Abstract
In mammals, polyadenylation of mRNA precursors (pre-mRNAs) by poly(A) polymerase (PAP) depends on cleavage and polyadenylation specificity factor (CPSF). CPSF is a multisubunit complex that binds to the canonical AAUAAA hexamer and to U-rich upstream sequence elements on the pre-mRNA, thereby stimulating the otherwise weakly active and nonspecific polymerase to elongate efficiently RNAs containing a poly(A) signal. Based on sequence similarity to the Saccharomyces cerevisiae polyadenylation factor Fip1p, we have identified human Fip1 (hFip1) and found that the protein is an integral subunit of CPSF. hFip1 interacts with PAP and has an arginine-rich RNA-binding motif that preferentially binds to U-rich sequence elements on the pre-mRNA. Recombinant hFip1 is sufficient to stimulate the in vitro polyadenylation activity of PAP in a U-rich element-dependent manner. hFip1, CPSF160 and PAP form a ternary complex in vitro, suggesting that hFip1 and CPSF160 act together in poly(A) site recognition and in cooperative recruitment of PAP to the RNA. These results show that hFip1 significantly contributes to CPSF-mediated stimulation of PAP activity.
Keywords: cleavage and polyadenylation specificity factor, polyadenylation, poly(A) site recognition, pre-mRNA 3′ end processing, upstream sequence elements
Introduction
The majority of eucaryotic mRNA precursors (pre-mRNAs) are processed at the 3′ end by addition of a poly(A) tail. Nuclear pre-mRNA 3′ end formation involves endonucleolytic cleavage at the poly(A) site, followed by polyadenylation of the newly generated 3′ end (reviewed in Zhao et al, 1999; Edmonds, 2002). Several cis-acting sequence elements on the pre-mRNA define the site of polyadenylation. In mammals, the highly conserved AAUAAA hexanucleotide, located about 10–30 nucleotides upstream of the cleavage site, is a core element for both steps of 3′ end processing. G/U- or U-rich downstream elements (DSEs), located about 30 nucleotides downstream of the cleavage site, are involved in the cleavage reaction only. Sequence elements located upstream of the conserved hexanucleotide (upstream sequence elements, USEs) were first identified in viral poly(A) sites, but were subsequently also detected in cellular genes (Moreira et al, 1995; Phillips and Virtanen, 1997; Brackenridge and Proudfoot, 2000; Aissouni et al, 2002). USEs are generally U-rich and were found to be required for full activity of their respective poly(A) site (Prescott and Falck-Pedersen, 1994; Gilmartin et al, 1995; Moreira et al, 1995; Graveley and Gilmartin, 1996; Lutz et al, 1996; Phillips and Virtanen, 1997; Brackenridge and Proudfoot, 2000; Aissouni et al, 2002).
The trans-acting protein factors required for reconstitution of mammalian 3′ end processing in vitro are poly(A) polymerase (PAP), cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), cleavage factors I and II (CF I, CF II), the carboxy-terminal domain of RNA polymerase II and nuclear poly(A)-binding protein 1 (reviewed in Zhao et al, 1999; Edmonds, 2002). The addition of the poly(A) tail to a precleaved pre-mRNA can be reconstituted with PAP and CPSF. PAP, which catalyzes the synthesis of the poly(A) tail at the 3′ end of the RNA, is a template-independent polymerase with high specificity for ATP as substrate. The basal polyadenylation activity of PAP is low, due to its weak and nonspecific binding to RNA (Wahle, 1991). In the presence of CPSF, however, PAP specifically and efficiently polyadenylates RNAs containing a poly(A) signal (Keller et al, 1991; Wahle, 1991). CPSF is a multimeric complex that binds to the AAUAAA signal (Keller et al, 1991) and also to USEs (Gilmartin et al, 1995; Graveley and Gilmartin, 1996; Brackenridge and Proudfoot, 2000). Four major polypeptides copurify with CPSF activity upon fractionation of cellular extracts: CPSF160, CPSF100, CPSF73 and CPSF30 (Bienroth et al, 1991; Murthy and Manley, 1992; Jenny et al, 1994; Jenny and Keller, 1995; Jenny et al, 1996; Barabino et al, 1997). CPSF160 interacts with the AAUAAA hexamer and with PAP (Murthy and Manley, 1995). However, CPSF160 alone is not able to stimulate PAP activity in vitro (Murthy and Manley, 1995), suggesting that additional subunits of CPSF are required for the CPSF-mediated stimulation of PAP.
Most protein factors involved in 3′ end formation are well conserved between mammals and yeast (reviewed in Shatkin and Manley, 2000). One yeast factor lacking a mammalian homolog is Fip1p (factor interacting with PAP), a subunit of CPF. FIP1 is essential and was identified in a two-hybrid screen for proteins interacting with yeast PAP. Extracts prepared from temperature-sensitive fip1 mutants are deficient in in vitro polyadenylation (Preker et al, 1995, 1997). Moreover, it was shown that yeast Fip1p inhibits PAP activity in vitro (Zhelkovsky et al, 1998; Helmling et al, 2001). Based on amino-acid sequence conservation, we have identified the human homolog of Fip1p. We show that human Fip1 (hFip1) is a subunit of CPSF that binds to PAP and to U-rich sequences on the pre-mRNA. hFip1 alone is sufficient to stimulate PAP activity in a U-rich sequence element-dependent manner. hFip1, CPSF160 and PAP form a ternary complex in vitro, suggesting that hFip1 and CPSF160 act together in poly(A) site recognition and in recruitment of PAP to the RNA.
Results
Isolation of a human cDNA encoding a protein with sequence homology to yeast Fip1p
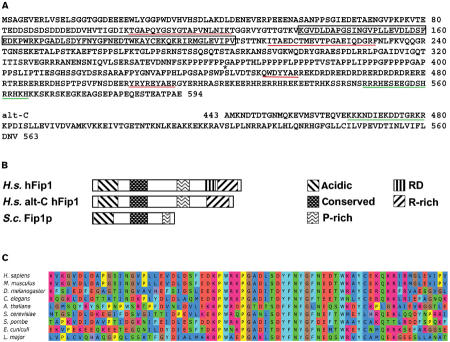
Database searches revealed the existence of several overlapping human ESTs encoding a partial open-reading frame (ORF) with sequence similarity to yeast Fip1p. PCR amplification of missing 5′ sequences on a random primed HeLa cDNA library led to the identification of another overlapping EST from human testis (GenBank accession number AL048910). This cDNA contained a complete ORF encoding a protein of 594 amino acids with a predicted molecular weight of 66 kDa (Figure 1A) that we named hFip1. These sequence data have been submitted to the GenBank database under the accession number AY366510. Fip1 proteins from yeast, mammals and other species share a similar domain organization, with an acidic N-terminus, a highly conserved region in the center of the proteins (48% identity, 72% similarity over 60 amino acids) followed by a proline-rich domain (Figure 1B and C; result not shown). Fip1 homologs from human, mouse, Caenorhabditis elegans and Drosophila melanogaster have a C-terminal extension containing a stretch of alternating arginines and aspartates (RD domain), followed by an arginine-rich region encompassing a predicted bipartite nuclear localization signal (Figure 1A and B; result not shown). The gene encoding hFip1 lies on chromosome 4q12 and is composed of 18 exons. EST analysis and RT–PCR on HeLa cell poly(A)+ RNA revealed different, possibly alternatively spliced forms of hFip1 missing exons 2, 9 or 11 (result not shown). A fourth alternative form of hFip1 missing exon 16 has a different C-terminus, which is also rich in basic amino acids, but does not contain an RD domain (Figure 1A and B, alt-C hFip1).
