Abstract
Notch and bone morphogenetic protein signaling pathways are important for cellular differentiation, and both have been implicated in vascular development. In many cases the two pathways act similarly, but antagonistic effects have also been reported. The underlying mechanisms and whether this is caused by an interplay between Notch and BMP signaling is unknown. Here we report that expression of the Notch target gene, Herp2, is synergistically induced upon activation of Notch and BMP receptor signaling pathways in endothelial cells. The synergy is mediated via RBP-Jκ/CBF-1 and GC-rich palindromic sites in the Herp2 promoter, as well as via interactions between the Notch intracellular domain and Smad that are stabilized by p/CAF. Activated Notch and its downstream effector Herp2 were found to inhibit endothelial cell (EC) migration. In contrast, BMP via upregulation of Id1 expression has been reported to promote EC migration. Interestingly, Herp2 was found to antagonize BMP receptor/Id1-induced migration by inhibiting Id1 expression. Our results support the notion that Herp2 functions as a critical switch downstream of Notch and BMP receptor signaling pathways in ECs.
Keywords: angiogenesis, BMP, Notch, signal transduction, Hairy-related proteins
Introduction
Notch signaling is involved in regulating the balance between cell differentiation and stem cell proliferation during the development of numerous tissues. Interference with the Notch pathway results in dramatic effects in the formation of many organs, including muscle, nervous system and the vascular system (Iso et al, 2003b). Notch encodes a transmembrane receptor that is proteolytically cleaved after binding to a transmembrane ligand, that is, Delta or Jagged, thereby releasing the Notch intracellular domain (NICD) (Artavanis-Tsakonas et al, 1999; Kadesch, 2000). The released NICD translocates into the nucleus. NICD has a transcriptional activation domain, but has no intrinsic DNA binding domain. In the nucleus, NICD associates with the transcription factor RBP-Jκ, also known as CBF-1 (Artavanis-Tsakonas et al, 1999; Kadesch, 2000), which binds to specific DNA binding sites (GTGGGAA). In the absence of NICD, RBP-Jκ/CBF-1 interacts with a corepressor complex, and inhibits transcription (Kao et al, 1998). In contrast, complex formation between RBP-Jκ/CBF-1 and NICD can transactivate primary target genes, such as the HES (Enhancer of Split) and HES-related repressor (HERP families of transcriptional repressors, also called Hey/Hesr/Hrt/CHF/gridlock) (Iso et al, 2003b). HES and HERP are basic helix–loop–helix (bHLH) proteins that negatively regulate the expression of downstream targets such as tissue-specific transcriptional activators. A gridlock mutant (homolog of HERP1 in mammals) in zebrafish revealed a defect in the formation of dorsal aorta (Zhong et al, 2000). Furthermore, both the overexpression and downregulation of HERP2 in endothelial cells (ECs) are known to block capillary-like network formation (Henderson et al, 2001; Taylor et al, 2002).
BMPs, like other members of the TGF-β superfamily, regulate cellular processes by binding to specific heteromeric complexes of type I and type II serine/threonine kinase receptors (Massagué, 1998). The type I receptor, also known as activin-like kinase (ALK), acts downstream of the type II receptors and propagates the signal to the nucleus by phosphorylating specific members of the receptor-regulated (R-) Smads at their extreme C-terminal residues. Whereas Smad2 and Smad3 act downstream of TGF-β and activin type I receptors, BMP type I receptors, that is ALK2, ALK3 and ALK6, stimulate the phosphorylation of Smad1, Smad5 and Smad8. Activated R-Smads form heteromeric complexes with common partner (Co)-Smads, that is, Smad4, and accumulate in the nucleus, where they participate in the transcriptional regulation of target genes. Activated Smad complexes can bind directly or indirectly via other transcription factors to the promoters of target genes (Derynck et al, 1998). An important direct target gene for BMPs is Id1 (inhibitor of DNA binding). BMPs were shown to promote EC migration and tube formation through induction of Id1 protein (Valdimarsdottir et al, 2002). Id1 was recently shown to turn off the expression of thrombospondin-1 (TSP-1), a well-known negative regulator of angiogenesis (Volpert et al, 2002).
Angiogenesis, the formation of new blood vessels from pre-existing vessels, is a multistep process that involves degradation of extracellular matrix, proliferation and directed migration of ECs followed by lumen formation and functional maturation. The transition from activation to resolution phase is determined by an intricately regulated balance between inducers, such as vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF) and inhibitors, including angiopoitins and thrombospondin (Carmeliet, 2000). Recently, in vitro studies, genetic studies in mice and genetic evidence from human vascular disorders have demonstrated that, in addition to many other cell types, Notch (Leong et al, 2002; Iso et al, 2003a; Liu et al, 2003) and BMPs (Lane et al, 2000; Valdimarsdottir et al, 2002) play important roles in determining the fate of vascular cells. Although both Notch and BMP signaling pathways are important for proper vascular development, little is known about the mechanism and a possible interaction between Notch and BMP signaling. To address this, we have investigated the role of HERP2, a gene acting downstream of both Notch and BMP, in ECs. While the two pathways have been shown to act similarly in, for example, the inhibition of myogenic differentiation, our results indicate that the synergistic upregulation of Herp2 expression by Notch and BMP antagonizes BMP-induced EC migration.
Results
The Herp2 promoter is synergistically induced upon activation of the Notch and BMP receptor pathways
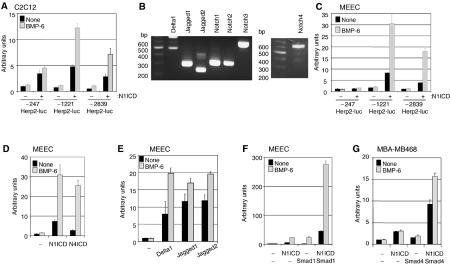
Mesenchymal cells are promoted to differentiate into osteoblasts and are inhibited in their myoblast differentiation by BMPs (Katagiri et al, 1994) or activated Notch (Tezuka et al, 2002), and this may be mediated, in part, via upregulation of Herp2. Herp2 has been reported to function as a transcriptional repressor of myogenesis by forming an inactive complex with MyoD (Sun et al, 2001). Using a gene microarray approach in C2C12 myoblast cells, Herp2 was identified as a target gene of constitutively active (ca) BMPR-IA (also termed ALK3) (Korchynskyi et al, 2003). Herp2 is known to be a primary target of Notch signaling in C2C12 cells (Iso et al, 2002). Therefore, we analyzed the effect of Notch and BMP on the activation of the Herp2 promoter. As can be seen in Figure 1A, three different lengths of mouse Herp2 promoter conjugated to the luciferase reporter (termed −2839Herp2-luc, −1221Herp2-luc and −247Herp2-luc) were significantly activated by Notch1 intracellular domain (N1ICD) in C2C12 cells. Interestingly, BMP had little effect by itself, but it strongly potentiated the activity of −1221Herp2-luc (and to a lesser extent that of −2839Herp2-luc) when N1ICD was present (Figure 1A). Similar results were obtained in HepG2 cells (data not shown). Recently, it has been demonstrated that both Notch and BMP signaling pathways have pivotal roles in angiogenesis (Leong et al, 2002; Valdimarsdottir et al, 2002; Iso et al, 2003a; Liu et al, 2003), but the molecular and cellular mechanisms involved have remained largely unclear. Therefore, we examined whether Herp2 is a common target of both Notch and BMP in ECs as well. First, we examined whether mouse embryonic endothelial cells (MEECs; Goumans et al, 2002; Valdimarsdottir et al, 2002) that are known to express endothelial markers and BMP signaling components also express Notch receptors and their ligands. We detected transcripts for Delta1, Jagged1, 2 and Notch1–4 (Figure 1B). Subsequently, we examined whether Herp2 is a common target of both Notch and BMPs in ECs as well. We found that −2839Herp2-luc and −1221Herp2-luc, but not −247Herp2-luc reporter, were activated by N1ICD and this response was strongly enhanced when costimulated with BMP-6 in MEECs (Figure 1C). Notch1, Notch3 and Notch4 are known to be expressed in vessels. In particular, Notch4 is primarily expressed in the endothelium and the endocardium (Uyttendaele et al, 1996). Therefore, we compared the effect of Notch1 with that of Notch4 on −1221Herp2-luc reporter activity in the absence or presence of BMP-6. Similar to N1ICD, Notch4 intracellular domain (N4ICD) could synergize with BMP-6 in inducing the Herp2 reporter in MEECs (Figure 1D). −1221Herp2-luc activity was stimulated by cotransfection with expression constructs for the Notch ligands Delta1, Jagged1 or Jagged2. This reporter activity was significantly enhanced further by subsequent treatment with BMP-6 (Figure 1E). BMP-6 alone had very little or no effect on the promoter activity of Herp2 in MEECs (Figure 1A and C–E). A synergistic response between N1ICD and BMP was greatly enhanced when the BMP signal transducer Smad1 was cotransfected (Figure 1F). To investigate whether the common mediator Smad4 is required for BMP-6-mediated synergy with N1ICD on −1221Herp2-luc, we used MDA-MB468 epithelial cells which are deficient in Smad4. In these cells, BMP-6 has no promoting effect on N1ICD-induced activation of −1221Herp2-luc unless Smad4 is ectopically expressed (Figure 1G). Taken together, these results indicate that in ECs Notch signaling can activate the Herp2 promoter, and that BMP-6, in a Smad-dependent manner, synergistically cooperates with this response.
