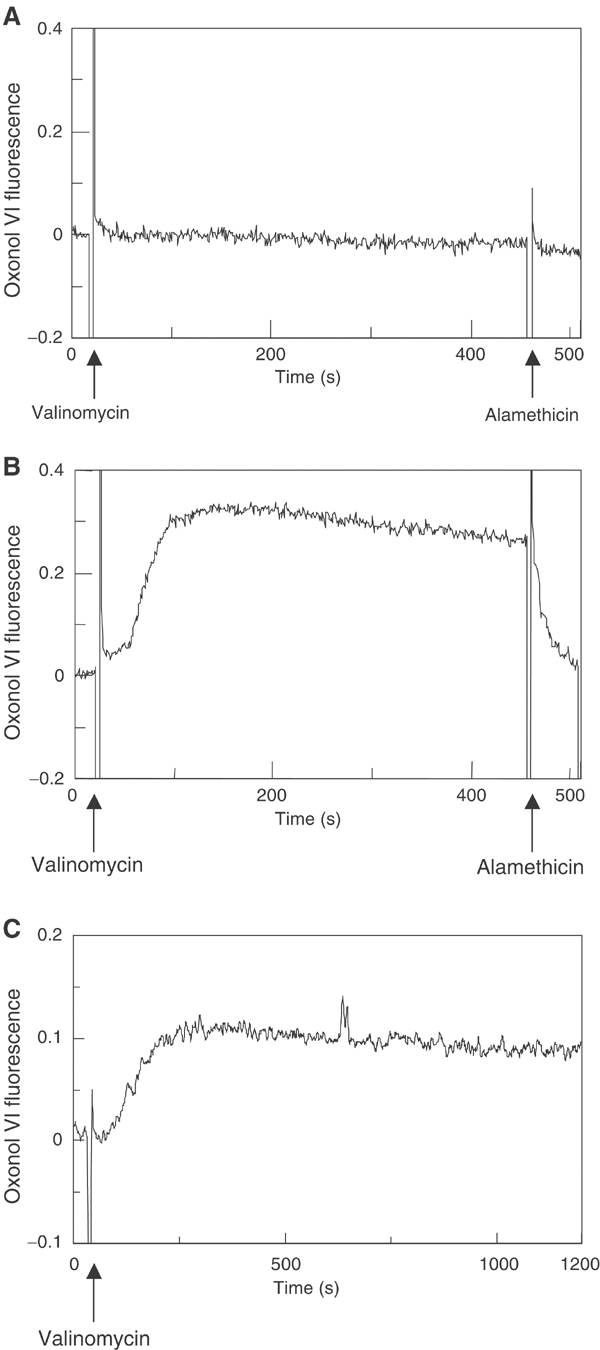
Generation of a membrane potential across liposomes. Extruded liposomes generated in 100 mM Na2SO4 were sedimented and resuspended in 100 mM Na2SO4 (A) or in 100 mM K2SO4 (B). To the liposomes, 0.1 μM oxonol VI was added and the fluorescence at an excitation/emission of 599/634 nm was recorded. After addition of 0.25 μM valinomycin, a transmembrane potential was generated as measured by the increase of the oxonol fluorescence. The membrane potential was abolished by the addition of 3 μg/ml alamethicin. The membrane potential was also monitored with YidC proteoliposomes (C).
Previous studies investigating the membrane insertion pathway of the Pf3 coat protein have shown that a mutant with an extended hydrophobic region (3L-Pf3 coat) was inserted into the E. coli membrane independent of the electrochemical membrane potential and independent of the two negatively charged amino-acid residues located in the N-terminal region, in contrast to the wild-type protein (Kiefer and Kuhn, 1999). This suggested that 3L-Pf3 inserts into the membrane using a different pathway than the wild-type Pf3 coat protein, which requires a negatively charged residue as well as the membrane potential and YidC (Kiefer et al, 1997; Chen et al, 2002). We therefore tested whether 3L-Pf3 can insert into liposomes by itself. 3L-Pf3 was purified to homogeneity and added to LUVs without and with the membrane potential (Figure 1C and D). The results show that the 3L-Pf3 is efficiently inserted into the liposomes and is protected from protease digestion regardless of a membrane potential. A small shift of the bands results from the clipped C-terminal tail of the inserted protein.
Membrane insertion of Pf3 and 3L-Pf3 coat proteins in vivo
The in vitro results show that Pf3 coat inserts into liposomes containing a membrane potential with a low efficiency, whereas the 3L-Pf3 coat is inserted with a high efficiency. The low efficiency of the wild-type protein is consistent with previous results showing that the membrane insertion of Pf3 coat protein requires YidC in vivo (Chen et al, 2002). Therefore, we were interested to know whether the membrane insertion of 3L-Pf3 also depends on the presence of YidC in the cell. To analyse the YidC requirement, we used the YidC depletion strain, JS 7131, where YidC expression is under the control of an araBAD promoter and operator (Samuelson et al, 2000). YidC expression was induced with arabinose and tightly repressed in the presence of glucose. To deplete YidC, the cells were grown for 3 h with glucose and then analysed for membrane insertion of Pf3 coat and 3L-Pf3 coat proteins (Figure 3). The cells were labelled with [35S]-methionine for 3 min, converted to spheroplasts and analysed for protease mapping. Under both conditions, 3L-Pf3 readily inserted into the membrane and was digested by proteinase K that had been added to the outside of the spheroplasts (Figure 3B). Wild-type Pf3 coat protein was only membrane inserted into the cells that were grown in the presence of arabinose (Figure 3A). For a control, M13 procoat protein was pulse labelled for 1 min and analysed for processing and translocation (Figure 3C). When the cells were grown in glucose, M13 procoat processing was inhibited and the protein accumulated in a nontranslocated form. In contrast, the minor amount of the processed protein was accessible to the protease. These results show that the 3L-Pf3 coat protein does not require the function of YidC in vivo, in contrast to the wild-type Pf3 protein and M13 procoat, which are only inserted into the membrane in the presence of YidC.