Figure 1.
hFip1 is homologous to yeast Fip1p. (A) Amino-acid sequence of hFip1. The conserved central region (amino acids 135–204) is framed, tryptic peptides identified by MALDI-TOF MS are underlined in red and predicted bipartite nuclear localization signals are underlined in green. The asterisk indicates the exon 15–16 junction beween amino acids 442 and 443. The C-terminal amino-acid sequence (starting from amino acid 443) of the alternative hFip1 form (alt-C) lacking exon 16 is shown below. (B) Schematic alignment of human hFip1, human hFip1 alt-C and S. cerevisiae Fip1p. The patterns used for the acidic N-terminus, the conserved central region, the proline-rich domain, the mixed charged domain (RD) and the arginine-rich regions are depicted on the right. (C) Multiple sequence alignment of the conserved central region of hFip1 (amino acids 136–204) and homologous proteins identified in the database. Swissprot or Trembl accession numbers are as follows: Homo sapiens: tr∣Q9H077; Mus musculus: tr∣Q9DBB2; D. melanogaster: tr∣Q9VN31; C. elegans: tr∣O16293; A. thaliana: tr∣Q9C834; S. cerevisiae: sp∣P45976; Schizosaccharomyces pombe: sp∣Q09801; Encephalitozoon cuniculi: tr∣Q8SU23; Leishmania major: tr∣Q9BHY5. The alignment was generated with the PILEUP program (GCG package). Amino acids in single letter code are on the colored background. Color code blue indicates hydrophobic, red basic, orange acidic, green stands for hydrophilic residues, yellow background was used with prolines, ochre with glycines, cyan with aromatic and purple with histidines.
hFip1 is a subunit of mammalian CPSF
In order to investigate whether hFip1 is involved in 3′ end processing, we raised a polyclonal antibody against the conserved region of the protein. In a Western blot analysis of HeLa cell nuclear extract, the α-hFip1 antiserum recognized four to five proteins with molecular weights between 65 and 80 kDa, as well as a protein of 95 kDa (Figure 2A, lane 1). The 65–80 kDa proteins were also detected in a Western blot of CPSF purified from HeLa cell nuclear extract, but not in any other purified 3′ end processing factors (Figure 2A, lanes 3–6). Furthermore, the α-hFip1 antiserum recognized proteins with molecular weights between 55 and 65 kDa in CPSF purified from calf thymus (Figure 2A, lane 7). This suggested that hFip1 is a common, so far unrecognized subunit of CPSF. Upon purification of CPSF from HeLa cell nuclear extract over six consecutive columns (Bienroth et al, 1991), the 65–80 kDa proteins recognized by α-hFip1 cofractionated with CPSF in perfectly overlapping peaks throughout all fractionation steps (results not shown) and became enriched in purified CPSF (Figure 2B). In contrast, the 95 kDa protein separated from CPSF upon DEAE chromatography (Figure 2B, compare lanes 2 to 3). On the final gel filtration column, the 65–80 kDa proteins coeluted with CPSF activity and with the four known CPSF subunits (Figure 2C). MALDI-TOF MS peptide fingerprinting of a peak fraction of the gel filtration (fraction 3, Figure 2C) confirmed that the 65–80 kDa proteins were identical to hFip1 (tryptic fragments identified by MALDI-TOF MS are underlined in Figure 1A). These results indicated that hFip1 is stably associated with CPSF. The size heterogeneity of hFip1 could result from alternative splicing, protein modification or proteolysis during fractionation. Recombinant His6-hFip1 had an apparent size of 80 kDa (result not shown), which corresponds to the largest form of hFip1 detectable in CPSF. A C-terminal fragment of hFip1 also showed a heterogeneous size distribution (see Figure 6A, lower panel), suggesting that this part of the protein might be susceptible to proteolysis. The size difference between the human and bovine protein could be due to species-specific isoforms of hFip1. The 95 kDa protein possibly crossreacted nonspecifically with the α-hFip1 antiserum.
Figure 2.
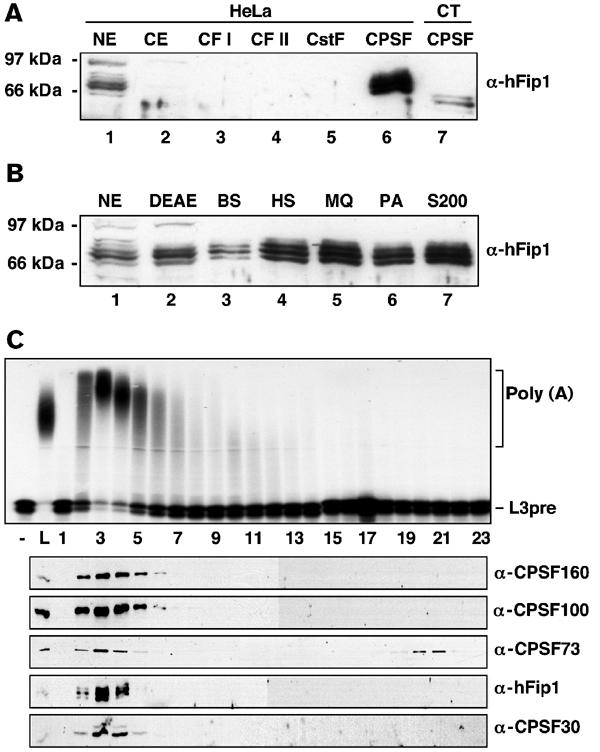
hFip1 is a subunit of CPSF. (A) Western blot analysis of HeLa cell nuclear and cytoplasmic extract and purified 3′ end processing factors. Proteins were separated by 10% SDS–PAGE, blotted onto nitrocellulose and detected with polyclonal α-hFip1 antiserum. NXT and CXT, HeLa cell nuclear and cytoplasmic extract, respectively; CT, calf thymus. (B) Western blot analysis of peak CPSF fractions of each chromatographic step during purification from HeLa cell nuclear extract. Proteins were analyzed as in (A). DEAE, DEAE-sepharose; BS, blue-sepharose; HS, heparin-sepharose; MQ, Mono Q; PA, Poly(A)-sephrose; S200, gel filtration on Superdex 200. (C) Separation of CPSF on an S200 gel filtration column. Individual fractions were tested for in vitro polyadenylation activity (upper panel), and for the presence of CPSF160, CPSF100, CPSF73, CPSF30 and hFip1 by Western blot analysis (lower panels). Polyadenylation assays were carried out with 1 ng recombinant PAP and 3 μl of the load of the column (L) or of the S200 fractions indicated at the bottom of the gel. For Western blot analysis, proteins were separated by SDS–PAGE, blotted onto nitrocellulose and detected with polyclonal antibodies against CPSF160, CPSF100, CPSF73, CPSF30 and hFip1, as indicated.
Figure 6.