Figure 1.
The Herp2 promoter is synergistically induced upon activation of the Notch and BMP receptor pathways. (A) C2C12 cells or (C) MEECs were transfected with each reporter construct in the absence or presence of N1ICD. (B) Detection of Notch receptors and their ligands in MEECs. The corresponding cDNA fragment is a main band in each lane. (D) MEECs were transfected with −1221Herp2-luc in the absence or presence of N1ICD or N4ICD. (E) −1221Herp2-luc was cotransfected with Delta1, Jagged1 or Jagged2 in MEECs. (F) −1221Herp2-luc was cotransfected with N1ICD and/or Smad1 in MEECs. (G) MDA-MB468 cells were transfected with −1221Herp2-luc in the presence of N1ICD and/or Smad4.
RBP-Jκ/CBF-1 and GC-rich palindromic sites are needed for the activation of the Herp2 promoter by Notch and BMP receptor signaling pathways
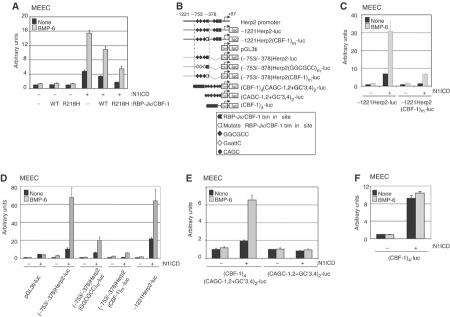
The N1ICD associates with the transcription factor RBP-Jκ/CBF-1 (Kadesch, 2000). BMP R-Smads and Co-Smad have been shown to require Smad Binding Elements (SBE; 5′-AGAC-3′), 5′-CAGC-3′ and/or 5′-GGCGCC-3′ sequence motifs (termed GC-rich palindromic sites) to activate gene expression efficiently (Korchynskyi and ten Dijke, 2002). Comparison of the mouse and human Herp2 promoter sequences revealed multiple conserved RBP-Jκ/CBF-1, GGCGCC and CAGC sequence elements, but no conserved SBEs. Ectopic expression of a dominant-negative RBP-Jκ/CBF-1 mutant, RBP(R218H), attenuated the effect of N1ICD alone as well as the synergistic effect of N1ICD and BMP-6 on the activation of −1221Herp2-luc in MEECs (Figure 2A). Thus, RBP-Jκ/CBF-1 is an indispensable factor for enabling both Notch and BMP signaling pathways to activate the Herp2 gene. The partially inhibitory effect of wild-type RBP-Jκ/CBF-1 on the activity of −1221Herp2-luc by N1ICD is probably caused by the recruitment of corepressor(s) with RBP-Jκ/CBF-1, as reported previously (Beatus et al, 1999; Wang et al, 2002). Mutation of the conserved RBP-Jκ/CBF-1 binding site (TTCCCACG to ggtgCACG) from −421 to −414 (Figure 2B) in −1221Herp2-luc strongly attenuated the activation by Notch alone and Notch and BMP together (Figure 2C). Thus, consistent with previous results (Maier and Gessler, 2000), the Notch-induced activation of the Herp2 promoter in ECs is critically dependent on RBP-Jκ/CBF-1.
Figure 2.
RBP-Jκ/CBF-1 and GC-rich palindromic sites are needed for the activation of the Herp2 promoter by Notch and BMP receptor signaling pathways. (A) Dominant-negative RBP-Jκ/CBF-1 interferes with synergy between Notch and BMP signaling pathways. −1221Herp2-luc was cotransfected with N1ICD, RBP-Jκ and/or RBP(R218H). (B) Cartoon of the luciferase constructs used in Figure 3. t+i: adenovirus major late promoter; luc: luciferase. Closed symbols: wild-type motif; open symbols: mutated motif. Small letters indicate mutated nucleotides. (C) RBP-Jκ/CBF-1 site in Herp2 promoter is important for activation by Notch and BMP signaling pathways. MEECs were transfected with indicated reporters in the absence or presence of N1ICD. (D) MEECs were transfected with indicated reporters in the absence or presence of N1ICD. (E) CBF-1 binding and GC-rich palindromic sites mimic synergy by BMP and Notch signaling pathways. MEECs were transfected with indicated reporters in the absence or presence of N1ICD. (F) The activity of (CBF-1)4-luc in the presence of N1ICD can not be enhanced by BMP-6 in MEECs. MEECs were transfected with (CBF-1)4-luc in the absence or presence of N1ICD.
A conserved Herp2 promoter fragment (−753 to −378) subcloned in pGL3ti was highly BMP responsive in the presence of N1ICD (Figure 2D). Mutation of the three GGCGCC palindromic sequence motifs in the Herp2 promoter severely inhibited but did not eliminate the synergistic activation of this reporter by N1ICD and BMP-6 (Figure 2D), whereas disruption of the RBP-Jκ/CBF-1 binding site in this promoter essentially eliminated activation of luciferase activity by Notch and/or BMP-6 (Figure 2D). The reason for the incomplete inhibition of synergistic activation upon mutation of the palindromic sites is that within the −753 and −378 fragment, several CGCC (nonpalindromic) and CAGC sites remain present, which can also support (albeit weaker than GGCGCC palindromic site) BMP-induced promoter activation. These results suggest that both RBP-Jκ/CBF-1 binding sites and BMP-6 responsive sites are essential for full synergistic activation of the promoter by N1ICD and BMP-6. To test whether the conserved RBP-Jκ/CBF-1, GGCGCC and CAGC sequence motifs are sufficient to mediate this synergy between Notch and BMP signaling pathways, we generated two synthetic reporter constructs containing a minimal promoter with two repeats of CAGC and GGCGCC motifs (Korchynskyi and ten Dijke, 2002) with or without four binding sites for RBP-Jκ/CBF-1. As anticipated, we could recapitulate a strong synergistic activation by N1ICD and BMP-6 by using a reporter containing two repeats of CAGC and GGCGCC motifs with four binding sites for RBP-Jκ/CBF-1. Furthermore, and as expected, a synthetic reporter carrying just 4 × CBF-1 sites is activated by N1ICD (Figure 4F), but is BMP-6 unresponsive. However, the reporter carrying just the GGCGCC and CAGC motifs was stimulated neither by N1ICD, nor BMP-6, nor their combination (Figure 2E and F). Thus, the RBP-Jκ/CBF-1 and GC-rich palindromic sites are both needed for the most efficient activation of the Herp2 promoter by Notch and BMP receptor signaling pathways.
