Abstract
A counting process senses the X chromosome/autosome ratio and ensures that X chromosome inactivation (XCI) initiates in the female (XX) but not in the male (XY) mouse embryo. Counting is regulated by the X-inactivation centre, which contains the Xist gene. Deleting 65 kb 3′ to Xist in XO embryonic stem (ES) cells affects counting and results in inappropriate XCI upon differentiation. We show here that normal counting can be rescued in these deleted ES cells using cre/loxP re-insertion, and refine the location of elements controlling counting within a 20 kb bipartite domain. Furthermore, we show that the 65 kb deletion also leads to inappropriate XCI in XY differentiated ES cells, which excludes the involvement of sex-specific mechanisms in the initiation of XCI. At the chromatin level, we have found that the Xist gene corresponds to a peak of H3 Lys-4 dimethylation, which is dramatically and specifically affected by the deletion 3′ to Xist. Our results raise the possibility that H3 Lys-4 dimethylation within Xist may be functionally implicated in the counting process.
Keywords: counting, histone dimethylation, X chromosome inactivation, Xist
Introduction
X chromosome inactivation (XCI), which occurs only in female (XX) and not in male (XY) embryos, ensures dosage compensation of X-linked genes between the sexes. The initial steps in XCI involve a ‘counting process', which senses the X chromosomes/autosomes ratio and restricts XCI to female mouse embryos. Initiation of XCI in the embryo proper also includes the random choice of the active (Xa) and the inactive (Xi) Xs in each cell. The selected Xi becomes the target of a chromosome-wide mechanism of transcriptional silencing, which constitutes an exciting paradigm for epigenetic regulations and confirms interest in the molecular dissection of the X-inactivation centre (Xic), a locus on the X chromosome that contains the Xist gene and the elements involved in counting, choice and silencing.
The random form of XCI occurs around the time of implantation of the late female blastocyst, in the differentiating epiblast that derives from the inner cell mass (ICM) and that will give rise to the embryo proper. Mouse embryonic stem (ES) cells, which are derived from the ICM, constitute a useful ex vivo system for the study of random-XCI (Rastan et al, 1985; Panning and Jaenisch, 1996; Penny et al, 1996; Clerc and Avner, 1998). Female ES cells possess two active X chromosomes and recapitulate random XCI when induced to differentiate. Under the same differentiation conditions, male ES cells maintain their single X in the active state. Xist is expressed at very low levels from every X chromosome in both undifferentiated male and female ES cells. A transcript antisense to Xist, termed Tsix (Lee et al, 1999a), is initiated mainly 12 kb downstream of Xist at the DXPas34 locus, and more weakly 28 kb downstream of Xist (Sado et al, 2001). Tsix is biallelically expressed in undifferentiated female ES cells and controlled in part by the Xite locus, a cluster of intergenic transcription start sites and DNaseI hypersensitive sites (Ogawa and Lee, 2003). At the onset of XCI in differentiated female ES cells, Tsix is silenced on the elect inactive X chromosome, which becomes rapidly coated by Xist RNA (Panning et al, 1997; Sheardown et al, 1997) and transcriptionally silenced. In parallel, Tsix persists transiently on the Xa, where it represses Xist upregulation (Lee et al, 1999a, 1999b; Luikenhuis et al, 2001; Morey et al, 2001; Sado et al, 2001; Stavropoulos et al, 2001).
Several covalent histone modifications are sequentially put in place following the accumulation of Xist RNA on the future Xi (H3 Lys-27 and Lys-9 hypermethylation and H3 Lys-4 hypomethylation, followed by hypoacetylation of both H3 and H4 (Heard et al, 2001; Mermoud et al, 2002; Plath et al, 2003; Silva et al, 2003)). These modifications are clearly major players in the establishment, spreading and maintenance of transcriptional silencing on the Xi. However, much less data are available on the role of histone modifications within the Xic in regulating the initiation of XCI. A H3 Lys-9 dimethylation hotspot spanning at least 150 kb upstream of the Xist gene is constitutively present in male and female ES cells (Heard et al, 2001). H3 Lys-9 dimethylation is generally correlated with regions of silent inactive chromatin (Rice and Allis, 2001). This has led to the proposal that the H3 Lys-9 dimethylation hotspot located 5′ to the Xist gene acts as a nucleation centre for the spreading of silencing along the Xi (Heard et al, 2001). Interestingly, a 120 kb region upstream to Xist has also been shown to present elevated levels of H4 acetylation in wild-type female ES cells but not in female cells bearing a heterozygous deletion of Xist (O'Neill et al, 1999). These data clearly point to the domain lying 5′ to Xist as a potential target for regulatory mechanisms, and calls for analysis of other histone modifications within the Xic prior to and at the onset of the initiation of XCI.