Figure 3.
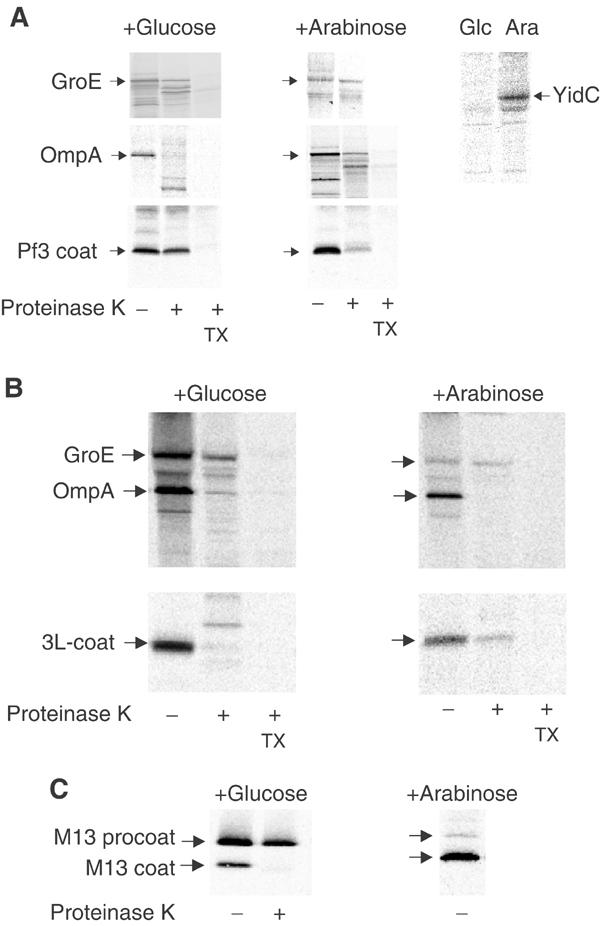
Membrane insertion of the Pf3 coat protein in vivo. Protease mapping of the Pf3 coat protein (A) and the mutant 3L-Pf3 (B) expressed in JS 7131 cells depleted of YidC (+glucose, left panels) or expressing YidC (+arabinose, right panels). Exponentially growing cells bearing the respective plasmids were induced for 10 min with 1 mM IPTG and pulse labelled with [35S]-methionine for 3 min, converted to spheroplasts and digested with 200 μg/ml proteinase K either in the absence or presence of 2% Triton X-100. The samples were immunoprecipitated with antibodies to Pf3 phage (A, B, lower panels) or to OmpA, GroEL and YidC (upper panels) and analysed by SDS–PAGE and phosphorimaging. For a control, the processing and protease accessibility of the M13 procoat protein was analysed (C).
Reconstitution of YidC into proteoliposomes
The 61 kDa E. coli YidC protein with a C-terminal hexahistidine tag was purified to homogeneity by affinity and ion exchange chromatography (Figure 4, lane 2). To investigate the function of YidC, the purified protein was reconstituted into lipid vesicles. A solubilized dry film of E. coli lipids (lane 1) was resuspended in 100 mM Na2SO4, Hepes (pH 8.0) buffer, mixed with purified YidC protein and passed through an extruder to obtain the proteoliposomes of a mean diameter of 0.25 μm. The proteoliposomes were collected by centrifugation and resuspended in 100 mM K2SO4. As can be seen by examining the Coomassie-stained SDS–polyacrylamide gel (Figure 4, M, lanes 1–4), all the YidC is bound to the lipid vesicles (lane 4 pellet, lane 3 supernatant).
Figure 4.
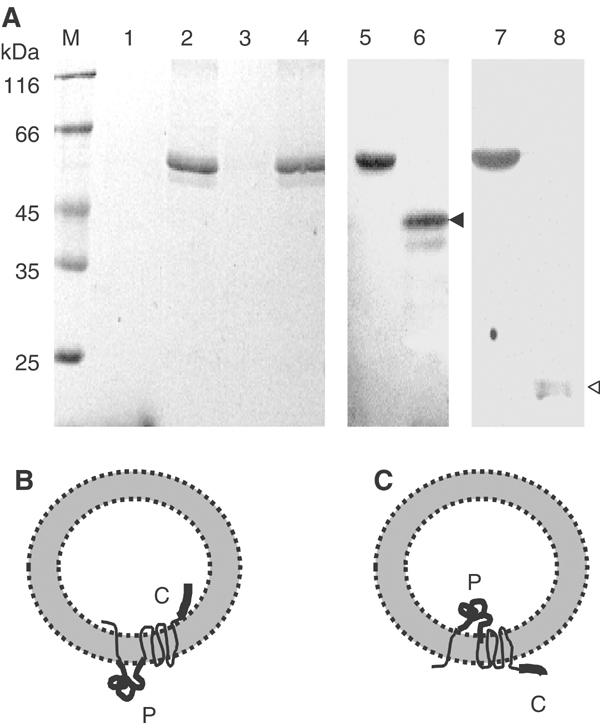
Reconstitution of YidC into proteoliposomes. (A) Purified YidC protein (lane 2) was mixed with E. coli lipids to form proteoliposomes. The proteoliposomes were pelleted in an airfuge (lane 4). The absence of the protein in the supernatant (lane 3) showed that it was efficiently integrated into the YidC-containing proteoliposomes. The samples were analysed by SDS–PAGE and Coomassie stained. For reference, molecular weight marker (lane M) and E. coli lipid (lane 1) were applied on the gel. The samples were analysed by SDS–PAGE and immunoblotted with an antibody directed to the periplasmic region of YidC (lanes 5 and 6) or to the C-terminal tail of YidC (lanes 7 and 8). Trypsin treatment of the proteoliposomes generated a 42 kDa fragment (lane 6, black arrowhead) or a 20 kDa fragment (lane 8, white arrowhead), respectively. (B, C) Two possible topologies of the reconstituted YidC. The large periplasmic domain (P) and the C-terminal tail (C) are highlighted.