Figure 4.
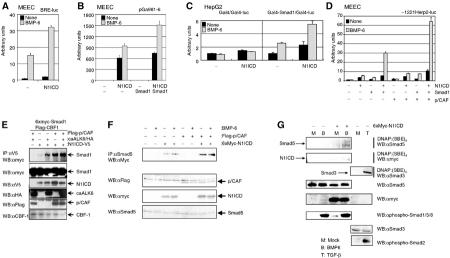
Notch and BMP signaling pathways cooperate in transcriptional responses via protein–protein interactions that also involve p/CAF. (A) Notch pathway affects BMP-6-induced BRE-luc activity. MEECs were transfected with N1ICD and BRE-luc. (B) BMP/Smad pathway potentiates activated Notch/RBP-Jκ/CBF-1-driven transcriptional response. MEECs were transfected with N1ICD, Smad1 and pGa981-6. (C) N1ICD potentiates transcriptional activity of Smad1 independent of direct binding of Smad1 to DNA. HepG2 cells were transfected with combinations of N1ICD, Gal4, Gal4-Smad1 and Gal4-M1-Luc. (D) p/CAF potentiates the synergy between Notch and BMP signaling pathways. MEECs were transfected with N1ICD, Smad1, p/CAF and −1221Herp2-luc. (E) Activated BMP receptor and p/CAF enhance the interaction between Smad1 and N1ICD. 6xMyc-Smad1, Flag-CBF-1, Flag-p/CAF, caALK6/HA and N1ICD-V5 were transfected in COS7 cells. The cell lysates were immunoprecipitated with anti-V5 antibody. The immunoprecipitates were subjected to Western blotting with anti-myc antibody (upper panel). Using total lysates, the expressions of 6xmyc-Smad1 (second panel), N1ICD-V5 (third panel), caALK6/HA (fourth panel), Flag-p/CAF (fifth panel) and Flag-CBF-1 (lower panel) are shown. (F) BMP-6 and/or p/CAF enhance the interaction between Smad5 and N1ICD. 6xMyc-N1ICD and Flag-p/CAF were transfected in MEECs. The cells were stimulated with 100 ng/ml BMP-6 2 h before lysis. The cell lysates were immunoprecipitated with anti-Smad5 antibody. The immunoprecipitates were subjected to Western blotting with anti-myc antibody (upper panel). Using total lysates, the expressions of Flag-p/CAF (second panel), 6xMyc-N1ICD (third panel) and endogenous Smad5 (lower panel) are shown. (G) N1ICD potentiates the ability of Smad5 to bind to SBE. 6xMyc-N1ICD was transfected in MEECs. The cells were stimulated with 100 ng/ml BMP-6 2 h before lysis. After DNAP, Western blotting was performed with anti-Smad5 (upper panel) or anti-myc antibody (second panel). Using total lysates, the expressions of endogenous Smad5 (fourth panel), 6xMyc-N1ICD (fifth panel), phospho-Smad1/5/8 (sixth panel), endogenous Smad3 (seventh panel) and phospho-Smad2 (lower panel) are shown. As a positive control, the lysates from MEECs stimulated with 5 ng/ml TGF-β for 2 h were used. Then, association of Smad3 with SBE was observed using anti-Smad3 antibody (third panel).
Notch and BMP receptor signaling pathways synergize to induce Herp2 mRNA expression
To investigate whether the induction of the Herp2 reporter indeed resulted in an increase in transcription, we performed Northern blot analysis of MEECs following the infection of the cells by various combinations of adenoviruses encoding N1ICD and ALKs. N1ICD stimulated the expression of Herp2 mRNA in MEECs (Figure 3A). Herp2 transcripts were weakly induced by ectopic expression of caALK6, but not by constitutively active (ca) TGF-β type I receptor (TβR-I)/caALK5 (Figure 3A). The weak induction may be mediated by endogenous Notch ligands that are expressed in MEECs (Figure 1B). Importantly, and consistent with the results of the Herp2 promoter activation in MEECs, coinfection of N1ICD and caALK6 gave a significant more than additive increase in Herp2 mRNA compared to N1ICD alone (Figure 3A). The expression of the various adenoviruses was confirmed by Western blotting (Figure 3B). Since BMP-6 can induce Herp2 mRNA, we next determined whether Herp2 is a direct target gene in BMP signaling (Figure 3C). Herp2 mRNA expression was clearly induced 1 h after BMP-6 stimulation even in the presence of cycloheximide (CHx) (Figure 3C), indicating that no de novo protein synthesis is required for this response. This result strongly supports the notion that Herp2 is a direct BMP target.
Figure 3.
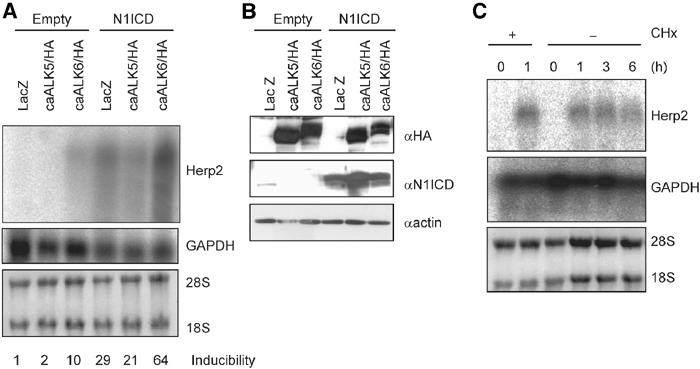
Notch and BMP signaling pathways synergize to induce Herp2 mRNA expression. (A) Northern blot analysis of Herp2 mRNA expression in MEECs challenged with LacZ, caALK5/HA and caALK6/HA in the absence or presence of N1ICD adenoviruses (upper panel). The DNA fragment encoding the C-terminal part of mouse Herp2, which has less identity among other Herp family, was used as a probe. Equal loadings of RNA samples are shown by expression of GAPDH mRNA (middle panel) and ethidium bromide staining pattern of 28S and 18S ribosomal RNAs before Northern blotting (lower panel). Relative intensity of bands corresponding to Herp2 mRNA was normalized using the intensity of bands for GAPDH mRNA. Each value is shown at the bottom. (B) Expression of ectopically expressed HA-tagged caALKs (upper panel) and N1ICD (lower panel). As a loading control, the same blot was stained with anti-actin antibody (lower panel). (C) Herp2 is a direct target gene for BMP. Northern blot analysis of Herp2 mRNA in MEECs stimulated with 100 ng/ml BMP-6 for the indicated times in the absence or presence of 5 μg/ml CHx (upper panel). Equal loadings of RNA samples are shown by expression of GAPDH mRNA (middle panel) and ethidium bromide staining pattern of 28S and 18S ribosomal RNAs before Northern blotting (lower panel).