The differential programming of XCI between male and female cells is poorly understood. It has been shown to depend on a counting process that senses the ratio of X chromosomes versus autosomes and ensures that every X is inactivated, with the exception of a single X per diploid set of autosomes (for a review, see Gartler and Riggs (1983)). Neither the mechanism of sensing nor the effector pathway acting downstream of it have been clearly identified. A simple model for counting would be that a ‘blocking factor' binds to a specific region of the Xic referred to as the ‘counting element' (Rastan et al, 1985). This blocking factor would be produced in a limiting quantity just sufficient to interact with a single counting element per diploid cell and to prevent the initiation of XCI in cis. A recent model additionally postulated the existence of a ‘competence factor' that would be produced by cells bearing more than one X chromosome such as female cells (Lee et al, 1999b) and would be required to allow the initiation of XCI. Male/female differential XCI programming may also involve chromosome-wide epigenetic marks. Indeed, X-linked genes have been shown to present different levels of histone modifications in female ES cells as compared to male ES cells (O'Neill et al, 2003), while differential H4 acetylation of a region upstream of Xist has been reported (O'Neill et al, 1999). Another uncertainty related to counting concerns the moment in mouse embryonic development when the process occurs, either prior to the ICM stage or concurrently with epiblast differentiation.
The ‘blocking factor' model for counting, which implies the existence of a mechanism capable of repressing the initiation of XCI, is supported by observations made on a 65 kb deletion targeted 3′ to Xist (Clerc and Avner, 1998). This deletion, which spans the Tsix antisense transcription, the DXPas34 element and the Xite locus, was targeted to one of the Xs in an XX ES cell line and resulted in a complete skewing of random X inactivation in favour of the mutated X. Essentially XO ES cells carrying the targeted Xic were derived from the XX mutant ES cell line (D102 ES cell line) through selection of a spontaneous truncation of the unmutated X proximal to the Hprt locus. Interestingly, the targeted X in the XO ES cells (XLD ES cell line; Clerc and Avner, 1998) was able to efficiently initiate XCI upon differentiation, despite the fact that a single Xic is present in these cells. This result identifies a repressor associated with the 65 kb region lying 3′ to Xist, which mediates counting. This element is likely to be distinct from Tsix and Xite since individual mutations of these loci did not result in significant initiation of X inactivation in XY differentiated ES cells (Lee et al, 1999b; Luikenhuis et al, 2001; Sado et al, 2001, 2002; Ogawa and Lee, 2003). However, definitive elucidation of the phenotypic differences between the 65 kb deletion in XO ES cells as compared to the Tsix and Xite mutations in XY ES cells clearly requires testing of the 65 kb deletion in an XY ES cell line.
Here, we have investigated the counting process through both site-specific transgenesis using our original deleted XO ES cell line and cre/loxP deletion of an XY ES cell line. We show that inappropriate XCI induced by differentiation in the deleted XO ES cells is prevented by re-inserting a 37 kb DNA sequence spanning Tsix and Xite. This phenotypic complementation demonstrates that counting is not irreversibly established prior to ES cell differentiation, and refines the genomic span lying 3′ to Xist involved in counting. We also report that the 65 kb deletion results in efficient XCI upon differentiation of XY ES cells, similar to its effect in XO ES cells. The control exerted on the initiation of XCI by the region lying 3′ to Xist must therefore be able to override sex-specific marks on the X chromosome. As a further step towards characterizing the Xic, we have analysed the function of the region lying 3′ to Xist using chromatin immunoprecipitation (ChIP). We have found that the hotspot of H3 Lys-9 methylation lying 5′ to Xist is unaffected by the 65 kb deletion, suggesting that the hotspot does not act in the counting pathway downstream of the region 3′ to Xist. We have also analysed H3 Lys-4 dimethylation and found elevated levels of this histone modification within the Xist gene. We show that the deletion 3′ to Xist, which affects counting, has a dramatic and specific effect on H3 Lys-4 dimethylation within the Xist gene itself.