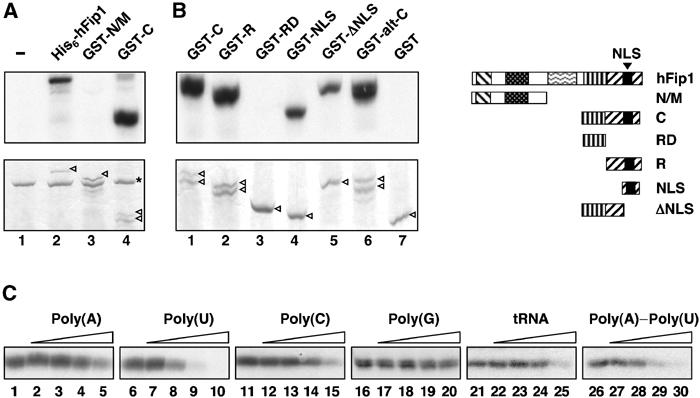
The C-terminal arginine-rich domain of hFip1 binds to RNA. (A) His6-hFip1 (lane 2), GST-N/M (lane 3) or GST-C (lane 4) were UV crosslinked to internally labeled L3 RNA. Lane 1 contains no hFip1. Proteins were separated by 10% SDS–PAGE, stained with Coomassie brilliant blue (lower panel) and visualized by autoradiography (upper panel). The hFip1 polypeptides are indicated by arrowheads in the lower panel, and BSA, which was included in the crosslinking reactions, is indicated by an asterisk. (B) GST-tagged C-terminal fragments of hFip1 were UV crosslinked to internally labeled L3-1 RNA and analyzed as in (A), except that proteins were separated by 12% SDS–PAGE. The hFip1 polypeptides are indicated by arrowheads in the lower panel. The hFip1 fragments tested in (B) are shown schematically on the right, with the same patterns as in Figure 1B, and with the NLS indicated by a black box. GST-C: amino acids (aa) 443–594 (lane 1); GST-R: aa 489–594 (lane 2); GST-RD: aa 455–490 (lane 3); GST-NLS: aa 545–578 (lane 4) and GST-ΔNLS: aa 443–546 (lane 5). GST-alt-C contained amino acids 443–563 of alt-C hFip1 (lane 6). Lane 7: GST alone. (C) Competition UV crosslinking experiments with 300 ng (6 pmol) GST-C, 300 fmol L3pre-RNA and 1 pmol (lanes 2, 7, 12, 17, 22 and 27), 6 pmol (lanes 3, 8, 13, 18, 23 and 28), 45 pmol (lanes 4, 9, 14, 19, 24 and 29) or 300 pmol (lanes 5, 10, 15, 20, 25 and 30) of competitor RNA. Lanes 1, 6, 11, 16 21 and 26 contained no competitor.
To provide more evidence for hFip1 being a component of the mammalian 3′ end formation machinery, we performed co-immunoprecipitation experiments with HeLa cell nuclear extract. The α-hFip1 antiserum co-immunoprecipitated CPSF160, CPSF100 and CPSF73 (Figure 3A, lane 1, upper panel). Likewise, hFip1 was co-immunoprecipitated by antibodies directed against CPSF100 and CPSF30 (lanes 3 and 4, upper panel), but not with antibodies directed against hPcf11, a subunit of CF II (result not shown). This is further evidence that hFip1 is a bona fide subunit of CPSF. Antibodies against hFip1, CPSF100 and CPSF30 also co-immunoprecipitated CstF64 (lanes 1, 3 and 4, lower panel), showing that CPSF and CstF are bound to each other in unfractionated HeLa cell nuclear extract, an observation that has been made before (Takagaki and Manley, 2000).
Figure 3.
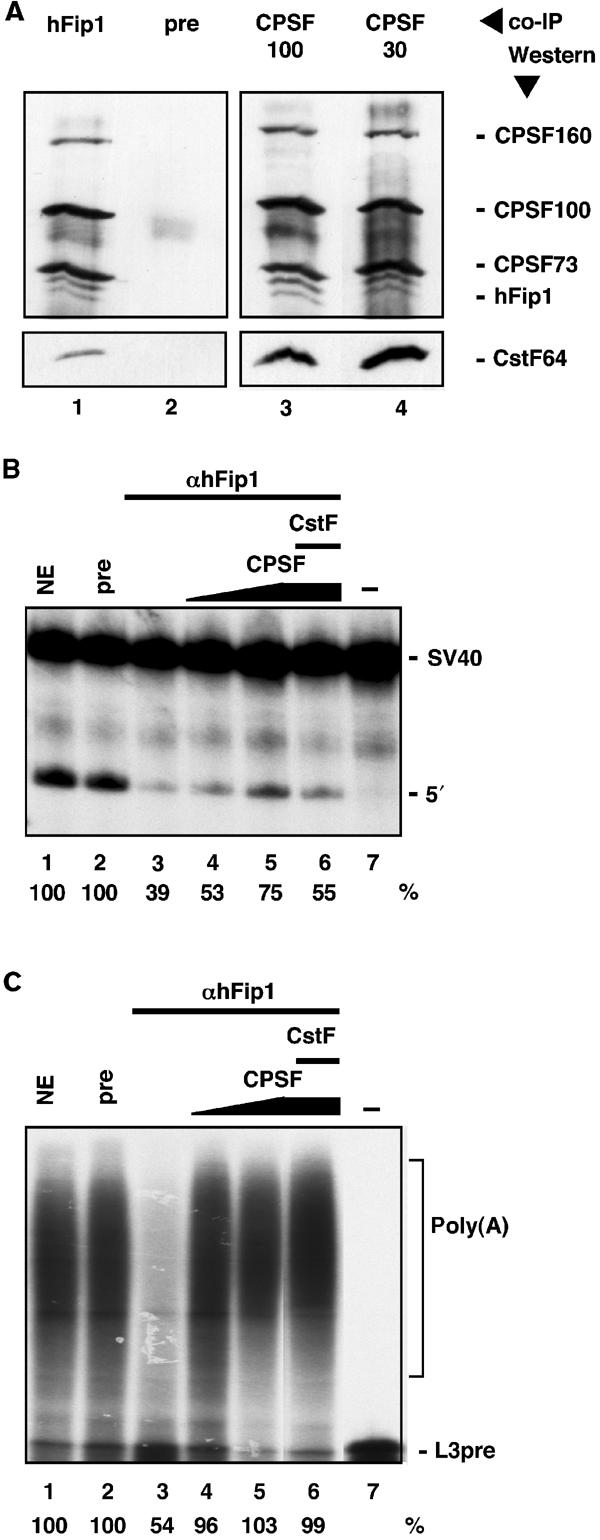
hFip1 is involved in in vitro 3′ end processing. (A) Co-immunoprecipation of hFip1 with CPSF and CstF. HeLa cell nuclear extract was subjected to immunoprecipitation with polyclonal antisera against hFip1 (lane 1), preimmune serum (pre, lane 2), CPSF100 (lane 3) and CPSF30 (lane 4). Bound proteins were detected by Western blotting with an antibody against CstF64 (lower panel) and, after stripping the membrane, with antisera against CPSF160, CPSF100, CPSF73 and hFip1 (upper panel). (B, C) HeLa cell nuclear extract was depleted with preimmune serum (pre, lane 2) or α-hFip1 serum (lanes 3–6) and tested for in vitro cleavage of SV40 (B) and for in vitro polyadenylation of precleaved L3pre (C) RNAs. Depleted extract was complemented with an increasing amount of purified CPSF (lanes 4 and 5), or CPSF and CstF (lane 6). The positions of the unreacted RNA substrate (SV40 and L3pre, lane 7), the 5′ cleavage product (5′) and the polyadenylated RNA (poly(A)) are indicated. Processing activities, as determined by accumulation of cleavage or polyadenylated product, respectively, are given in percentages below the panels. The activity of the untreated extact (NE, lane 1) was set at 100%.
Immunodepletion of HeLa cell nuclear extract with α-hFip1 reduces in vitro cleavage and polyadenylation activity
HeLa cell nuclear extract was depleted with α-hFip1 antiserum and tested for in vitro cleavage of SV40 and polyadenylation of precleaved L3pre-mRNAs. α-hFip1-depleted extract was reduced in cleavage and in polyadenylation (Figure 3B and C, lane 3). Cleavage activity was partially restored by adding back purified CPSF (Figure 3B, lanes 4 and 5). The addition of CstF on top did not increase processing efficiency (Figure 3B, lane 6). In contrast, polyadenylation activity was efficiently restored upon supplying the extract with purified CPSF (Figure 3C, lanes 4 and 5). Taken together, these results indicate that hFip1 participates in pre-mRNA 3′ end processing.
hFip1 interacts with PAP, CPSF30, CPSF160 and CstF77
To identify proteins interacting with hFip1 and to map interaction domains, we carried out GST pull-down experiments with fragments of hFip1 expressed as GST fusion proteins in Escherichia coli and in vitro translated and 35S-labeled 3′ end processing factor subunits. Full-length GST-hFip1 was not included in this experiment, since we were not able to express it in sufficient amounts. RNase A was added to the reactions to avoid indirect interactions mediated by RNA. As shown in Figure 4A, hFip1 interacted with PAP, CPSF30, CPSF160 and CstF77. PAP bound to the acidic N-terminus (Figure 4A, lane 3), CPSF30 to the highly conserved domain (lane 4), and CPSF160 and CstF77 bound to a fragment spanning the entire N-terminal region of hFip1 (lane 2). In addition, CPSF160 also interacted with the C-terminus of hFip1 (lane 5). No binding could be detected between any of the hFip1 fragments and CPSF100, CPSF73, hClp1, CFI 68 and PABPN1 (results not shown). However, we cannot rule out further interactions, possibly involving the proline-rich domain of hFip1 or the full-length protein.