To analyse whether the reconstituted YidC is correctly oriented in the proteoliposomes, trypsin was added to the outside. After incubating at 0°C for 1 h, a 42 kDa fragment was generated (lane 6, see the black arrow), which was recognized by the antiserum directed to a periplasmic YidC peptide, but not by the antiserum to a C-terminal YidC peptide (lane 8). If the YidC protein is inserted right-side-out (Figure 4B), the generation of a 20 kDa fragment should be detectable by the serum to the C-terminal region (lane 8, see the white arrow). If the YidC protein is inserted in the inverted orientation (Figure 4C), the generation of a 42 kDa fragment is expected. The result shows that most of the YidC protein is in the inverted orientation, with the large periplasmic domain of YidC localized in the lumen of the proteoliposomes.
Insertion of purified Pf3 protein into YidC proteoliposomes
To the reconstituted YidC proteoliposomes, 8 μg of purified Pf3 protein (Figure 5A, lane 1) was added and the sample was incubated at 37°C for 60 min. Following the incubation, the proteoliposomes were sedimented. The majority of the Pf3 coat was found bound to the lipid vesicles (lane 3) and not in the supernatant (lane 2). The Pf3 coat bound to the YidC lipid vesicles was analysed with proteinase K to determine the amount of the protein that was membrane inserted (lane 4). Most of the Pf3 coat protein was resistant to the added proteinase K, showing that it was efficiently inserted into the membrane. Disruption of the proteoliposomes by detergent (lane 5) resulted in a complete accessibility of the protein to the proteinase.
Figure 5.
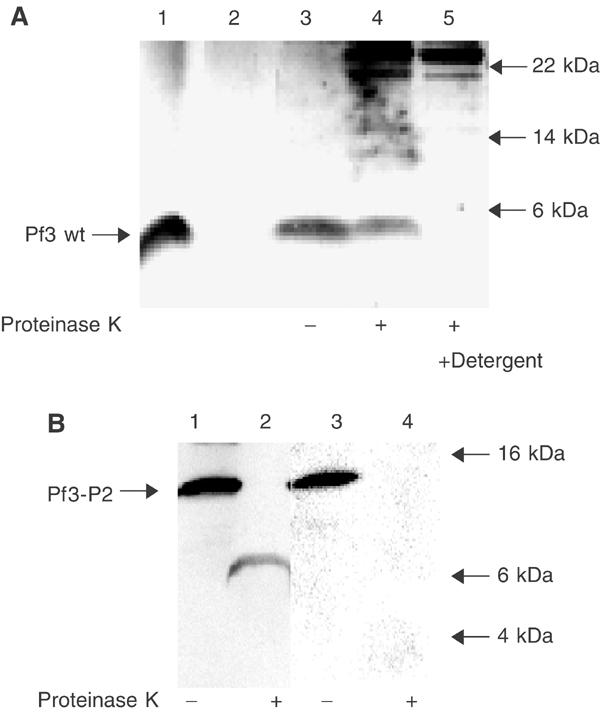
Membrane insertion of Pf3 and Pf3-P2 coat proteins into YidC proteoliposomes in vitro. (A) Purified Pf3 coat protein (lane 1) was mixed with YidC proteoliposomes (lanes 2–5), incubated for 1 h at 37°C (lanes 3–5) and sedimented by centrifugation. Essentially all the protein was bound to the proteoliposomes (lane 2 supernatant, lane 3 pellet). An aliquot of the sample was digested with 0.5 mg/ml proteinase K for 1 h in the absence (lane 4) or presence of detergent (lane 5). The samples were acid precipitated, and analysed by SDS–PAGE and silver staining. The insertion efficiency of the Pf3 coat protein was quantified as 74% on calculating the difference between the Pf3 coat protein bands in lanes 3 and 4 using a Kodak imager. (B) Purified Pf3-P2 was added to YidC proteoliposomes as in (A) (lanes 1 and 3), and analysed by proteinase K (lanes 2 and 4). The samples were applied to SDS–PAGE and immunoblotted with antiserum to the N-terminal region of the Pf3 protein (lanes 1 and 2) or to leader peptidase P2 region (lanes 3 and 4). For reference, the positions of a molecular weight marker are indicated by arrows.