Results
Site-specific re-insertion of a 37 kb region 3′ to Xist in Δ65 XO ES cells re-establishes normal transcriptional and chromatin features
Using the cre site-specific recombinase, we have re-inserted into the deleted locus of the XLD ES cell line (Δ65 XO ES cells of 129/C3 H.Pgk1a background; Clerc and Avner, 1998) a 37 kb DNA sequence extending 3′ to Xist (Figure 1A). This region includes the terminal Xist exons, the DXPas34 minisatellite (Debrand et al, 1999), the major and minor antisense start sites (Lee et al, 1999a; Sado et al, 2001), and the Xite locus (Ogawa and Lee, 2003). The genomic structure of the re-inserted ES cell clones was verified by Southern blot analysis (Figure 1B). Two independently complemented XLDp37 ES cell clones (XLDp37#5 and XLDp37#6) were systematically analysed. As both gave essentially similar results for all the experiments described in this paper, data for only a single clone are presented.
Figure 1.
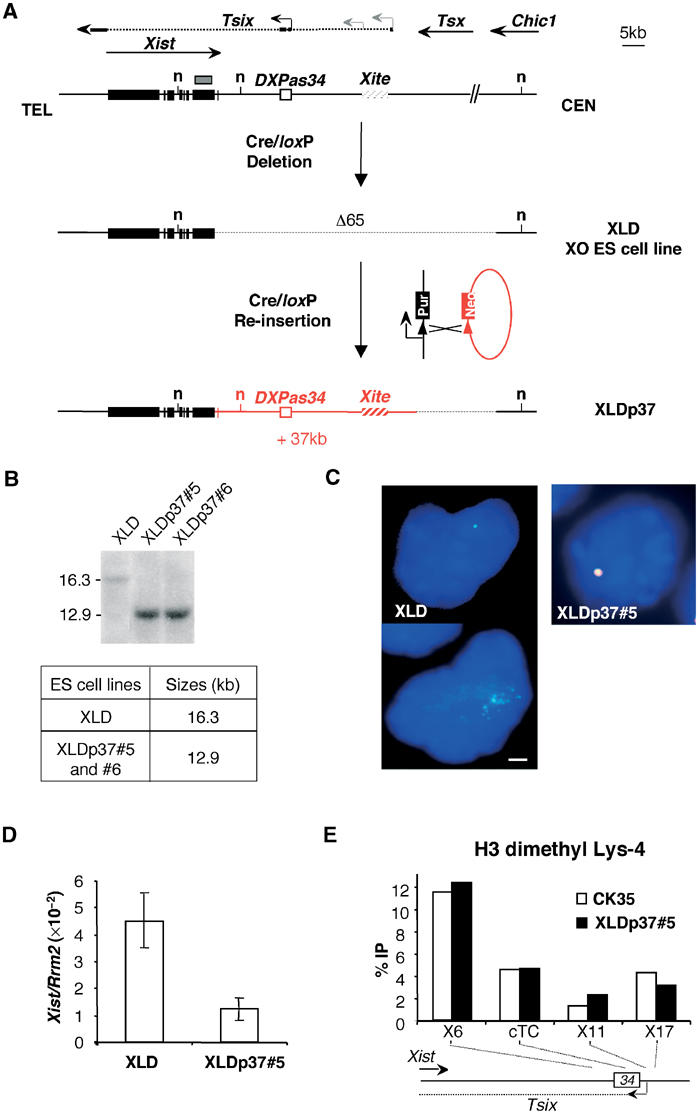
The 37 kb re-inserted DNA is refolded into a functional chromatin structure and restores normal Xist expression in undifferentiated XO ES cells. (A) Deletion and complementation strategies (Morey et al, 2001) in XO ES cells. The resulting 65-kb-deleted and 37-kb-complemented alleles are depicted underneath the map of the Xist locus. Arrows indicate the direction of transcription and arrowheads identify promoter sequences. Solid boxes, Xist exons; the plain and dotted lines show the exonic and intronic Tsix structure with major and minor Tsix promoters symbolized as black and grey arrowheads; hatched box, the Xite element; small arrow, intergenic antisense transcription constitutive of the Xite element (Ogawa and Lee, 2003); grey box, DNA probe; n, NdeI restriction sites relevant to the analysis in (B). (B) Southern-blot analysis and expected sizes of the NdeI restriction fragments detected with the DNA probe depicted in (A). Two re-inserted ES cell clones XLDp37#5 and XLDp37#6 were analysed. The structure of the XLDp37#5 and XLDp37#6 ES cell lines was also confirmed by PCRs at the boundaries of the 37 kb re-inserted region. (C) Two-colour RNA-FISH analysis for Xist (L510, green) and DXPas34/Tsix (Debrand et al, 1999) (red) on DAPI-stained undifferentiated XLD and XLDp37#5 ES cells. Overlapping green and red signals are seen as yellow. The bar equals 5 μm. The XLD ES cells show a faint Xist pinpoint (upper left panel) or scattered dots (in 15% of nuclei, n=200, down left panel), whereas a normal Xist pinpoint signal superimposed on a restored DXPas34/Tsix expression is detected in the undifferentiated XLDp37#5 ES cell line. (D) Dual real-time quantitative RT–PCR for Xist (spanning exons 5 and 6) and Rrm2 (ribonucleoside diphosphate reductase M2) (Lee et al, 1999b) as a reporter in undifferentiated XLD and XLDp37#5 ES cell lines. Means±SD (n=4) of Xist transcripts are standardized over Rrm2 RNA and expressed in arbitrary units as previously described (Morey et al, 2001). Using the 18S ribosomal RNA as an alternative reporter gave essentially similar results in all the experiments of this paper (data not shown). The elevated Xist level in XLD ES cells is restored to a normal low level in the XLDp37#5 ES cell