Figure 4.
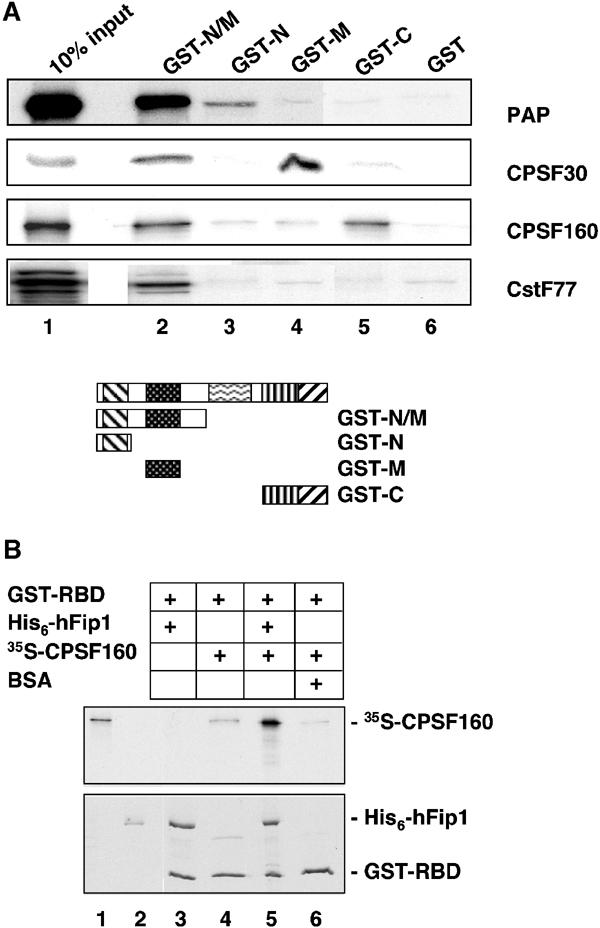
Interactions between hFip1 and other 3′ end processing components. (A) GST pull-down experiment with GST-tagged hFip1 fragments and in vitro translated 35S-labeled polyadenylation factor subunits, as indicated on the right of the panel. The GST-tagged hFip1 fragments used in this experiment are depicted schematically below the full-length protein, with the same patterns as in Figure 1B. GST-N/M, amino acids 1–355 (lane 2); GST-N, amino acids 1–111 (lane 3); GST-M, amino acids 137–243 (lane 4); GST-C, amino acids 443–594 (lane 5). Lane 6 contained GST alone. Lane 1 shows 10% of the input of the in vitro translated proteins. (B) GST pull-down experiment with GST-tagged RNA-binding domain of PAP (GST-RBD, amino acids 366–513, lanes 3–6), recombinant purified His6-hFip1 (lanes 3 and 5) and in vitro translated 35S-labeled CPSF160 (lanes 4–6). Lane 6 contained BSA instead of His6-hFip1. Lanes 1 and 2 show 10% of the input of in vitro translated CPSF160 or His6-hFip1, respectively. RNase A was added to the reactions to rule out RNA-mediated effects. Proteins were separated by SDS–PAGE, stained with Coomassie brilliant blue (lower panel) and visualized by autoradiography (upper panel).
hFip1, CPSF160 and PAP form a ternary complex in vitro
In order to map the binding site of hFip1 on PAP, we performed pull-down experiments with GST fragments of PAP and hFip1. Baculovirus-expressed His6-hFip1 bound to an N-terminal fragment of PAP (amino acids 53–214; result not shown) and to the RNA-binding domain of PAP (GST-RBD, amino acids 366–513, Figure 4B, lane 3). CPSF160 binds to PAP as well (Murthy and Manley, 1995), and this interaction also involves the RBD of PAP (Figure 4B, lane 4). In order to know whether hFip1 and CPSF160 were mutually influencing their binding to PAP, we added all three proteins to a GST pull-down reaction. Under these conditions, both hFip1 and CPSF160 were retained in a complex with GST-RBD (Figure 4B, lane 5), showing that PAP, hFip1 and CPSF160 can interact simultaneously with each other.
hFip1 stimulates the in vitro polyadenylation activity of PAP
Native CPSF stimulates the activity of PAP. Since we found that hFip1 is associated with CPSF and interacts with PAP, we tested the effect of hFip1 on PAP activity in in vitro polyadenylation reactions. To this end, recombinant PAP was incubated together with recombinant hFip1 and precleaved L3pre-RNA in the presence of Mg2+. In order to minimize the reassociation of PAP with the same primer molecules, we used a 30-fold molar excess of RNA over PAP. Under these conditions, the activity of PAP is low due to its low affinity for RNA (Wahle, 1991). As expected, PAP alone elongated only a small fraction of the primer with a few adenosines (Figure 5, lanes 1–7). In the presence of hFip1, poly(A) synthesis by PAP was stimulated (Figure 5, lanes 8–14). The length of the poly(A) tails, as well as the amount of precursor elongated, increased gradually with time. After 30 min, a fraction of the RNA primer was elongated to heterogeneous lengths between 80 and 600 nucleotides. In the presence of purified CPSF though, the majority of precursor RNA was polyadenylated to a length between 300 and 600 adenosines (Figure 5, lanes 22–28). Most importantly, however, poly(A) synthesis in the presence of recombinant hFip1 did not require an AAUAAA signal, because an RNA carrying a U to G mutation in the consensus hexamer was elongated as efficiently as wild-type RNA (Figure 5, lanes 29–35). Stimulation of polyadenylation by recombinant hFip1 was also observed with other RNAs (SV40, human C2), and in the presence of Mn2+ instead of Mg2+ (results not shown). Stimulation of polyadenylation was dependent on the amount of hFip1 added and required at least equimolar amounts of hFip1 and RNA. When no PAP was added to the reaction, poly(A) synthesis was not detectable, indicating that the stimulation was not caused by contaminating PAP (results not shown). Thus, hFip1 alone was able to stimulate PAP activity, although not with the same efficiency and specificity as native CPSF.
Figure 5.