Notch and BMP receptor signaling pathways cooperate in transcriptional responses via protein–protein interactions
One possibility for how Notch and BMP signaling pathways could synergize is via protein–protein interactions. To explore this possibility, we analyzed the effect of the N1ICD on a BMP-6-inducible transcriptional reporter containing two repeats of SBE conjugated with the GC-rich palindromic site but lacking RBP-Jκ/CBF-1 binding sites (BRE-luc). Also, the effect of BMP/Smads on a transcriptional reporter lacking SBEs and containing only 12 repeats of the RBP-Jκ/CBF-1 binding site (pGa981-6) was tested. As expected, BMP-6 strongly induced BRE-luc, while N1ICD alone marginally increased the activity of BRE-luc. Despite the absence of an RBP-Jκ/CBF-1 binding site, the capacity of BMP-6 to induce the activity of BRE-luc was potentiated in the presence of N1ICD (Figure 4A). In agreement with this observation, the BMP/Smad signaling pathways were capable of enhancing the N1ICD/RBP-Jκ/CBF-1-mediated transcriptional response of the pGa981-6 reporter (Figure 4B). In contrast, we did not observe BMP-mediated enhancement of (CBF-1)4-luc in the presence of N1ICD (Figure 2F), suggesting that the interactions between Notch and BMP signaling proteins are weak. When we uncoupled the direct binding of Smad1 to DNA by making a Gal4-Smad1 fusion, we observed that N1ICD could also upregulate the transcriptional activity of Gal4-Smad1, indicative of a direct interaction between Smad1 and N1ICD. On the other hand, Gal4 alone was influenced by neither Notch nor BMP-6 (Figure 4C).
p/CAF was identified as a binding partner for both N1ICD (Kurooka and Honjo, 2000; Beatus et al, 2001) and R-Smads (Itoh et al, 2000). Interestingly, p/CAF strongly potentiated the synergy between the N1ICD and BMP/Smad signaling pathways (Figure 4D). To test the interaction between Notch and BMP signaling components, we coexpressed Smad1, N1ICD and RBP-Jκ/CBF-1 with or without p/CAF and/or caALK6 in COS7 cells, and subjected the cell lysates to immunoprecipitation for N1ICD followed by immunoblotting for Smad1 (Figure 4E). Interaction between N1ICD and Smad1 was greatly stimulated upon the cotransfection of caALK6 or p/CAF. Furthermore, we transfected N1ICD and/or p/CAF in MEECs, and the cell lysates were subjected to immunoprecipitation with anti-Smad5 antibody followed by Western blotting with anti-myc antibody. Interaction between exogenous N1ICD and endogenous Smad5 was stimulated when MEECs were co-transfected with p/CAF (Figure 4F). Thus, consistent with the results obtained in luciferase assays, p/CAF enhanced the association of N1ICD and Smad5. Taken together, these data indicate that synergy between the Notch and BMP receptor signaling pathways might be mediated by interactions of their intracellular nuclear effector proteins.
Next, we examined whether N1ICD can affect the ability of Smad5 to bind to DNA upon BMP-6 stimulation. We performed DNA affinity precipitation (DNAP) using biotinated SBE (Jonk et al, 1998) with lysates from MEECs, followed by Western blotting with anti-Smad5 and anti-myc antibodies to detect Smad5 and N1ICD, respectively. As seen in Figure 4G, upon BMP stimulation, N1ICD interacts with Smad5 and potentiates the ability of Smad5 to bind to SBE. Thus, interaction of Smad5 with the N1ICD stimulates Smad5 binding to DNA.
Herp2 antagonizes BMP receptor-induced migration of ECs by inhibiting Id1 expression
To study the biological consequences of the synergy in Herp2 expression between the Notch and BMP receptor signaling pathways, we used migration assay as a biological readout. Consistent with the previous findings (Henderson et al, 2001; Taylor et al, 2002), the activation of Notch1 or ectopic expression of Herp2 inhibited MEEC migration (Figure 5A and B). Furthermore, activated Notch was capable of inhibiting the BMP-induced migratory response (Figure 5A). Thus, activation of the Notch and BMP pathways has opposite effects on MEEC migration but the effect of the Notch pathway is dominant over that of BMP.
Figure 5.
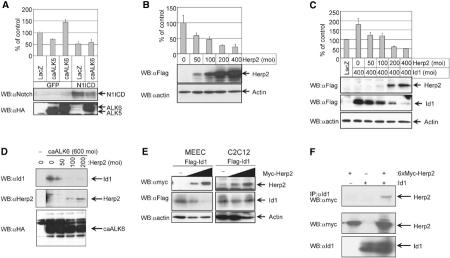
Activation of Notch and BMP receptors and their downstream targets regulate EC migration. (A) Effect of N1ICD in the absence or presence of caALK6 or caALK5 on the migration of MEECs. MEECs infected with the indicated adenoviruses were used. The cells were infected with the same amounts of adenoviruses (a moi of 500). The value for the migration in MEECs infected with both GFP and LacZ was set as 100% (upper panel). Using total lysates, the expressions of N1ICD (middle panel) and caALKs (lower panel) are shown. (B) Effect of Herp2 on the migration of MEECs. MEECs infected with adenoviruses expressing LacZ and/or Flag-Herp2 were used. The cells were infected with the same amounts of adenoviruses (a moi of 400). Using adenovirus expressing LacZ, total amounts of adenoviruses were adjusted. The value for the migration in MEECs infected with LacZ alone was represented as 100% (upper panel). Using total lysates, the expressions of Herp2 (middle panel) and actin (lower panel) are shown. (C) Herp2 inhibits Id1-induced cell migration. MEECs infected with adenoviruses expressing LacZ, Flag-Herp2 and Flag-Id1 were used in a migration assay. The cells were infected with the same amounts of adenoviruses (a moi of 800). Using adenovirus expressing LacZ, total amounts of adenoviruses were adjusted. The number of cells migrated in MEECs infected with LacZ alone was set at 100% (upper panel). Cell lysates of MEECs infected with adenoviruses expressing Flag-Herp2 (second panel) and Flag-Id1 (third panel) were subjected to Western blotting with anti-Flag M5 antibody. To show equal loading of each sample, anti-actin antibody (lower panel) was used. (D) Overexpression of Herp2 decreases caALK6-induced Id1 expression. MEECs infected with adenoviruses expressing caALK6/HA and Flag-Herp2 were used. The cells were infected with the same amounts of adenoviruses (a moi of 800). Using adenovirus expressing LacZ, total amounts of adenoviruses were adjusted. Cell lysates of MEECs infected with adenoviruses were subjected to Western blotting with anti-Id1 (upper panel), anti-Herp2 (middle panel) and anti-HA (lower panel) antibodies, respectively. (E) Effect of Herp2 on Id1 expression in MEECs or C2C12 cells. MEECs (left panel) or C2C12 cells (right panel) were transfected with Flag-Id1 and different amounts of Myc-Herp2. Total cell lysates were used for the detection of Myc-Herp2 (upper panel), Flag-Id1 (middle panel) and actin (lower panel). (F) Interaction of Herp2 with Id1 in COS7 cells. 6xMyc-Herp2 and Id1 were transfected in COS7 cells, and the cell lysates were immunoprecipitated with anti-Id1 antibody. The immunoprecipitates were subjected to Western blotting with anti-myc antibody (upper panel). Using total lysates, the expressions of 6xMyc-Herp2 (second panel) and Id1 (lower panel) are shown.