line. This level is not significantly different from the amount of Xist transcripts originating from the same allele in the parental XX ES cell line. Similar results were obtained in at least two independent ES cell cultures for each ES cell line. (E) ChIP analysis of H3 Lys-4 dimethylation within the 37 kb re-inserted region in the undifferentiated XLDp37#5 ES cell line. Immunoprecipitated DNA was analysed by real-time quantitative PCR. The location of the primer pairs is represented on the small map below the graph. A 5 kb region around the DXPas34 locus, which has a crucial function in the regulation of the Xist locus, was analysed. The graphs show the %IP obtained for each primer pairs and each ES cell line. CK35 and XLDp37 harbour very similar H3 Lys-4 dimethylation profiles indicative of an accurate chromatin refolding of the 37 kb region in the XLDp37#5 ES cell line. All experiments described in this figure have been carried out in parallel on the XLDp37#6 ES cell line with essentially identical results (data not shown).
We first addressed the effect of the 37 kb re-insertion on Xist expression in undifferentiated ES cells. In the original 65-kb-deleted XLD ES cell line, the lack of antisense transcription was associated with a faint Xist pinpoint (Figure 1C, upper left panel) or with a diffusion of the Xist transcripts in the vicinity of their transcription site in about 15% of the nuclei (n=200) (Figure 1C, down left panel). The XLD ES cell line also showed elevated steady-state levels of Xist transcripts (real-time RT–PCR; Figure 1D), which were increased approximately three- to five-fold as compared to the untargeted allele of the parental XX ES cell line (not shown; Morey et al, 2001). In the XLDp37 ES cell lines, a restored Tsix/DXPas34 signal colocalized with a normal Xist pinpoint in 80% of the nuclei (n=200), as visualized using RNA-FISH (XLDp37#5, Figure 1C, right panel; XLDp37#6, data not shown). In parallel, Xist RNA amounts were restored to normal in these cell lines (Figure 1D). These results probably reflect the repressive effect of Tsix on the steady-state levels of Xist RNA in undifferentiated ES cells, which has been previously reported (Luikenhuis et al, 2001; Morey et al, 2001; Sado et al, 2001; Stavropoulos et al, 2001).
We also wished to investigate whether the 37 kb DNA sequence, when re-inserted as naked DNA in our ES cell line, is capable of reconstituting a native chromatin structure. To address this issue, we chose to analyse the distribution of histone H3 dimethylated on Lys-4, which marks transcriptionally active genomic regions (Turner, 2002), using ChIP and real-time quantitative PCR (Figure 1E). We focused our analysis on the DXPas34 locus and the major initiation site for Tsix, because of the primary importance of this region in the regulation of XCI. No difference in the H3 Lys-4 methylation profile of this region in the XLDp37 ES cell lines was seen when compared to that of the wild-type XY CK35 ES cell line (XLDp37#5; Figure 1E; XLDp37#6, data not shown). This result validates our site-specific transgenesis approach by showing that the transgenic chromatin structure of the DXPas34/Tsix locus can be accurately reconstituted in complemented ES cells.