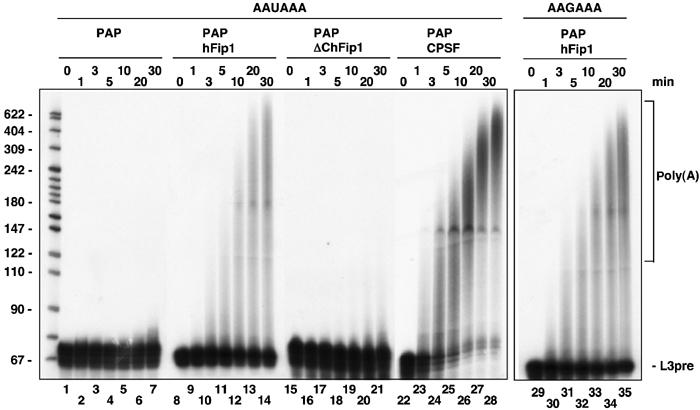
hFip1 stimulates the polyadenylation activity of PAP. Internally labeled RNA (60 fmol) was incubated with 2 fmol His6-PAP either alone (lanes 1–7) or together with 600 fmol His6-hFip1 (lanes 8–14 and 29–35), 600 fmol His6-ΔChFip1 (lacking amino acids 456–594; lanes 15–21) or 25 fmol purified CPSF (lanes 22–28) in the presence of 2 mM Mg2+. At the time points indicated on top of the panel, the reactions were stopped and the samples were analyzed on a denaturing 10% polyacrylamide gel. Lanes 1–28: AAUAAA-L3pre; Lanes 29–35: AAGAAA-L3pre. The positions of the unreacted RNA substrate (L3pre) and of the polyadenylated RNA (poly(A)) are indicated on the right, and the size of DNA markers in nucleotides is shown on the left.
hFip1 contains a C-terminal extension rich in charged amino acids that is missing in Saccharomyces cerevisiae Fip1p. A truncated form of hFip1 lacking the last 138 amino acids (ΔC-hFip1) was no longer able to stimulate the activity of PAP. After 30 min, only a small fraction of the primer was elongated with few adenosines as in the absence of hFip1 (Figure 5, lanes 15–21). This indicates that the C-terminal domain of hFip1 is necessary for stimulation of polyadenylation activity. A fragment of hFip1 containing the C-terminus only was not sufficient to stimulate PAP (result not shown), suggesting that also the N-terminal part of hFip1, which harbors the PAP-binding site, was required. The extreme C-terminus of mammalian PAP has been implicated in the modulation of polyadenylation activity by trans-acting proteins (Gunderson et al, 1994; Gunderson et al, 1998). A PAP deletion mutant lacking this domain (Martin and Keller, 1996) could still be stimulated by hFip1, although the resulting poly(A) tails were slightly shorter than those generated with full-length PAP (result not shown). Therefore, the C-terminus of PAP was not needed for the stimulation by hFip1.
The C-terminal arginine-rich domain of hFip1 binds to RNA
One possible mechanism by which PAP could be stimulated by hFip1 is by recruitment to the RNA. This assumption prompted us to investigate the RNA-binding properties of hFip1 with UV crosslinking experiments. Indeed, full-length His6-hFip1 could be crosslinked to L3 RNA (Figure 6A, lane 2). The same result was obtained with a C-terminal GST fragment of hFip1, but not with an N-terminal fragment (Figure 6A, lanes 4 and 3, respectively), indicating that the RNA-binding activity of hFip1 is located within the C-terminus of the protein. To define more precisely the RNA-binding domain of hFip1, further C-terminal deletions of hFip1 were tested in UV crosslinking experiments. A fragment containing only the arginine-rich domain still bound to RNA (Figure 6B, lane 2), whereas the RD domain did not (lane 3). A shorter fragment of the arginine-rich region containing the NLS was sufficient for crosslinking to RNA (lane 4). However, deletion of this region decreased RNA binding only weakly (lane 5), indicating that the entire arginine-rich region of hFip1 contributes to RNA binding. Interestingly, the C-terminus of the alternative hFip1 form (hFip1 alt-C), which is also rich in arginines and lysines (see Figure 1A), could be crosslinked to RNA too (Figure 6B, lane 6).
hFip1 binds to U-rich RNA sequences
hFip1 crosslinked strongly to L3 and SV40 RNAs and less efficiently to L3pre-RNA (result not shown). The apparent KD values for these interactions were 2 nM for L3 and 4 nM for L3pre, as determined with filter-binding experiments (results not shown). To further characterize the RNA-binding properties of hFip1, we performed competition UV crosslinking experiments with radiolabeled L3pre and RNA homopolymers as competitors. Binding of hFip1 to L3pre was most efficiently competed by single-stranded poly(U) and to a minor extent by double-stranded poly(A)–poly(U) (Figure 6C, lanes 6–10 and 26–30). In contrast, single-stranded poly(A), poly(C), poly(G) and tRNA were less efficient (Figure 6C, lanes 1–5, 11–15, 16–20 and 21–25). Therefore, hFip1 appeared to have a preference for poly-uridines, both in single- and double-stranded RNA. However, we cannot rule out that poly(A)–poly(U) also contained partially single-stranded poly(U).
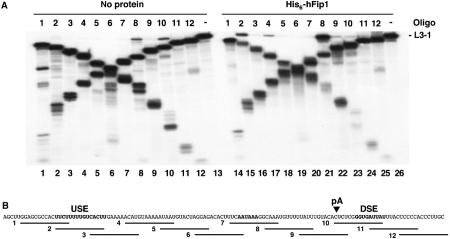
In order to locate the binding site of hFip1 on the polyadenylation substrates, we performed RNase H footprinting experiments (Dichtl and Keller, 2001). Internally labeled L3-1 RNA was preincubated either alone or in the presence of His6-hFip1. After preincubation, DNA oligonucleotides complementary to the RNA (oligos 1–12, see Figure 7B) and RNase H were added. In the absence of protein, the RNA was accessible to the oligonucleotides, leading to the formation of RNA–DNA hybrids, which were degraded by RNase H (Figure 7A, lanes 1–12). Oligos 8 and 10 (lanes 8 and 10) were slightly less efficient, as can be seen from residual amounts of full-length L3-1 RNA, which could be due to the secondary structure of RNA. In contrast, preincubation of RNA with hFip1 resulted in protection of the regions complementary to oligos 2 and 8 from RNase H degradation, as deduced from increased amounts of full-length L3-1 RNA (Figure 7A, lanes 15 and 21). This indicates that hFip1 binds to the USE and to the region between the AAUAAA and the poly(A) site. Both regions are particularly rich in uridines (Figure 7B). The same result was obtained with C-terminal fragments of hFip1 (GST-C, GST-R; results not shown). Weak protection was also observed with oligos 4, 9 and 11 (Figure 7A, lanes 17, 22 and 24). However, these results showed some variation between different experiments. The region complementary to oligo 10 became more accessible to RNase H-dependent cleavage in the presence of hFip1 (compare lanes 10 and 23). This suggests that binding of hFip1 might also alter the secondary structure and therefore the accessibility of the oligonucleotides to the RNA. However, the two major regions protected by hFip1, the USE and the region between the AAUAAA and the poly(A) site, are consistent with the preferential binding of hFip1 to U-rich sequences. Interestingly, both these sequence elements have also been shown to be required for efficient 3′ end processing of L3pre-mRNA in vivo (Prescott and Falck-Pedersen, 1994).
Figure 7.
hFip1 binds to U-rich RNA sequence elements. (A) RNase H protection analysis with His6-hFip1 and L3-1 RNA. Internally labeled L3-1 RNA was preincubated without (lanes 1–13) or with 1 μg of His6-hFip1 (lanes 14–26). The oligos used for protection are indicated on top of the gel. The reactions in lanes 13 and 26 contained no oligo and show the uncleaved L3-1 RNA. (B) Nucleotide sequence of L3-1 RNA showing the annealing positions of the 14-mer antisense DNA oligos. The USE, the AAUAAA signal and the DSE are shown in bold and the arrow marks the poly(A) site (pA).