BMP was shown to promote EC migration by upregulating Id1 expression (Valdimarsdottir et al, 2002). We therefore tested whether Herp2 would inhibit Id1 action as well. Indeed, Herp2 inhibited Id1-induced MEEC migration in a dose-dependent fashion (Figure 5C). Interestingly, we noted that the Id1 expression levels were reduced in response to increasing levels of Herp2 (Figure 5C). Moreover, in response to increasing levels of Herp2, a dose-dependent reduction of the endogenous Id1 expression induced by caALK6 was observed although the expression of caALK6 was equal (Figure 5D). Therefore, it appears that Herp2 blocks caALK6- or Id1-induced migration at least in part by promotion of the degradation of Id1 protein by Herp2.
It has been reported that both Notch and BMP signaling pathways contribute to osteoblast differentiation (Tezuka et al, 2002) and the inhibition of myoblast differentiation, and that these effects are, in part, mediated via their downstream target genes Herp2 and Id1, respectively (Jen et al, 1992; Sun et al, 2001). However, our results on EC migration indicate that the Notch/Herp2 pathway and BMP receptor/Id1 pathways function in opposite manners. To investigate the possibility that Herp2 decreases the expression of Id1 in a cell-type-specific manner, we cotransfected Id1 with increasing amounts of Herp2 in MEECs as well as C2C12 cells and measured Id1 (and Herp2) expression levels. Whereas Herp2 affected the level of Id1 protein in MEECs, the level of Id1 in C2C12 cells was stable despite an increase in Herp2 expression (Figure 5E). Thus, the effect of Herp2 on Id1 protein levels in a specific cell type correlates with either an antagonistic or a synergistic response between the Notch and BMP receptor signaling pathways.
Herp2 might associate with Id1 to promote the degradation of Id1. To further test this possibility, we transfected Herp2 and Id1 in COS7 cells, and cell lysates were subjected to immunoprecipitation with anti-Id1 antibody, followed by Western blotting with anti-myc antibody. As seen in Figure 5F, Herp2 did interact with Id1.
To investigate in detail whether degradation of Id1 can be accelerated by Herp2, we analyzed the half-life of Id1 in the absence or presence of Herp2. The half-life of Id1 without Herp2 in MEECs was approximately 60 min while that with Herp2 was around 30 min (Figure 6A and B). After 2–4 h, most of Id1 disappeared in the presence of Herp2. Consistent with the result in Figure 5C, the intensity of the band corresponding to Id1 in the presence of Herp2 at 0 min was relatively lower than that in the absence of Herp2 because the excess of Herp2 promoted degradation of Id1. We also asked whether proteasome pathway contributed to degradation of Id1 by Herp2. MG-132, a proteasome inhibitor, could prolong the Id1 expression in the presence of Herp2 (Figure 6A and C). Thus, the proteasome pathway is involved in the Herp2-mediated Id1 degradation.
Figure 6.
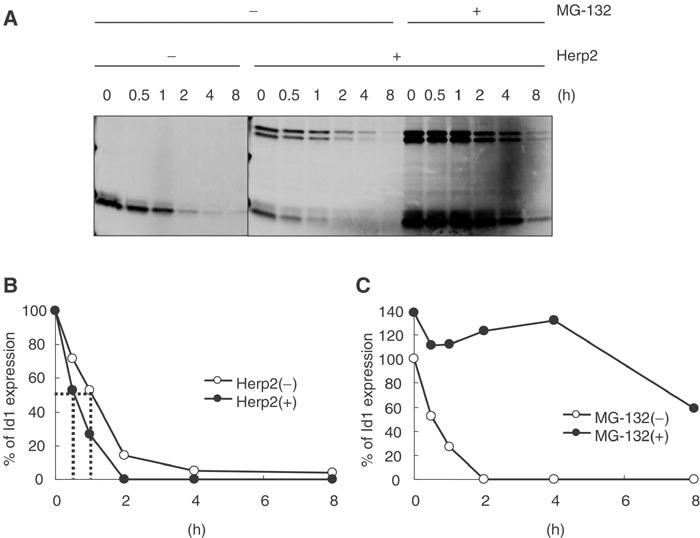
Herp2 accelerates degradation of Id1 in MEECs. (A) MEECs infected with Flag-Herp2 and/or Flag-Id1 adenoviruses in the absence and/or presence of 10 μM MG-132 were subject to be labeled with Trans[35S-Label]. The cells were lysed at the indicated times. (B) The intensity of bands corresponding to Id1 in (A) without MG-132 was measured using the phosphoimager. (C) The intensity of bands corresponding to Id1 in (A) with MG-132 was measured using the phosphoimager.
Herp2 acts a critical switch gene for ECs downstream of Notch and BMP receptor signaling pathways
To demonstrate the importance of Herp2 in the synergy between the Notch and BMP receptor signaling pathways, we designed short interfering RNA (siRNA) to knock down selectively the expression of Herp2. Treatment of MEECs with Herp2 siRNAs, but not control siRNA, partially inhibited the N1ICD/caALK6-induced Herp2 expression levels. The levels of β-actin mRNA were unaffected by the Herp2 siRNA treatment (Figure 7C). Even though the Herp2 levels in N1ICD-treated cells transfected with control siRNA were comparable to that in N1ICD/caALK6-treated cells transfected with Herp2 siRNA, the migration in the latter case is much more potent (Figure 7A). This is consistent with our model that Id1 is induced by caALK6, and also contributes in part to the rescue of the EC migration response.
Figure 7.
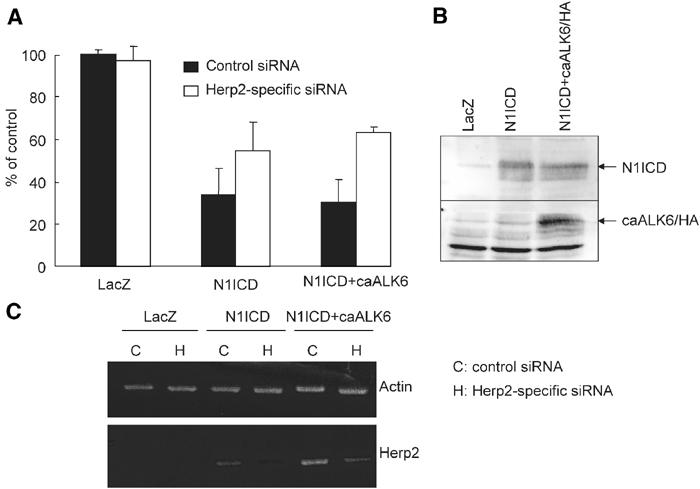
Herp2 acts as a critical switch gene for ECs downstream of Notch and BMP signaling pathways. (A) Effect of Herp2-specific siRNA on the migration of MEECs challenged with adenoviruses expressing N1ICD and/or caALK6/HA. MEECs infected with adenoviruses expressing LacZ, N1ICD and caALK6/HA were used in a migration assay. The cells were infected with 200 moi of caALK6 and/or 400 moi of N1ICD. Using adenovirus expressing LacZ, total amounts of adenoviruses were adjusted. The value for the migration in MEECs infected with LacZ alone when treated with the control siRNA was represented as 100%. (B) The expression of N1ICD and caALK6 in MEECs. Anti-N1ICD (upper panel) and anti-HA antibodies (lower panel) were used to detect the expression of N1ICD and caALK6, respectively. (C) Detection of Herp2 mRNA from the cells treated with Herp2-specific siRNAs. RT–PCR was performed with primers specific for Herp2 and actin.