As a second way to show that the Pf3 coat protein can insert in a YidC-dependent manner, we examined the insertion of a Pf3 coat derivative with an extended C-terminal tail of 102 residues (Chen et al, 2002). When this 15 kDa protein inserts into the membrane, it is expected that a 6 kDa fragment is protected from the protease by the membrane (the Pf3 coat N-terminal tail and transmembrane region) and the extended C-terminal cytoplasmic region is degraded. The purified protein is detectable by antibodies to Pf3 and also to leader peptidase recognizing the extended C-tail (Figure 5B, lanes 1 and 3). Membrane insertion into the YidC proteoliposomes was followed by protease treatment (lanes 2 and 4). The protease digested the C-terminal tail of Pf3-P2, and only a small fragment of about 6 kDa was protected (lane 2). This fragment was recognized by the antiserum to the Pf3 protein but not by the serum to leader peptidase. We conclude from this result that Pf3-P2 has a defined transmembrane topology with the N-terminal region in the lumen of the proteoliposomes and the C-terminus exposed in the cytoplasm.
To analyse the amount of YidC that is critical for Pf3 protein insertion, different dilutions of the purified YidC protein were used for the reconstitution experiments. We found that an average of 25 molecules of YidC per liposome (protein:lipid ratio 1:25 000) was most efficient in promoting the membrane insertion of Pf3 (Figure 6). The translocation rate reached 70% of the applied Pf3 protein. Liposomes with less than five molecules of YidC (protein:lipid ratio 1:200 000) did not efficiently insert the Pf3 coat protein. The amount of YidC proteins per liposome was increased up to 120 molecules per liposome (protein:lipid ratio 1:5000). However, the efficiency of Pf3 insertion slightly decreased at the higher YidC concentrations. The raw data of the protected Pf3 coat used in Figure 6A are shown in Figure 6B.
Figure 6.
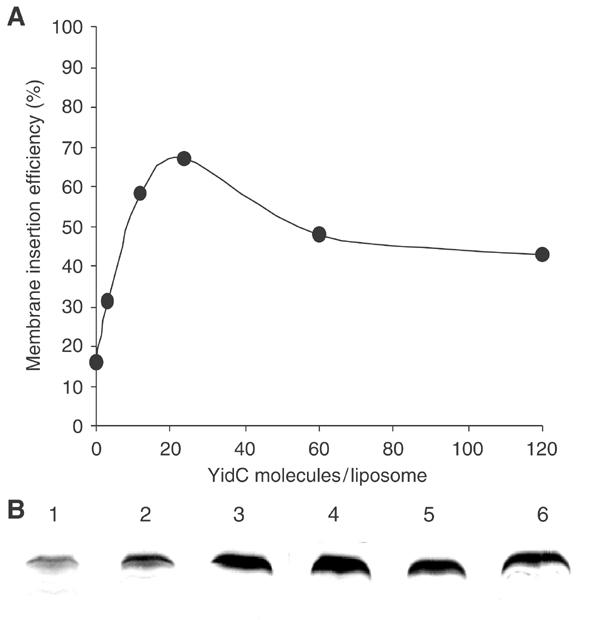
Proteoliposomes with different YidC contents. Proteoliposomes were generated that had YidC:lipid ratios varying from 1:5000 to 1:200 000. Assuming 650 000 lipid molecules per liposome, this gives a range from 3 to 120 YidC molecules per liposome (A). Liposomes without YidC (B, lane 1) and proteoliposomes of YidC:lipid ratios of 1:200 000 (lane 2), 1:50 000 (lane 3), 1:25 000 (lane 4), 1:10 000 (lane 5) and 1:5000 (lane 6) were used to examine the membrane insertion of Pf3 coat following the identical protocol as in Figure 5. The efficiency of membrane insertion was quantified using a Kodak imager.
For the proteoliposomes that had an average of about 50 molecules of YidC, the kinetic of the insertion process was analysed (Figure 7). The purified Pf3 protein was added to the energized YidC proteoliposomes and the reaction was stopped by chilling and the addition of proteinase K. Membrane insertion of the Pf3 protein into the YidC proteoliposomes was monitored for various times up to 30 min of incubation (Figure 7A (black columns) and Figure 7C). When no YidC protein was present, only low amounts of the Pf3 protein were resistant to the proteinase K (Figure 7A (white columns) and Figure 7B). When the YidC proteoliposomes had no membrane potential, a slightly lower amount of the Pf3 coat protein was inserted as compared to the proteoliposomes with potential (data not shown), suggesting that in vitro the insertion of Pf3 coat mainly occurs independent of the potential. This is reminiscent of the Sec-mediated protein translocation, where a transmembrane potential is not required in the reconstituted system (Bassilana and Wickner, 1993; Hanada et al, 1994).
Figure 7.