U-rich sequence elements increase hFip1-mediated stimulation of PAP
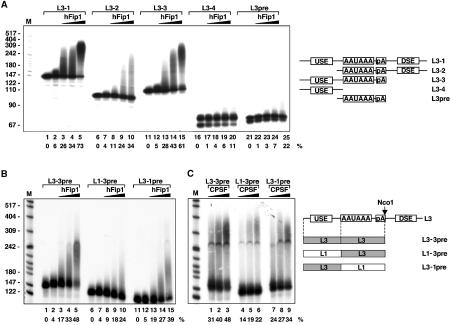
To investigate whether U-rich sequence elements contributed to the hFip1-mediated stimulation of PAP, we prepared RNAs containing different combinations of poly(A) signals, as shown schematically in Figure 8A. These RNAs were tested in in vitro polyadenylation experiments containing PAP and increasing amounts of hFip1. hFip1 stimulated polyadenylation of L3pre, which contains the AAUAAA signal and the U-rich poly(A) site (Figure 8A, lanes 23–25). L3-1 RNA, which contains in addition a USE and a DSE, was more efficiently polyadenylated in the presence of hFip1 (lanes 3–5). Deletion of the USE (L3-2 RNA) resulted in a decrease of hFip1-mediated stimulation of PAP activity (lanes 8–10), whereas deletion of the DSE (L3-3 RNA) had little or no effect (lanes 13–15). Therefore, the USE appears to be a critical determinant of the hFip1-mediated stimulation of PAP activity. However, an RNA containing only the USE (L3-4) was not efficiently polyadenylated (lanes 18–20). This result suggests that both sequence elements, the USE and the region surrounding the poly(A) site, contributed to most efficient hFip1-dependent stimulation of PAP activity. This assumption was further tested with chimeric L3/L1 RNAs carrying sequences from the inefficient L1 poly(A) site (Prescott and Falck-Pedersen, 1994). Stimulation of polyadenylation by hFip1 was efficient with L3-3pre, which contains both binding sites of hFip1 (Figure 8B, lanes 3–5). Replacing the USE of L3 with corresponding sequences of the L1 poly(A) site, which lacks an USE (L1-3pre), reduced stimulation by hFip1 (lanes 8–10). Exchanging the U-rich region surrounding the poly(A) site with corresponding sequences from L1 (L3-1pre) also resulted in a decrease of hFip1-dependent stimulation of PAP (lanes 13–15). Similar effects were observed with purified CPSF (Figure 8C). However, under these conditions, the poly(A) tails were longer compared to those seen in the presence of hFip1, indicating that polyadenylation was more efficient with native CPSF. In summary, these results show that both U-rich sequence elements contribute to stimulation of polyadenylation by hFip1. Moreover, they indicate that the sequence dependence of hFip1 could also be observed with purified CPSF. However, native CPSF is more efficient than hFip1 alone and is in addition specific for the AAUAAA signal.
Figure 8.
Stimulation of PAP activity by hFip1 depends on U-rich sequence elements. (A) In all, 60 fmol RNA and 2 fmol His6-PAP were incubated either alone (lanes 2, 7, 12, 17 and 22) or together with 60 fmol His6-hFip1 (lanes 3, 8, 13, 18 and 23), 200 fmol His6-hFip1 (lanes 4, 9, 14, 19 and 24) or 600 fmol His6-hFip1 (lanes 5, 10, 15, 20 and 25) for 30 min. Lanes 1, 6, 11, 16 and 21 show the RNA incubated in the absence of proteins. The RNAs used in this experiment are derivatives of L3 (Humphrey et al, 1987) and are depicted schematically on the right of the panel. pA stands for the poly(A) site. Analysis of the samples and labeling are as in Figure 6. The extent of polyadenylation activities in percentages is given below the panel. (B) As in (A), but with L3-3pre-, L1-3pre- or L3-1pre-RNAs (Prescott and Falck-Pedersen, 1994), as shown schematically on the right of the panel. Sequences derived from L3 poly(A) sites are depicted in gray, and sequences derived from L1 in white. (C) In all, 2 fmol His6-PAP and 60 fmol RNA were incubated with 10 fmol (lanes 1, 4 and 7), 20 fmol (lanes 2, 5 and 8) or 40 fmol (lanes 3, 6 and 9) purified CPSF for 30 min. RNAs are as in (B).
Discussion
We have isolated the human homolog of S. cerevisiae Fip1p and have found that hFip1 is a genuine subunit of CPSF. Our results reported here indicate that hFip1 contributes to poly(A) site recognition and to CPSF-dependent stimulation of polyadenylation. hFip1 binds to U-rich sequence elements on the pre-mRNA and is able to stimulate the activity of PAP.
CPSF has originally been described as a tetrameric complex containing the subunits CPSF160, CPSF100, CPSF73 and CPSF30, based on their stoichiometric cofractionation with CPSF activity. These polypeptides are related to four subunits of the yeast polyadenylation factor CPF (Yhh1p, Ydh1p, Ysh1p and Yth1p). However, CPF contains additional subunits (Preker et al, 1997; Dichtl et al, 2002a; Gavin et al, 2002; Walsh et al, 2002), one of which is Fip1p. Based on sequence conservation, we have identified the human homolog of yeast Fip1p and have shown that hFip1 is an additional, so far unrecognized subunit of CPSF. Most likely, hFip1 has not been detected before because of its substoichiometric appearance upon gel electrophoresis of purified CPSF. This could be either because hFip1 is poorly stained or because it is present in nonequimolar amounts compared to the other subunits. In addition, the size heterogeneity of hFip1 might have made it difficult to identify it as a distinct component of CPSF. Yeast CPF contains further subunits whose mammalian homologs have not been identified. For most of these, a metazoan protein with sequence similarity can be found in the databases, and it is likely that these proteins are also components of the 3′ end processing machinery. Accordingly, it was shown recently that the Arabidopsis thaliana protein FY, which is homologous to yeast Pfs2p, is involved in 3′ end formation in plants (Simpson et al, 2003).
CPSF confers specificity and efficiency to PAP. Earlier studies have shown that CPSF160 binds to PAP and to the AAUAAA signal (Keller et al, 1991; Murthy and Manley, 1995). However, CPSF160 alone binds only inefficiently to RNA and is not able to stimulate PAP activity in vitro (Murthy and Manley, 1995). This suggests that additional polypeptides of CPSF are involved in mediating the function of native CPSF (Murthy and Manley, 1995). We found that hFip1 was able to stimulate PAP activity in a U-rich element-dependent manner. A critical determinant of this stimulation is the C-terminus of hFip1, which binds to RNA. Therefore, the hFip1-mediated stimulation of polyadenylation may result from recruiting the polymerase to the RNA. hFip1 also interacts with CPSF160, and together with PAP these proteins form a ternary complex in vitro. This possibly contributes to cooperative protein–protein and protein–RNA interactions in the assembly of the 3′ end processing complex. However, neither hFip1 and CPSF160 together, nor in combination with recombinant CPSF100, CPSF73 and CPSF30, were sufficient for reconstitution of AAUAAA-dependent polyadenylation with PAP in vitro (I Kaufmann, S Dettwiler and W Keller, unpublished result). Therefore, additional proteins might be required to modulate or complement the activities of these CPSF subunits.
hFip1 bound to the USE and the U-rich region surrounding the poly(A) site on L3-1 RNA. The presence of both of these sequence elements also led to most efficient hFip1-dependent stimulation of PAP activity. In accordance, both elements are required for efficient 3′ end formation of L3pre-mRNA in vivo (Prescott and Falck-Pedersen, 1994). Interestingly, the complement C2 and lamin B2 genes also have in addition to the USE a U-rich element surrounding the poly(A) site (Moreira et al, 1995; Brackenridge and Proudfoot, 2000).