Discussion
The formation of a vascular network is a complex event that requires correct vessel branching as well as the guidance of the new sprout within the target tissue. Recent studies have demonstrated that both the Notch and BMP signaling pathways, important for regulating cell fate, are involved in multiple aspects of vascular development. Notch, after interacting with its transmembrane ligand Delta or Jagged, and BMPs, via binding to their specific serine/threonine kinase receptors, elicit their cellular effects by regulating gene transcription. Whereas Hes and Herp transcriptional repressor proteins have been implicated as pivotal primary target genes of activated Notch, important effectors of BMP signaling are the Id proteins, inhibitors of basic HLH transcription factors. Interestingly, BMP was found to potently induce the expression of Herp2, a direct target gene for Notch signaling, in the presence of activated Notch (Figures 1 and 3). Since Herp2 is downstream of Notch and BMP signaling pathways, we investigated the role of Herp2 to gain more insight into the mechanisms by which Notch and BMP signaling pathways regulate vascular development. Herp2 expression mimicked the effect of activated Notch in which it inhibited EC migration (Figure 5). Consistent with our results, Herp2 was found to inhibit EC migration when ectopically expressed (Henderson et al, 2001). Moreover, Herp2 was found to antagonize the BMP/Id1-induced migration by lowering the expression of Id1. Therefore, we propose that after BMP stimulation, an EC migrates until it contacts a cell expressing Delta or Jagged capable of activating Notch, upon which Herp2 is potently induced and migration is stopped (Figure 8).
Figure 8.
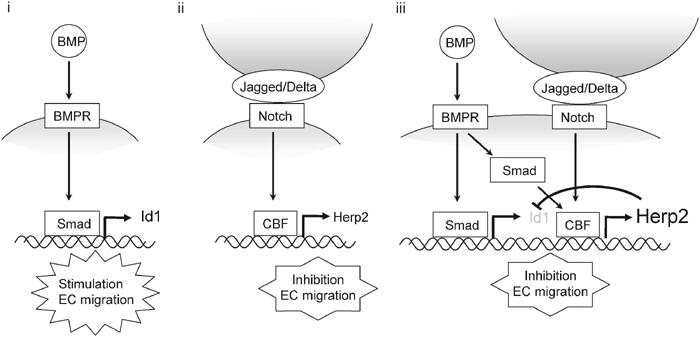
Schematic model of the functional role of Herp2 and Id1 in the Notch and BMP receptor signaling pathways in ECs. (i) BMP signaling alone induces Id1 expression, which activates EC migration. (ii) Notch signaling alone induces Herp2 expression. Subsequently, Herp2 inhibits EC migration. (iii) When ECs simultaneously receive both BMP and Notch signaling, Herp2 expression is synergistically induced. Then, Herp2 is able to promote the degradation of Id1. As a consequence, EC migration is efficiently blocked.
A soluble form of Jagged1, acting in a dominant-negative manner, was shown to stimulate angiogenesis. Furthermore, Leong et al (2002) recently reported that activated Notch4 inhibits EC migration, proliferation and sprouting, in part, by promoting β1-integrin-mediated adhesion to the underlying matrix. The observed potent inhibition of EC migration by activated Notch suggests a role for Notch in the maturation phase of angiogenesis.
While BMP itself had little effect on Herp2 promoter activation (Figure 1), the induction of Herp2 by activated Notch was potently induced by BMP. Consistent with these findings, we observed that Herp2 mRNA expression was induced by Notch, and could be further enhanced by costimulation with BMP. BMP by itself, weakly and directly, induced Herp2 expression. We cannot exclude the possibility that the latter response is dependent on endogenous Notch signaling in ECs. Characterization of the mouse Herp2 promoter revealed that the synergy between the two pathways is mediated via both RBP-Jκ/CBF-1 and GC-rich palindromic sites that are located in a region that is highly conserved between the mouse and human promoters (data not shown); both sequence elements were found to be needed for efficient activation of the Herp2 promoter by activated Notch and BMP receptor. RBP-Jκ/CBF-1 is known to associate with NICD to activate gene transcription. GC-rich palindromic sites can interact with activated BMP–Smad complexes. Analysis of the BMP-inducible Id1 promoter has shown that SBEs, GC-rich palindromic and CAGC sequence motifs are needed to activate the promoter efficiently (Korchynskyi and ten Dijke, 2002). Whereas multiple conserved GC-rich palindromic and CAGC elements are present, no conserved SBEs were identified in the Herp2 promoter. The latter could explain why this promoter is not efficiently activated by BMP. Interaction of Smads with the strong DNA binding transcription factor RBP-Jκ/CBF-1 may be needed for BMPs to activate this promoter. Indeed, activated Notch and activated Smads were found to interact with each other in the presence of RBP-Jκ/CBF-1. The physical interaction and functional synergy could be further enhanced by the cotransfection with the transcriptional coactivator p/CAF. p/CAF was previously shown to interact with N1ICD as well as R-Smads, and may be important for stabilizing the complex (Itoh et al, 2000; Kurooka and Honjo, 2000; Beatus et al, 2001). Thus, synergy between Notch/RBP-Jκ/CBF-1 and BMP receptor/Smad appears to be mediated via DNA and protein–protein interactions on the Herp2 promoter.
In agreement with the finding that the inhibition of EC migration by activated Notch is dominant over the BMP/Id1-induced stimulation of migration, we found that ectopic expression of Herp2 alone was capable of downregulating (ectopically or endogenously) expressed Id1 protein in MEECs. Since both Id1 and Herp2 possess an HLH domain, we examined the possibility that these two proteins can interact with each other. When both proteins were ectopically expressed, Herp2 was found to be capable of making a heteromeric complex with Id1 (Figure 5F). In mesenchymal cells where Notch and BMP and their targets Herp2 and Id1, respectively, synergize on promoting osteoblast differentiation and inhibiting myoblast differentiation, we found that Herp2 does not induce instability of Id1 protein. A likely explanation for the opposite results obtained in MEECs versus C2C12 cells is that Herp2 and Id1 interact with each other in MEECs, but that in C2C12 cells the proteins do not interact with each other but rather with bHLH proteins, like MyoD and myogenenin. Since the Herp2-induced degradation of Id1 was inhibited upon pretreatment of MEECs with a proteasome inhibitor (Figure 6), Herp2 may recruit an E3 ligase(s) to the Id1 protein and promote its degradation.
Intriguingly, the notion that blood vessels and nerves use the same families of proteins for directed cell migration might implicate cross-talk between the Notch and BMP signaling pathways in the differentiation of the neuronal stem cells. BMP has strong neurogenic activity for the multipotent neural crest (Morrison et al, 2000), whereas Notch has been shown to be dominant over BMP and to inhibit neurogenesis and instead promote glial differentiation. Interestingly, the differentiation of neural crest cells to myofibroblasts is promoted by BMP, and can be potentiated by activating Notch. These antagonistic and synergistic effects between Notch and BMP with respect to neuronal and myofibroblast differentiation are reminiscent of the opposite effects we observed on MEECs and mesenchymal C2C12 cells. Herp2 is a strong candidate for an elusive feedback signal induced by Notch that promotes a switch from neuronal to glial differentiation. When neural crest cells invade the pharyngeal arches and differentiate into mesenchymal cells, Herp2 expression shows its peak in the pharyngeal arch precursors, just prior to the neural crest invasions (Nakagawa et al, 1999), suggesting a similar role for Notch and BMP in inhibiting neural crest cell migration as the model we propose for EC migration. An interesting topic for future research will be to determine the function of Herp2 and Id during neural crest cell differentiation.