A 37 kb site-specific transgene rescues the counting process in Δ65 X0 ES cells
In order to investigate whether the 37 kb transgene was able to prevent the inappropriate initiation of XCI occurring in the differentiated XLD ES cells (Clerc and Avner, 1998), we analysed Xist expression in differentiated cells of both the deleted and complemented ES cell lines. A near-complete inhibition of the Xist upregulation was observed using real-time quantitative RT–PCR in embryoid bodies of the XLDp37 ES cell lines as compared to embryoid bodies of the parental XLD ES cell line (XLDp37#5, Figure 2A; XLDp37#6, data not shown). To confirm this result using another differentiation method, we differentiated the cells using low-density cell culture conditions and performed quantitative RT–PCR and RNA-FISH analysis for Xist RNA in parallel at the same differentiation time point. After 3 days of differentiation under low cell density, the XLDp37 ES cell lines showed reduced steady-state levels of Xist RNA (XLDp37#5, Figure 2B; XLDp37#6, data not shown) as well as a dramatically decreased number of nuclei with a Xist domain (9% in XLDp37#5, n=200, Figure 2C; XLDp37#6, data not shown), as compared to the original XLD ES cell line (60%, n=200; Figure 2C).
Figure 2.

Inappropriate X-inactivation in the differentiated XLD XO ES cell line is prevented upon re-insertion of 37 kb of DNA lying 3′ to Xist. (A) Quantification of Xist RNAs (as described in Figure 1D) during differentiation into embryoid bodies. The steady-state level of Xist RNAs is upregulated during ES cell differentiation in XLD, but not in the XLDp37#5 ES cell line. (B) Quantification of Xist RNA standardized over Rrm2 in the XLD and XLDp37#5 ES cells differentiated for 3 days in low-density cell culture. (C) In the same differentiation experiment as (B), Xist expression was assessed by RNA-FISH with the L510 probe (green) on DAPI-stained nuclei. The bar equals 25 μm. Respectively 60 and 9% of total nuclei exhibited a Xist domain (n=200) in XLD and XLDp37#5 ES cells differentiated for 3 days, consistent with a repression of Xist accumulation consecutive to the 37 kb complementation. Two independent differentiations of XLDp37#5 and of XLDp37#6 ES cell lines using the two differentiation systems gave similar results.
Taken together, our results show that the inappropriate initiation of XCI occurring upon differentiation in the XLD ES cell line is prevented through site-specific reinsertion of a 37 kb DNA sequence encompassing Tsix, DXPas34 and Xite. This indicates that elements involved in counting are located within this region. Abnormal counting has been suppressed in our XO ES cell line without prior passage through the germ line or the early stages of embryonic development. This suggests that counting is not irreversibly established in developmental stages preceding the ICM, and occurs in ES cells concurrently with or just prior to cell differentiation.
Construction of a 65 kb deletion 3′ to Xist in an XY ES cell line
It was of crucial importance to assess the role of the region 3′ to Xist in the counting pathway in ES cells since differences in X chromosome chromatin structure between XY and XX ES cells have been reported previously (O'Neill et al, 1999, 2003). We therefore decided to reproduce the original 65 kb deletion in the CK35 XY ES cells of 129/Sv origin through double targeting of loxP sites and cre expression (Figure 3A). Deleted clones were selected by the reconstitution of a functional puromycin gene flanked by frt sites, which was subsequently excised by expressing eflp, leaving a single 48 bp frt site (Schaft et al, 2001). The genomic structures of the deleted clones were confirmed in a Southern blot analysis (Figure 3B). In this study, two deleted clones with and two without the puromycin cassette were analysed and gave the same results. Data for a clone without the cassette are presented here.
Figure 3.