USEs were originally identified in viruses, but were subsequently also found in cellular genes (Moreira et al, 1995; Phillips and Virtanen, 1997; Brackenridge and Proudfoot, 2000; Aissouni et al, 2002). A recent computational survey detected a significant accumulation of uridines upstream of the AAUAAA signal in a large number of human genes (Legendre and Gautheret, 2003), indicating that USEs are a common feature of human 3′ end processing signals. It is assumed that poly(A) site definition involves multiple sequence elements in addition to the AAUAAA hexamer, which are recognized by individual 3′ end processing proteins. Hence, the strength of a poly(A) site is the sum of these interactions. It has been shown previously that the USEs of HIV-1, adenovirus EIA and human lamin B2 pre-mRNAs increase the stability of the CPSF–RNA complex (Gilmartin et al, 1995; Graveley and Gilmartin, 1996; Brackenridge and Proudfoot, 2000). Moreover, the presence of a USE in HIV-1 and L3pre-RNAs enhances the efficiency of CPSF-dependent polyadenylation in vitro (Gilmartin et al, 1995, and this study). Our results suggest that hFip1 participates in mediating these USE-dependent stimulatory effects of CPSF. The USEs of the SV40 and of the human C2 genes are recognized by U1A protein or polypyrimidine tract-binding protein, respectively (Lutz and Alwine, 1994; Moreira et al, 1998). hFip1 also stimulated polyadenylation of these pre-mRNAs (I.K. and W.K., result not shown). Further experiments are required to test whether hFip1 also binds to the USEs of these and possibly other transcripts. Moreover, it remains to be shown whether the alternative form of hFip1, which has a different C-terminal RNA-binding domain, has the same RNA preference. It is conceivable that the two hFip1 forms regulate 3′ end processing efficiency of distinct pre-mRNAs.
The RNA-binding activity of hFip1 lies within the arginine-rich C-terminus of the protein. Arginine-rich RNA-binding motifs (ARMs) are found in a variety of RNA-binding proteins, including the antiterminator protein N of bacteriophage λ and the trans-activator protein Tat of HIV-1 (Burd and Dreyfuss, 1994). Interestingly, binding of HIV-1 Tat and λN to their target sites on the nascent transcript ultimately results in an increase of processivity or in a termination-resistant form of the transcribing polymerase (Friedman and Court, 1995; Karn, 1999). ARMs have originally been described as small domains with a core of six to eight basic amino acids, typically arginines, flanked by nonbasic polar or charged amino acids (Lazinski et al, 1989). Nucleic acid-binding domains often overlap with NLSs (LaCasse and Lefebvre, 1995). Consistently, a partial fragment of the arginine-rich domain of hFip1 containing just the NLS was sufficient for RNA binding. However, deletion of the NLS only slightly impaired interaction with the RNA, suggesting that the entire arginine-rich domain of hFip1 contributes to RNA binding and possibly contains multiple independent ARMs. ARMs often recognize a particular RNA structure rather than a sequence (Burd and Dreyfuss, 1994). Structural elements have also been proposed to be critical determinants of poly(A) site recognition (Graveley et al, 1996; Klasens et al, 1999; Hans and Alwine, 2000). Accordingly, the RNA target of hFip1 could acquire a specific conformation.
The observation that Fip1 regulates PAP activity has also been reported with yeast Fip1p. However, it was shown that yeast Fip1p inhibits the activity of Pap1p in vitro (Zhelkovsky et al, 1998; Helmling et al, 2001). The different effect of the human and the yeast protein on their respective polymerases could be explained by the C-terminal RNA-binding domain of hFip1, which is absent in yeast Fip1p. The hFip1-mediated recruitment of PAP to the RNA could be a specific feature of polyadenylation in higher eucaryotes. In yeast, this might be performed by other CPF subunits like Yhh1p, Ydh1p and Yth1p, which have been shown to bind to U-rich sequences surrounding the poly(A) site (Barabino et al, 2000; Dichtl and Keller, 2001, 2002b). Importantly, however, purified CPF, which contains Fip1p, stimulates the polyadenylation activity of Pap1p in vitro (Preker et al, 1997) and recombinant Fip1p restores the polyadenylation defect of a fip1 mutant extract (Helmling et al, 2001), suggesting that in the presence of other CPF subunits yeast Fip1p can also have a positive effect on Pap1p.
The gene encoding hFip1 lies on chromosome 4q12 upstream of the PDGFRα gene. It has recently been reported that a genetic rearrangement, caused by the chromosomal deletion of 800 kb and fusion of the hFip1 and PDGFRα genes, might be the cause of idiopathic hypereosinophilic syndrome, a severe hematologic disorder (Cools et al, 2003; Griffin et al, 2003). The corresponding chimeric protein, hFip1-PDGFRα, contained the N-terminus of hFip1 and the C-terminal kinase domain of PDGFRα. Expression of the hFip1-PDGFRα fusion protein in hematopoietic cells resulted in constitutive kinase activity and transformation of the cells. Fusion of amino acids 1–29 of hFip1 to PDGFRα was found to be sufficient for kinase activation and transformation (Cools et al, 2003). The N-terminal domain of hFip1 could mediate growth factor-independent dimerization and thus activation of the fusion protein, or alternatively could directly stimulate the kinase domain. Further analysis will be required to evaluate these possibilities.
Materials and methods
Isolation of hFip1 cDNAs, plasmids and buffers used in this study, expression of recombinant proteins, generation of polyclonal hFip1 antiserum, purification of CPSF from HeLa cell nuclear extract and MALDI-TOF MS analysis
See Supplementary data.
Co-immunoprecipitations and immunodepletions of HeLa cell nuclear extract
Co-immunoprecipitations and immunodepletions with HeLa cell nuclear extract were performed as described (Barabino et al, 1997).
Preparation of RNA substrates
L3pre was prepared from pSP6L3pre (Humphrey et al, 1987). pGEM3L3-1 and pGEM3L3-2 (nucleotides 111–234 or 151–234 of L3 RNA) are PCR derivatives of pSP6L3. L3-1 and L3-2 were in vitro transcribed from EcoR1-linearized pGEM3L3-1 and pGEM3L3-2, respectively. L3-3 and L3-4 were in vitro transcribed from pGEM3L3-1 linearized with Ava1 or Rsa1, respectively. L3-3pre, L1-3pre and L3-1pre were in vitro transcribed from Nco1-linearized pGEM2L333, pGEM2L133 or pGEM2L313, respectively (Prescott and Falck-Pedersen, 1994).
Cleavage and polyadenylation assays
In vitro cleavage, polyadenylation and time-course polyadenylation experiments were carried out as described (Bienroth et al, 1991, 1993; Rüegsegger et al, 1996).
Protein–protein interactions
In vitro translation was performed with a TNT coupled transcription–translation rabbit reticulocyte system (Promega). In all, 1 μg purified GST protein and 2–5 μl of the in vitro translation reaction were incubated at 25°C for 1 h in binding buffer containing 0.1 mg/ml RNase A. Proteins were bound to Glutathione Sepharose 4B (Pharmacia), washed four times with IPP150 buffer containing 0.1 mg/ml RNase A, eluted with SDS sample buffer and analyzed by SDS–PAGE and autoradiography.
Protein–RNA interactions
UV crosslinking experiments were performed with 500 ng protein and 300 fmol RNA in XL buffer as described (Martin and Keller, 1996). For competition experiments, six-fold reactions were preincubated at 25°C for 5 min. After addition of competitor E. coli tRNA, poly(A) (Boehringer), poly(U) (Pharmacia), poly(C), poly(G) or poly(A)–poly(U) (Sigma), reactions were further incubated for 5 min and crosslinked. RNase H protection experiments were performed as described (Dichtl and Keller, 2001).
Supplementary Material
Supplementary Material
Acknowledgments
We thank Sabine Dettwiler for pGEM3L3-1 and pGEM3L3-2, John Prescott and Erik Falck-Pedersen for pGEM2L333, pGEM2L133, pGEM2L313, Alexandra Moreira and Nick Proudfoot for pGEMC2 and Clinton MacDonald for the anti-CstF64 antibody. We are grateful to Bernhard Dichtl for helpful discussions and critically reading the manuscript. This work was supported by the University of Basel, the Swiss National Science Fund and the Louis-Jeantet Foundation for Medicine.