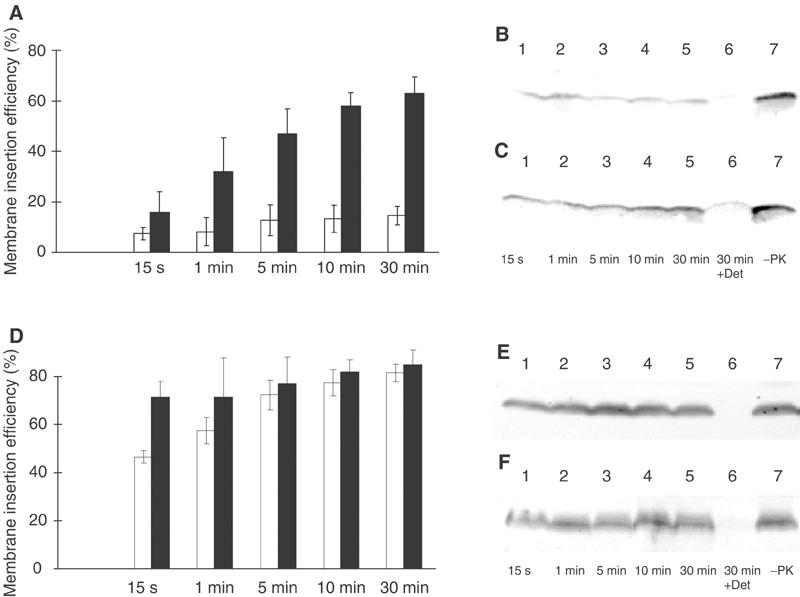
Kinetics of YidC-mediated membrane insertion. Reconstituted YidC proteoliposomes (A, D: black columns; panels C, F) or liposomes (A, D: white columns; panels B, E) were mixed with Pf3 coat protein (A–C) or the mutant 3L-Pf3 (D–F), and incubated at 37°C for the indicated times. The reaction was stopped by chilling on ice and the addition of 0.5 mg/ml proteinase K. The proteinase reaction was continued for 1 h at 0°C. The samples were then acid precipitated and analysed by a Western immunoblot with an antiserum to Pf3 phage. The bands were quantified as described in Figure 6. The mean values of five independent experiments were calculated (A, D).
The 3L-Pf3 protein was inserted into the YidC proteoliposomes more efficiently than the wild-type protein (Figure 7D, compare with Figure 7A). However, the insertion of 3L-Pf3 protein into the liposomes lacking YidC was slightly slower (Figure 7D (white columns) and Figure 7E) compared to its insertion into the YidC proteoliposomes (Figure 7D (black columns) and Figure 7F). This shows that YidC kinetically supports also the insertion of 3L-Pf3 protein.
Discussion
We show here that the YidC protein of E. coli reconstituted into liposomes can efficiently support the membrane insertion of chemical amounts of purified Pf3 coat protein (Figures 5, 6 and 7). In the absence of YidC, only low amounts were found to insert into the liposomes (Figures 1, 6 and 7). This suggests that YidC on its own acts as an enzyme catalysing the membrane insertion of a protein that does not use the classical Sec pathway.
The analysis of the reconstituted YidC proteoliposomes showed that the protein was oriented with the periplasmic region inside as judged from the appearance of a trypsin-resistant fragment of 42 kDa (Figure 4). This fragment includes the large periplasmic domain between the first two transmembrane regions that is recognized by a peptide-specific antibody. A 42 kDa protease-resistant fragment is also found in inverted membrane vesicles after protease treatment (Koch et al, 2002).
The identification of YidC as an essential component for the membrane insertion of the Sec-independent Pf3 coat protein (Chen et al, 2002) suggested that YidC constitutes a novel translocase. Indeed, we found that micrograms of the purified coat protein were inserted into the liposomes within minutes (Figure 7). Efficient membrane insertion was observed when more than five molecules of YidC were present in each liposome (Figure 6). In this experiment, about 150 Pf3 coat protein molecules had been inserted per YidC molecule, suggesting that YidC catalytically supports the insertion reaction. The most efficient insertion occurred with a density of about 25 YidC molecules per liposome (Figure 6), which corresponds to a protein:lipid ratio of 1:25 000.
In the absence of YidC, the Pf3 coat protein inserted into the liposomes, although with a low efficiency (Figure 1B). This indicates that the protein can, in principle, autonomously insert into the membrane bilayer, but to make this reaction efficient YidC is needed for enzymatic support. Whether autonomous membrane insertion occurs also in vivo is questionable, since pulse-chase experiments show that most of the Pf3 coat protein accumulates nontranslocated in the absence of YidC (Chen et al, 2002). In addition, the lipid composition and the lipid arrangement within liposomes may be different from the lipid arrangement found in the cell membrane and may affect the efficiency of membrane insertion (Ridder et al, 2000, 2001).
The occurrence of autonomous membrane insertion of the Pf3 coat protein is mainly suggested by the results obtained with the 3L mutant. We found that this protein is readily exposed to the periplasmic face in vivo even under conditions when YidC was depleted (Figure 3). The 3L-Pf3 mutant has previously been shown to insert independently of the membrane potential by in vitro translocation studies with membrane vesicles (Kiefer and Kuhn, 1999; Ridder et al, 2000). In accordance with these results, the 3L protein was efficiently inserted, even when the liposomes had no membrane potential (Figure 1C). Interestingly, when YidC was incorporated into lipid vesicles, the 3L mutant was inserted into the proteoliposomes with a slightly faster kinetic compared to pure liposomes (Figure 7). This suggests that YidC enzymatically supports the insertion of 3L-Pf3, although it is not absolutely required.
Previous experiments have shown that the Sec-independent proteins M13 procoat and Pf3 coat hydrophobically interact with the membrane in the absence of YidC (Samuelson et al, 2001; Chen et al, 2002). This leads to a partial partitioning of the protein into the membrane bilayer without the translocation of the hydrophilic domain. The enzymic activity of YidC is to support the translocation event and to promote the folding of the hydrophobic region into a transmembrane configuration. The substrate for YidC is a non-membrane-spanning membrane-bound protein and the product is the transmembrane protein in the bilayer. We propose that YidC functions as a membrane chaperone in supporting these folding reactions within the membrane (Kuhn et al, 2003). This might be achieved by YidC interacting with the hydrophobic parts of the protein, which has been shown to occur using crosslinking techniques (Chen et al, 2002) and, secondly, by shielding the hydrophilic parts of a translocating protein chain from the lipid phase of the membrane. We think that YidC protein provides the required amphiphilic surface within the membrane that allows the membrane transfer of polar regions of the protein.