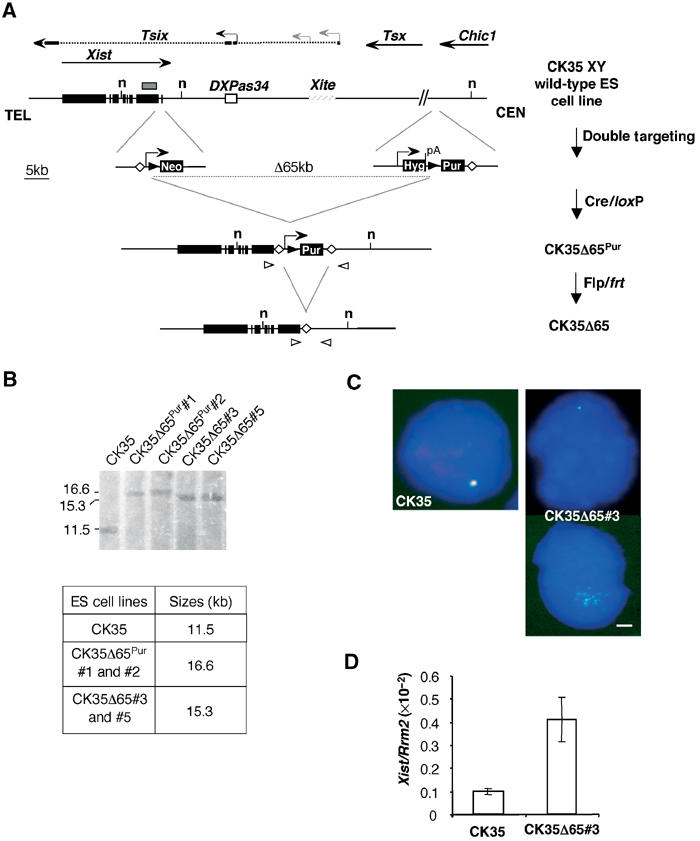
Deleting 65 kb 3′ of Xist in ES cells results in Xist RNA upregulation and alters Xist RNA distribution within the nuclei of undifferentiated ES cells. (A) Strategy for the 65 kb deletion in CK35 XY ES cells. Targeting vectors and the structures resulting after Cre and Flp expression are shown. Black triangles, loxP sites; open diamonds, frt sites. Open triangles indicate the primer pair (DC651Up-Del1Lo) used for the PCR screening of the deleted clones. Arrowheads represent promoter sequences and pA, a polyadenylation signal. (B) Southern-blot analysis of the CK35Δ65 ES cells. The analysis was performed using the NdeI restriction enzyme (n, NdeI restriction sites) and a probe located in Xist exon 7 (grey box). The flp/frt cassette excision was carried out on two independent cre/loxP-deleted clones CK35Δ65Pur#1 and CK35Δ65Pur#2, and gave rise, respectively, to cell lines CK35Δ65#3 and CK35Δ65#5. The Southern-blot analysis confirmed the genomic structure of the deleted alleles. (C) RNA-FISH for Xist (green) and DXPas34/Tsix (red) on CK35 and CK35Δ65#3 on DAPI-stained nuclei of undifferentiated ES cells. The bar equals 5 μm. CK35Δ65#3 ES cells exhibit a reduced Xist pinpoint (upper right panel) or faint Xist RNA signals diffused around the Xist transcription site (down right panel). (D) Quantification of Xist transcripts in undifferentiated CK35 and CK35Δ65#3 ES cell lines using quantitative real-time RT–PCR for Xist and the Rrm2 reporter gene. CK35Δ65#3 ES cells show a four-fold increase in Xist steady-state levels compared to the wild-type CK35 ES cell line. Similar results were obtained for the CK35Δ65#5 ES cell line.
Using RNA-FISH, Xist was detected as a faint pinpoint or as a collection of weak scattered signals in the CK35Δ65 undifferentiated ES cell lines (CK35Δ65#3, Figure 3C right panels; CK35Δ65#5, data not shown). Xist RNA steady-state levels were four-fold higher in the deleted CK35Δ65 ES cells compared to the wild-type CK35 ES cell line (Figure 3D), consistent with the de-repression of Xist consecutive to the lack of antisense transcription in CK35Δ65 ES cells. We note incidentally the low levels of Xist RNA in the CK35 XY ES cell line as compared to other wild-type ES cell lines, which likely results from differences in genetic backgrounds (data not shown). We conclude that undifferentiated ES cells deleted for 65 kb downstream to Xist show a phenotype essentially identical to the XLD ES cell line bearing the same deletion in an XO female context.