References
- Aissouni Y, Perez C, Calmels B, Benech PD (2002) The cleavage/polyadenylation activity triggered by a U-rich motif sequence is differently required depending on the poly(A) site location at either the first or last 3′-terminal exon of the 2′-5′ oligo(A) synthetase gene. J Biol Chem 277: 35808–35814 [DOI] [PubMed] [Google Scholar]
- Barabino SM, Hübner W, Jenny A, Minvielle-Sebastia L, Keller W (1997) The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev 11: 1703–1716 [DOI] [PubMed] [Google Scholar]
- Barabino SM, Ohnacker M, Keller W (2000) Distinct roles of two Yth1p domains in 3′ end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J 19: 3778–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienroth S, Keller W, Wahle E (1993) Assembly of a processive messenger RNA polyadenylation complex. EMBO J 12: 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienroth S, Wahle E, Suter-Crazzolara C, Keller W (1991) Purification of the cleavage and polyadenylation factor involved in the 3′ processing of messenger RNA precursors. J Biol Chem 266: 19768–19776 [PubMed] [Google Scholar]
- Brackenridge S, Proudfoot NJ (2000) Recruitment of a basal polyadenylation factor by the upstream sequence element of the human lamin B2 polyadenylation signal. Mol Cell Biol 20: 2660–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science 265: 615–621 [DOI] [PubMed] [Google Scholar]
- Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, Kutok J, Clark J, Galinsky I, Griffin JD, Cross NC, Tefferi A, Malone J, Alam R, Schrier SL, Schmid J, Rose M, Vandenberghe P, Verhoef G, Boogaerts M, Wlodarska I, Kantarjian H, Marynen P, Coutre SE, Stone R, Gilliland DG (2003) A tyrosine kinase created by fusion of the PDGFRa and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med 348: 1201–1214 [DOI] [PubMed] [Google Scholar]
- Dichtl B, Blank D, Ohnacker M, Friedlein A, Roeder D, Langen H, Keller W (2002a) A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol Cell 10: 1139–1150 [DOI] [PubMed] [Google Scholar]
- Dichtl B, Blank D, Sadowski M, Hübner W, Weiser S, Keller W (2002b) Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J 21: 4125–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Keller W (2001) Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J 20: 3197–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds M (2002) A history of poly A sequences: from formation to factors to function. Prog Nucleic Acid Res Mol Biol 71: 285–389 [DOI] [PubMed] [Google Scholar]
- Friedman DI, Court DL (1995) Transcription antitermination: the lambda paradigm updated. Mol Microbiol 18: 191–200 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch V, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edlemann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Gilmartin GM, Fleming ES, Oetjen J, Graveley BR (1995) CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev 9: 72–83 [DOI] [PubMed] [Google Scholar]
- Graveley BR, Fleming ES, Gilmartin GM (1996) RNA structure is a critical determinant of poly(A) site recognition by cleavage and polyadenylation specificity factor. Mol Cell Biol 16: 4942–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Gilmartin GM (1996) A common mechanism for the enhancement of mRNA 3′ processing by U3 sequences in two distantly related lentiviruses. J Virol 70: 1612–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JH, Leung J, Bruner RJ, Caligiuri MA, Briesewitz R (2003) Discovery of a fusion kinase in EOL-1 cells and idiopathic hypereosinophilic syndrome. Proc Natl Acad Sci USA 100: 7830–7835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson SI, Beyer K, Martin G, Keller W, Boelens WC, Mattaj IW (1994) The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell 76: 531–541 [DOI] [PubMed] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, Mattaj IW (1998) U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70 K and poly(A) polymerase. Mol Cell 1: 255–264 [DOI] [PubMed] [Google Scholar]
- Hans H, Alwine JC (2000) Functionally significant secondary structure of the simian virus 40 late polyadenylation signal. Mol Cell Biol 20: 2926–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmling S, Zhelkovsky A, Moore CL (2001) Fip1 regulates the activity of poly(A) polymerase through multiple interactions. Mol Cell Biol 21: 2026–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey T, Christofori G, Lucijanic V, Keller W (1987) Cleavage and polyadenylation of messenger RNA precursors in vitro occurs within large and specific 3′ processing complexes. EMBO J 6: 4159–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Hauri HP, Keller W (1994) Characterization of cleavage and polyadenylation specificity factor and cloning of its 100-kilodalton subunit. Mol Cell Biol 14: 8183–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Keller W (1995) Cloning of cDNAs encoding the 160 kDa subunit of the bovine cleavage and polyadenylation specificity factor. Nucleic Acids Res 23: 2629–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A, Minvielle-Sebastia L, Preker PJ, Keller W (1996) Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science 274: 1514–1517 [DOI] [PubMed] [Google Scholar]
- Karn J (1999) Tackling Tat. J Mol Biol 293: 235–254 [DOI] [PubMed] [Google Scholar]
- Keller W, Bienroth S, Lang KM, Christofori G (1991) Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J 10: 4241–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasens BI, Thiesen M, Virtanen A, Berkhout B (1999) The ability of the HIV-1 AAUAAA signal to bind polyadenylation factors is controlled by local RNA structure. Nucleic Acids Res 27: 446–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCasse EC, Lefebvre YA (1995) Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res 23: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazinski D, Grzadzielska E, Das A (1989) Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell 59: 207–218 [DOI] [PubMed] [Google Scholar]
- Legendre M, Gautheret D (2003) Sequence determinants in human polyadenylation site selection. BMC Genomics 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS, Alwine JC (1994) Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev 8: 576–586 [DOI] [PubMed] [Google Scholar]
- Lutz CS, Murthy KG, Schek N, O'Connor JP, Manley JL, Alwine JC (1996) Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev 10: 325–337 [DOI] [PubMed] [Google Scholar]
- Martin G, Keller W (1996) Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and a catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J 15: 2593–2603 [PMC free article] [PubMed] [Google Scholar]
- Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley JL, Proudfoot NJ (1998) The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev 12: 2522–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira A, Wollerton M, Monks J, Proudfoot NJ (1995) Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J 14: 3809–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy KG, Manley JL (1992) Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem 267: 14804–14811 [PubMed] [Google Scholar]
- Murthy KG, Manley JL (1995) The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′ end formation. Genes Dev 9: 2672–2683 [DOI] [PubMed] [Google Scholar]
- Phillips C, Virtanen A (1997) The murine IgM secretory poly(A) site contains dual upstream and downstream elements which affect polyadenylation. Nucleic Acids Res 25: 2344–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker PJ, Lingner J, Minvielle-Sebastia L, Keller W (1995) The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell 81: 379–389 [DOI] [PubMed] [Google Scholar]
- Preker PJ, Ohnacker M, Minvielle-Sebastia L, Keller W (1997) A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J 16: 4727–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J, Falck-Pedersen E (1994) Sequence elements upstream of the 3′ cleavage site confer substrate strength to the adenovirus L1 and L3 polyadenylation sites. Mol Cell Biol 14: 4682–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüegsegger U, Beyer K, Keller W (1996) Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J Biol Chem 271: 6107–6113 [DOI] [PubMed] [Google Scholar]
- Shatkin AJ, Manley JL (2000) The ends of the affair: capping and polyadenylation. Nat Struct Biol 7: 838–842 [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C (2003) FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113: 777–787 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL (2000) Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol 20: 1515–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E (1991) Purification and characterization of a mammalian polyadenylate polymerase involved in the 3′ end processing of messenger RNA precursors. J Biol Chem 266: 3131–3139 [PubMed] [Google Scholar]
- Walsh EP, Lamont DJ, Beattie KA, Stark MJ (2002) Novel interactions of Saccharomyces cerevisiae type 1 protein phosphatase identified by single-step affinity purification and mass spectrometry. Biochemistry 41: 2409–2420 [DOI] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelkovsky A, Helmling S, Moore C (1998) Processivity of the Saccharomyces cerevisiae poly(A) polymerase requires interactions at the carboxyl-terminal RNA binding domain. Mol Cell Biol 18: 5942–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material