The reconstitution studies described here show that YidC on its own is sufficient for the integration of a Sec-independent protein into a lipid bilayer. However, additional unidentified components may make the insertion reaction more efficient in vivo. As had been demonstrated in the reconstitution of the Sec translocase (Brundage et al, 1990; Akimaru et al, 1991), SecAYE was found as the minimal component in vitro, while additional components, including SecGDFYajC, made the translocation reaction more efficient (Nishiyama et al, 1993; Duong and Wickner, 1997).
Finally, the observation that YidC can function on its own to promote membrane insertion makes it understandable that Oxa1p in mitochondria can function without a Sec machinery. Whether the Oxa1p homologue in chloroplasts, Alb3, also functions without Sec is still an open question.
Materials and methods
Strains, plasmids and growth conditions
Cloning and mutagenesis experiments were performed with E. coli XL1-Blue recA1 thi supE44 endA1 hsdR17 gyrA96 relA1 lac F′ (proAB+ lacIq lacZΔM15 Tn10) (Stratagene, Heidelberg, Germany). The plasmids pT7-7 encoding the Pf3 coat protein and pET22 encoding Pf3-P2 were used to overexpress the coat proteins in E. coli BL21(DE3)pLysS, which encodes the T7 RNA polymerase under the inducible lacUV 5 promoter (Studier et al, 1990). YidC depletion experiments were performed in the JS7131 strain (Samuelson et al, 2000). Expression of Pf3 coat in JS7131 was induced from pUC19 derivatives, where the Pf3 coat sequence was cloned using the Xba1 and EcoR1 restriction sites.
Antibodies
The YidC polyclonal antibody directed against the periplasmic region was generated with a synthetic peptide (DEKYEKYKTIADNEC). The YidC antibody to the C-terminal region was raised with the peptide (CLEKRGLHSREKKK) and was a gift from Dr W Neupert (Munich). The Pf3 coat antibody was raised against the purified Pf3 phage.
Protease mapping assay
Cultures (1 ml) of E. coli JS 7131 with pUC19-Pf3 were grown for 2 h at 37°C to an optical density of about 0.2 at 600 nm in Luria broth containing ampicillin (100 μg/ml), spectinomycin (25 μg/ml) and either 0.2% arabinose or 0.2% glucose, respectively. Cells were harvested by centrifugation and washed 2 × in the M9 salt medium. Washed cells were resuspended in M9 minimal medium (Miller, 1972) with 0.2% arabinose or 0.2% glucose, respectively, and with 20 μg/ml of each amino acid but methionine. Cultures (0.5 ml) were continued for 1 h at 37°C with shaking to adapt the cells to the M9 medium. IPTG (1 mM) was added to the M9 medium to induce synthesis of the plasmid-encoded proteins. After 10 min, the cells were labelled with 30 μCi [35S]-methionine for 5 min, chilled on ice and centrifuged for 1 min at 4°C. The pellet was resuspended in 250 μl 0.1 M Tris-acetate, pH 8.2, 0.5 M sucrose and 5 mM EDTA, and was treated with 80 μg/ml lysozyme (Serva) and diluted with 250 μl of water. After 8 min, 150 μl of 200 mM MgSO4 was added to stabilize the spheroplasts and collected by centrifugation. The spheroplasts were resuspended in 300 μl of 50 mM Tris-acetate, pH 8.2, 250 mM sucrose and 10 mM MgSO4. An aliquot of 100 μl was removed and precipitated with TCA. Another aliquot of 100 μl was removed and treated with 2.5% Triton X-100 and 1 mg/ml proteinase K (Sigma) for 60 min. To the remaining sample, proteinase K was added and incubated on ice for 60 min. After quenching the protease with 5 mM PMSF, the samples were precipitated with TCA and immunoprecipitated with antibodies to Pf3 coat protein, YidC, OmpA or GroE. Samples were then analysed by SDS–PAGE and phosphorimaging.
Expression and purification of YidC protein
The gene encoding wild-type yidC (1647 bp) was cloned with a C-terminal hexahistidine tag into expression vector pBAD Myc-His A (Invitrogen, Paisley, UK). Plasmids were transformed into E. coli strain C43(DE3). Cells were grown in 2xYT medium at 37°C until an OD600 of ∼0.6 was reached, induced with 0.2% L-(+)-arabinose, and grown for an additional 3 h at 37°C. Cells were resuspended in TSB buffer (20 mM Tris–HCl/300 mM NaCl, 10% glycerol pH 8.0) and broken by two passes in a microfluidizer. The suspension was centrifuged for 50 min at 42 000 rpm (45 Ti rotor), and the membranes were extracted with 1% n-decyl β-D maltopyranoside (DM; Anatrace, Maumee, OH) in TSB for 30 min at 4°C. The supernatant was subsequently loaded on a Nickel affinity column (chelating sepharose; Amersham Pharmacia), washed with 0.2% DM/30 mm imidazole in TSB and eluted with 330 mM imidazole. YidC was purified to homogeneity by gel filtration (Superdex-200; Amersham Pharmacia) and ion exchange (Mono-Q; Amersham Pharmacia) at pH 9.0 (10 mM Tris/100 mM NaCl/10% glycerol/0.5 mM TCEP/0.1% DM). The protein was concentrated to 5–10 mg/ml (Centricon Plus-20; molecular weight cutoff 100 kDa), flash frozen and stored at −80°C.