Materials and methods
DNA constructs
(SBE)4-luc, BRE-luc, pGL-3ti, Flag-Smad1, Flag-Smad4, pSG424 (termed Gal4), Gal4-Smad1, Gal4-M1-Luc, (CAGC-1,2+GC′3,4)2-luc, Flag-p/CAF, Flag-Herp2 and Id1 have been described previously (Jonk et al, 1998; Itoh et al, 2000, 2001; Iso et al, 2001; Korchynskyi and ten Dijke, 2002). (CBF-1)4-luc, 6xMyc-N1ICD and 6xMyc-N4ICD were obtained from Dr Kopan (Saxena et al, 2001). −2839Hey1-luc, −1221Hey1-luc and −247Hey1-luc (termed −2839Herp2-luc, −1221Herp2-luc and −247Herp2-luc, respectively, in the text) were provided by Dr Gessler (Maier and Gessler, 2000). Flag-Delta 1, Flag-Jagged 1 and Flag-Jagged 2 were obtained from Dr Hirai (Shimizu et al, 2002). RBP-Jκ-myc and RBP(R218H) were gifted by Dr Honjo (Kato et al, 1997). Flag-CBF-1 and pGa981-6 were from Dr Kadesch (Kao et al, 1998) and Dr Strobl (Minoguchi et al, 1997), respectively. Other plasmids described were constructed by PCR-based amplification or QuikChange site-directed mutagenesis kit. After generation of all mutants, the sequences in each plasmid were confirmed.
Adenoviruses
Adenovirus expressing Flag-HERP2 was generated using pShuttle-CMV vector (He et al, 1998). After recombination of pShuttle-CMV-Flag-Herp2 with pAdEasy-1, the plasmid obtained was transfected into 293T cells, and then adenoviruses were amplified. Other adenoviruses have been described previously (Rangarajan et al, 2001; Goumans et al, 2002).
Cell culture and adenoviral infection
COS7, HepG2, MDA-MB468 and MEECs were cultured as described previously (Itoh et al, 2001; Goumans et al, 2002). MEECs were infected with adenoviruses as described previously (Goumans et al, 2002).
RNA isolation, Northern blotting and RT–PCR
Total RNA was isolated using RNeasy columns. Northern blotting and RT–PCR were performed as described previously (Goumans et al, 2002; Korchynskyi and ten Dijke, 2002). After RT–PCR, each fragment was excised from the agarose gel and sequenced to confirm the specificity (data not shown).
Western blotting and immunoprecipitations
Western blotting and immunoprecipitation were performed as described previously (Itoh et al, 2001). As primary antibodies, anti-Smad3 (Nakao et al, 1997), anti-Smad5 (Goumans et al, 2002), anti-phospho-Smad1/5/8 (Goumans et al, 2002), anti-phospho-Smad2 (Goumans et al, 2002), anti-Flag M5 (Sigma), anti-c-myc(9E10) (SantaCruz), anti-HA12C5 (Roche), anti-N1ICD (SantaCruz), anti-RBP-Jκ/CBF-1 (SantaCruz), anti-V5 (Invitrogen), anti-Id1 (SantaCruz), anti-actin (Chemicon International) and anti-Herp2 antibodies (Iso et al, 2002) were used.
DNAP
After MEECs stimulated with 100 ng/ml BMP-6 for 2 h were lysed with TNE buffer, cell lysates were precleared with biotinated 4xMu (Jonk et al, 1998) and streptavidin agarose for 30 min, and incubated with biotinated 4xWT (Jonk et al, 1998) for 2 h at 4°C. Subsequently, streptavidin agarose was added to the reaction mixture and incubated for 30 min at 4°C. After precipitates were washed with TNE buffer five times, precipitates and aliquots of total lysates were separated by SDS–PAGE. Then, proteins were transferred to the membrane. The membrane was incubated with the indicated primary antibodies. Primary antibodies were detected as described above.
Transfections and reporter assays
MEECs were transiently transfected using lipofectamine (Invitrogen) and plus reagent (Invitrogen). Fugene6 (Roche) was used for the transfections in C2C12 and MDA-MB468 cells. HepG2 cells were transfected by the method of calcium phosphate precipitation (Itoh et al, 2001). If necessary, the cells were stimulated with 100 ng/ml BMP-6 for 18 h. In all reporter assays, the β-galactosidase expression plasmid pCH110 served as an internal control to correct for transfection efficiency. The experiments were performed in triplicate at least three times. The representative data are shown (mean±s.d.).
EC migration assay and knockdown of Herp2 using siRNA
Migration (chemokinesis) was measured as described previously (Goumans et al, 2002). MEECs were transfected with a mixture of Herp2-specific siRNAs (5′-GACGGAGAGGCAUCAUCGAdTdT-3′/3′-dTdTCUGCCUCUCCGUAGUAGCU-5′ and 5′-GGUUAUUUUGACGCGCACGdTdT-3′/3′-dTdTCCAAUAAAACUGCGCGUGC-5′) (Qiagen) or control siRNA (5′-UUCUCCGAACGUGUCACGUdTdT-3′/3′-dTdTAAGAGGCUUGCACAGUGCA-5′) (Qiagen) using TransMessenger Transfection Reagent. The data show mean±s.d. (n=5).
Pulse-chase experiments
MEECs were infected with adenoviruses expressing Id1 and/or Herp2 for 24 h. Subsequently, 0.37 MBq/ml Trans[35S-Label] (ICN) was added to the medium and incubated for 1 h. Then, the cells were washed and cultured for the indicated times. After lysis, immunoprecipitation was performed with anti-Flag M5 antibody. If necessary, 10 μM MG-132 was added.
Acknowledgments
We thank Drs B Defize and C-H Heldin for valuable discussion. We are grateful to Dr Sampath for recombinant BMP-6. We thank Drs Kopan, Gessler, Hirai, Honjo, Kadesch, Strobl, Taga and Miyazono for useful reagents. This work was supported by grants from Dutch Cancer Society (NKI 2000-22117), Dutch Organization for Scientific Research (MW 902-16-295), the Netherlands Heart foundation (grant 99-046) and Ludwig Institute for Cancer Research.
References
- Artavanis-Tsakonas S, Rand SMD, Lake RJ (1999) Notch signaling: cell fate control and signal integration in development. Science 284: 770–776 [DOI] [PubMed] [Google Scholar]
- Beatus P, Lundkvist J, Öberg C, Lendahl U (1999) The notch 3 intracellular domain represses notch 1-mediated activation through Hairy/Enhancer of split (HES) promoters. Development 126: 3925–3935 [DOI] [PubMed] [Google Scholar]
- Beatus P, Lundkvist J, Öberg C, Pedersen K, Lendahl U (2001) The origin of the ankyrin repeat region in Notch intracellular domains is critical for regulation of HES promoter activity. Mech Dev 104: 3–20 [DOI] [PubMed] [Google Scholar]
- Carmeliet P (2000) Mechanisms of angiogenesis and arteriogenesis. Nat Med 6: 389–395 [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang Y, Feng X-F (1998) Smads: transcriptional activators of TGF-β responses. Cell 95: 737–740 [DOI] [PubMed] [Google Scholar]
- Goumans M-J, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, ten Dijke P (2002) Balancing the activation state of the endothelium via two distinct TGF-β type I receptors. EMBO J 21: 1743–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T-C, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B (1998) A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA 95: 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson AM, Wang S-J, Taylor AC, Aitkenhead M, Hughes CCW (2001) The basic helix–loop–helix transcription factor HESR1 regulates endothelial cell tube formation. J Biol Chem 276: 6169–6176 [DOI] [PubMed] [Google Scholar]
- Iso T, Chung G, Hamamori Y, Kedes L (2002) HERP1 is a cell type-specific primary target of Notch. J Biol Chem 277: 6598–6607 [DOI] [PubMed] [Google Scholar]
- Iso T, Hamamori Y, Kedes L (2003a) Notch signaling in vascular development. Arter Tromb Vas Biol 23: 543–553 [DOI] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y (2003b) HES and HERP families: multiple effectors of the notch signaling pathway. J Cell Physiol 194: 237–255 [DOI] [PubMed] [Google Scholar]
- Iso T, Sartorelli V, Chung G, Shichinohe T, Kedes Lamd Hamamori Y (2001) HERP, a new primary target of Notch regulated by ligand binding. Mol Cell Biol 21: 6071–6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh F, Asao H, Sugamura K, Heldin C-H, ten Dijke P, Itoh S (2001) Promoting bone morphogenetic protein signaling through negative regulation of inhibitory Smads. EMBO J 20: 4132–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh S, Ericsson J, Nishikawa J, Heldin C-H, ten Dijke P (2000) The transcriptional co-activator P/CAF potentiates TGF-β/Smad signaling. Nucleic Acids Res 28: 4291–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen Y, Weintraub H, Benezra R (1992) Overexpression of Id protein inhibits the muscle differentiation program: in vivo association of Id with E2A proteins. Genes Dev 6: 1466–1479 [DOI] [PubMed] [Google Scholar]
- Jonk LJ, Itoh S, Heldin C-H, ten Dijke P, Kruijer W (1998) Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer. J Biol Chem 273: 21145–21152 [DOI] [PubMed] [Google Scholar]
- Kadesch T (2000) Notch signaling: a dance of proteins changing partners. Exp Cell Res 260: 1–8 [DOI] [PubMed] [Google Scholar]
- Kao H-Y, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T (1998) A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev 12: 2269–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney JM, Fujisawa-Sehara A, Suda T (1994) Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol 127: 1755–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T (1997) Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development 124: 4133–4141 [DOI] [PubMed] [Google Scholar]
- Korchynskyi O, ten Dijke P (2002) Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem 277: 4883–4891 [DOI] [PubMed] [Google Scholar]
- Korchynskyi O, Dechering KJ, Sijbers A, Olijve W, ten Dijke P (2003) Gene array analysis of bone morphogenetic protein type I receptor-induced osteoblast differentiation. J Bone Miner Res 18: 1177–1185 [DOI] [PubMed] [Google Scholar]
- Kurooka H, Honjo T (2000) Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem 275: 17211–17220 [DOI] [PubMed] [Google Scholar]
- Lane KB, Machado RD, Pauciulo MW, Thomson JR, Phillips JA III, Loyd JE, Nichols, and Trembath (2000) Heterozygous germline mutations in BMPR2, encoding a TGF-β receptor, cause familial primary pulmonary hypertension. Nat Genet 26: 81–84 [DOI] [PubMed] [Google Scholar]
- Leong KG, Hu X, Li L, Noseda M, Larrivée B, Hull C, Hood L, Wong F, Karsan A (2002) Activated Notch4 inhibits angiogenesis: role of β1-integrin activation. Mol Cell Biol 22: 2830–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z-J, Shirakawa T, Li Y, Soma A, Oka M, Dotto GP, Fairman RM, Velazquez OC, Herlyn M (2003) Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol Cell Biol 23: 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier MM, Gessler M (2000) Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem Biophys Res Commun 275: 652–660 [DOI] [PubMed] [Google Scholar]
- Massagué J (1998) TGF-β signal transduction. Annu Rev Biochem 67: 753–791 [DOI] [PubMed] [Google Scholar]
- Minoguchi S, Taniguchi Y, Kato H, Okazaki T, Strobl LJ, Zimber-Strobl U, Bornkamm GW, Honjo T (1997) RBP-L, a transcription factor related to RBP-Jκ. Mol Cell Biol 17: 2679–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Perez SE, Qiao Z, Verdi JM, Hicks C, Weinmaster G, Anderson DJ (2000) Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell 101: 499–510 [DOI] [PubMed] [Google Scholar]
- Nakagawa O, Nakagawa M, Richardson JA, Olson EN, Srivastava D (1999) HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev Biol 216: 72–84 [DOI] [PubMed] [Google Scholar]
- Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin C-H, Miyazono K, ten Dijke P (1997) TGF-β receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J 16: 5353–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, Aster JC, Krishna S, Metzger D, Chambon P, Miele L, Aguet M, Radtke F, Dotto GP (2001) Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J 20: 3427–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena MT, Schroeter EH, Mumm JS, Kopan R (2001) Murine Notch homologs (N1–4) undergo presenilin-dependent proteolysis. J Biol Chem 276: 40268–40273 [DOI] [PubMed] [Google Scholar]
- Shimizu K, Chiba S, Saito T, Takahashi T, Kumano K, Hamada Y, Hirai H (2002) Integrity of intracellular domain of Notch ligand is indispensable for cleavage required for release of the Notch2 intracellular domain. EMBO J 21: 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Kamei CN, Layne MD, Jain MK, Liao JK, Lee M-E, Chin MT (2001) Regulation of myogenic terminal differentiation by the hairy-related transcription factor CHF2. J Biol Chem 276: 18591–18596 [DOI] [PubMed] [Google Scholar]
- Taylor KL, Henderson AM, Hughes CCW (2002) Notch activation during endothelial cell network formation in vitro targets the basic HLH transcription factor HESR-1 and downregulates VEGFR-2/KDR expression. Microvasc Res 64: 372–383 [DOI] [PubMed] [Google Scholar]
- Tezuka K, Yasuda M, Watanabe N, Morimura N, Kuroda K, Miyatani S, Hozumi N (2002) Stimulation of osteoblastic cell differentiation by Notch. J Bone Miner Res 17: 231–239 [DOI] [PubMed] [Google Scholar]
- Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J (1996) Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development 122: 2251–2259 [DOI] [PubMed] [Google Scholar]
- Valdimarsdottir G, Goumans M-J, Rosendahl A, Brugman M, Itoh S, Lebrin F, Sideras P, ten Dijke P (2002) Stimulation of Id1 expression by bone morphogenetic protein is sufficient and necessary for bone morphogenetic protein-induced activation of endothelial cells. Circulation 106: 2263–2270 [DOI] [PubMed] [Google Scholar]
- Volpert OV, Pili R, Sikder HA, Nelius T, Zaichuk T, Morris C, Shiflett CB, Devlin MK, Conant K, Alani RM (2002) Id1 regulates angiogenesis through transcriptional repression of thrombospondin-1. Cancer Cell 2: 473–483 [DOI] [PubMed] [Google Scholar]
- Wang W, Campos AH, Prince CZ, Mou Y, Pollman MJ (2002) Coordinate Notch3-hairy-related transcription factor pathway regulation in response to arterial injury. Mediator role of platelet-derived growth factor and ERK. J Biol Chem 277: 23165–23171 [DOI] [PubMed] [Google Scholar]
- Zhong TP, Rosenberg M, Mohideen M-APK, Weinstein B, Fishman MC (2000) glidlock, an HLHgene required for assembly of the aorta in zebrafish. Science 287: 1820–1824 [DOI] [PubMed] [Google Scholar]