Purification of Pf3 coat proteins
The cell pellets (5 g) were resuspended in 100 mM Tris–HCl (pH 8.5) buffer containing 8 M urea, treated by ultrasonication at 50% pulse intensity for 10 min and the debris were removed by centrifugation at 20 000 g for 30 min at 4°C. The crude extract was fractionated by reversed phase chromatography on an RPC-Source 15 column (Amersham Pharmacia, Buckinghamshire, UK) using a linear gradient from 5 to 80% isopropanol in 100 mM Tris–HCl (pH 7.5) containing 0.1% 1,1,1-trifluoroethanol. The fractions containing the protein were pooled, concentrated by ultrafiltration with an omega filter (1 kDa cutoff; Pall Gelman, Ann Arbor, MI) and further purified by size exclusion chromatography using first a Superdex 200 prep grade column and then a Superdex 75 prep grade column (Amersham Pharmacia). For both columns, the elution buffer was 100 mM Tris/HCl (pH 8.5) containing 10% isopropanol.
Preparation of liposomes and proteoliposomes
E. coli lipids (Avanti, Alabaster, AL) were dissolved in chloroform and evaporated under a stream of nitrogen. The dry lipid film was dissolved in 10 mM Hepes buffer (pH 8.0) containing 100 mM Na2SO4. The lipid emulsion was extruded 10 times through a 0.4 μm membrane (Avanti), resulting in liposomes with a mean particle size of 250 nm as determined by photon correlation spectroscopy (Coulter, Hialeah, FL). For preparing proteoliposomes, the YidC protein (10 mM Tris/100 mM NaCl/10% glycerol/0.5 mM TCEP/0.1% DM) was added to the lipid solution and extruded as described above.
For generating energized liposomes or proteoliposomes, the sedimented liposomes (10 min airfuge; Beckman, Stanford, CA) were resuspended in 10 mM Hepes buffer (pH 8.0) containing 100 mM K2SO4. After addition of 0.25 μM valinomycin, a transmembrane potential was generated as measured by oxonol VI fluorescence at a wavelength (excitation/emission) of 599/634 nm (Apell and Bersch, 1987; Venema et al, 1993). The proteins per liposome were calculated from the liposome surface area (200 000 nm2) and a lipid density of 6.5 lipid molecules/nm2 (Eisenhawer et al, 2001).
Acknowledgments
We thank Gerda Baer and Gisela Sury for technical assistance. We thank Dr W Neupert for providing the YidC antiserum and stimulating discussions, and Dr Tom Rapoport for helpful comments on the manuscript. This work was supported by grants from the DFG (Ku 749/2-1 and 749/3-1) to AK and NIH (GM63862) to RED.
References
- Akimaru J, Matsuyama S, Tokuda H, Mizushima S (1991) Reconstitution of a protein translocation system containing purified SecY, SecE, and SecA from Escherichia coli. Proc Natl Acad Sci USA 88: 654–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apell HJ, Bersch B (1987) Oxonol VI as an optical indicator for membrane potentials in lipid vesicles. Biochim Biophys Acta 903: 480–494 [DOI] [PubMed] [Google Scholar]
- Bassilana M, Wickner W (1993) Purified Escherichia coli preprotein translocase catalyzes multiple cycles of precursor protein translocation. Biochemistry 32: 2626–2630 [DOI] [PubMed] [Google Scholar]
- Beck K, Wu LF, Brunner J, Müller M (2000) Discrimination between SRP- and SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor. EMBO J 19: 134–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundage L, Hendrick JP, Schiebel E, Driessen AJM, Wickner W (1990) The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell 62: 649–657 [DOI] [PubMed] [Google Scholar]
- Carson MJ, Barondess J, Beckwith J (1991) Ths FtsQ protein of E. coli: membrane topology, abundance, and cell division phenotypes due to overproduction and insertion mutations. J Bacteriol 173: 2187–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Samuelson J, Jiang F, Müller M, Kuhn A, Dalbey RE (2002) The E. coli YidC can function distinct from the Sec translocase. J Biol Chem 277: 7670–7675 [DOI] [PubMed] [Google Scholar]
- Dalbey RE, Kuhn A (2000) Evolutionarily related insertion pathways of bacterial, mitochondrial, and thylakoid membrane proteins. Annu Rev Cell Dev Biol 16: 51–87 [DOI] [PubMed] [Google Scholar]
- De Gier J-WL, Scotti PA, Sääf A, Valent QA, Kuhn A, Luirink J, von Heijne G (1998) Differential use of the signal recognition particle translocase targeting pathway for inner membrane protein assembly in Escherichia coli. Proc Natl Acad Sci USA 95: 14646–14651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen AJ, Manting EH, van der Does C (2001) The structural basis of protein targeting and translocation in bacteria. Nat Struct Biol 8: 492–498 [DOI] [PubMed] [Google Scholar]
- Duong F, Wickner W (1997) The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J 16: 4871–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhawer M, Cattarinussi S, Kuhn A, Vogel H (2001) Fluorescence energy transfer shows a close helix–helix distance in the transmembrane M13 procoat protein. Biochemistry 40: 12321–12328 [DOI] [PubMed] [Google Scholar]
- Glick B, von Heijne G (1996) Saccharomyces cerevisiae mitochondria lack a bacterial-type Sec machinery. Protein Sci 5: 2651–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerlich D, Rapoport TA (1993) Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 75: 615–630 [DOI] [PubMed] [Google Scholar]
- Hanada M, Nishiyama K, Mizushima S, Tokuda H (1994) Reconstitution of an efficient protein translocation machinery comprising of Sec A and the three membrane proteins, Sec Y, Sec E and Sec G (p12). J Biol Chem 269: 23625–23631 [PubMed] [Google Scholar]
- Herrmann JM, Neupert W, Stuart RA (1997) Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J 16: 2217–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, Soll J (2001) Toc, Tic and chloroplast protein import. Biochim Biophys Acta 1541: 64–79 [DOI] [PubMed] [Google Scholar]
- Keegstra K, Froehlich JE (1999) Protein import into chloroplasts. Curr Opin Plant Biol 2: 471–476 [DOI] [PubMed] [Google Scholar]
- Kiefer D, Hu X, Dalbey RE, Kuhn A (1997) Negatively charged amino acid residues play an active role in orienting the Sec-independent Pf3 coat protein in the Escherichia coli membrane. EMBO J 16: 2197–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer D, Kuhn A (1999) Hydrophobic forces drive the spontaneous membrane insertion of the bacteriophage Pf3 coat protein without topological control. EMBO J 18: 6299–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch HG, Moser M, Schimz KL, Müller M (2002) The integration of YidC into the cytoplasmic membrane of E. coli requires the signal recognition particle, SecA and SecYEG. J Biol Chem 277: 5715–5718 [DOI] [PubMed] [Google Scholar]
- Koch H-G, Müller M (2000) Dissecting the translocase and integrase functions of the Escherichia coli SecYEG translocon. J Cell Biol 150: 689–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A (1995) Bacteriophage Pf3 and M13 as model systems for Sec-independent protein transport. FEMS Microbiol Rev 17: 185–190 [DOI] [PubMed] [Google Scholar]
- Kuhn A, Stuart R, Henry R, Dalbey RE (2003) The Alb3/Oxa1/YidC protein family: membrane-localized chaperones facilitating membrane protein insertion? Trends Cell Biol 13: 510–516 [DOI] [PubMed] [Google Scholar]
- Miller JH (1972) Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Moore M, Harrison MS, Peterson EC, Henry R (2000) Chloroplast Oxa1p homolog Albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J Biol Chem 275: 1529–1532 [DOI] [PubMed] [Google Scholar]
- Nargang FE, Preuss M, Neupert W, Herrmann JM (2002) The Oxa1 protein forms a homooligomeric complex and is an essential part of the mitochondrial export translocase in Neurospora crassa. J Biol Chem 277: 12846–12853 [DOI] [PubMed] [Google Scholar]
- Nishiyama K, Mizushima S, Tokuda H (1993) A novel membrane protein involved in protein translocation across the cytoplasmic membrane of Escherichia coli. EMBO J 12: 3409–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzer S, Dreier L, Hartmann E, Kostka S, Rapoport TA (1995) Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar 2. Cell 81: 561–570 [DOI] [PubMed] [Google Scholar]
- Pfanner K, Geissler A (2001) Versatility of the mitochondrial protein import machinery. Nat Rev Mol Cell Biol 2: 339–349 [DOI] [PubMed] [Google Scholar]
- Ridder AN, Kuhn A, Killian JA, de Kruijff B (2001) Anionic lipids stimulate Sec-independent insertion of a membrane protein lacking charged amino acid side chains. EMBO Rep 2: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridder AN, Morein S, Stam JG, Kuhn A, de Kruijff B, Killian JA (2000) Analysis of the role of interfacial tryptophan residues in controlling the topology of membrane proteins. Biochemistry 39: 6521–6528 [DOI] [PubMed] [Google Scholar]
- Roos T, Kiefer D, Hugenschmidt S, Economou A, Kuhn A (2001) Indecisive M13 procoat mutants bind to SecA but do not activate the translocation ATPase. J Biol Chem 276: 37909–37915 [DOI] [PubMed] [Google Scholar]
- Samuelson JC, Chen M, Jiang F, Möller I, Wiedmann M, Kuhn A, Phillips GJ, Dalbey RE (2000) YidC mediates membrane protein insertion in bacteria. Nature 406: 637–641 [DOI] [PubMed] [Google Scholar]
- Samuelson JC, Jiang F, Yi L, Chen M, Kuhn A, Dalbey RE (2001) Function of YidC for the insertion of M13 procoat protein in E. coli: translocation of mutants that show differences in their membrane potential dependence and Sec-requirement. J Biol Chem 276: 34847–34852 [DOI] [PubMed] [Google Scholar]
- Scotti PA, Urbanus ML, Brunner J, de Gier J-W, von Heijne G, van der Does C, Driessen AJ, Oudega B, Luirink J (2000) YidC, the E. coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J 19: 542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol 185: 60–89 [DOI] [PubMed] [Google Scholar]
- Urbanus ML, Scotti PA, Fröderberg L, Sääf A, de Gier J-WL, Brunner J, Samuelson JC, Dalbey RE, Oudega B, Luirink J (2001) Sec-dependent membrane protein insertion: sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep 2: 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema K, Gibrat R, Grouzis J-P, Grignon C (1993) Quantitative measurement of cation fluxes, selectivity and membrane potential using liposomes multilabelled with fluorescent probes. Biochim Biophys Acta 1146: 87–96 [DOI] [PubMed] [Google Scholar]