A 65 kb deletion located 3′ to Xist results in XCI in XY differentiated ES cells
In order to analyse the impact of the 65 kb deletion in ES cells on the counting process, CK35Δ65 ES cells were differentiated into embryoid bodies and analysed for Xist RNA expression. Using quantitative RT–PCR, a greater than 40-fold upregulation of Xist expression after 3 days of differentiation was observed (CK35Δ65#3, Figure 4A; CK35Δ65#5, data not shown). This corresponds to a 20-fold difference in the levels of Xist RNA between CK35Δ65#3 and parental CK35 differentiated cells at day 3 (Figure 4A). A highly significant increase in the steady-state level of Xist RNA in the CK35Δ65#3 cell line as compared to the CK35 ES cell line was also observed at day 3 of differentiation under low-density cell culture conditions (Figure 4B). We then determined the kinetics of appearance of Xist accumulations using RNA-FISH. Essentially no Xist accumulations could be detected in wild-type CK35 differentiated ES cells (Figure 4C and D). In contrast, in the CK35Δ65#3 ES cell line, 30% of the cells exhibited a mature Xist domain as early as day 2 of differentiation (CK35Δ65#3, Figure 4D; CK35Δ65#5, data not shown). Bearing in mind that the lack of synchronization of the cell differentiation process usually results in less than 50% of cells with a Xist domain at day 2 (see, e.g., Morey et al, 2001; Plath et al, 2003), this suggests that ES cells carrying the 65 kb deletion, differentiated for short times, have a capacity to initiate XCI comparable to that of wild-type XX ES cells.
Figure 4.
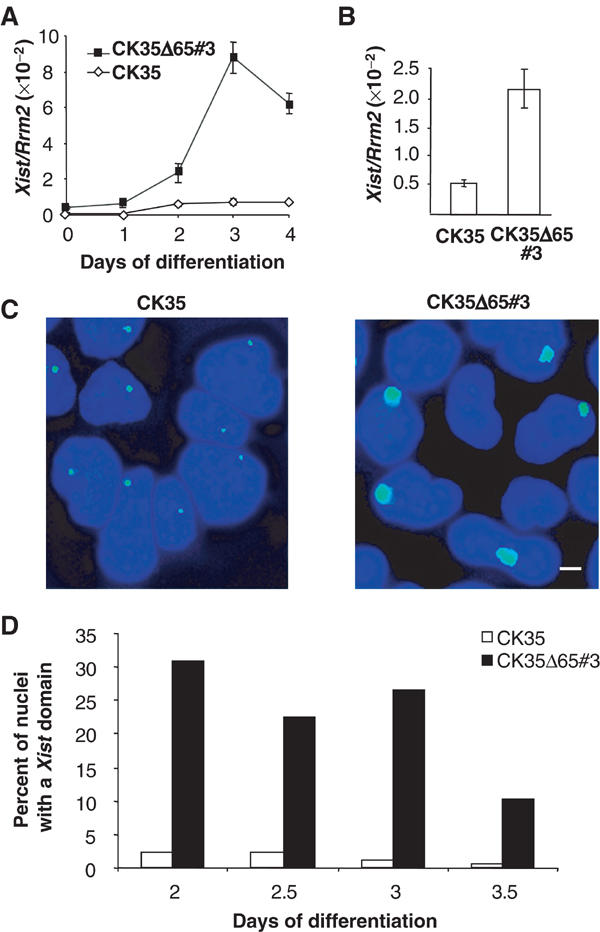
Inappropriate initiation of X-inactivation occurs in differentiated CK35Δ65 ES cells. (A) Xist expression measured in real-time quantitative RT–PCR during ES cell differentiation into embryoid bodies. CK35Δ65#3 ES cell line shows an increase in Xist RNA levels during differentiation into embryoid bodies. (B) Xist steady state levels measured by real-time RT–PCR and normalized using the Rrm2 reporter gene in CK35 and CK35Δ65#3 ES cells differentiated for 3 days in low-density cell culture. (C) In the same differentiation experiment, Xist RNA-FISH on DAPI-stained nuclei of CK35 and CK35Δ65#3 ES cell lines differentiated for 3 days. The bar equals 25 μm. High levels of Xist expression are associated with inappropriate Xist accumulation at the XΔ65 in the CK35Δ65#3 ES cell line. (D) Kinetics of Xist accumulation during ES cell differentiation under low-density cell culture conditions. The proportion of nuclei showing an accumulation of Xist RNA between days 2 and 3.5 of differentiation is illustrated. A total of 100–200 nuclei were scored for each differentiation time point. Two independent experiments for CK35Δ65#3 and CK35Δ65#5 and CK35Δ65Pur#1 and CK35Δ65Pur#2 using both differentiation systems gave similar results.
In order to test whether Xist accumulations on the XΔ65 were associated with transcriptional silencing, we performed RNA-FISH with probes for the X-linked genes Chic1 and Mecp2 located 12.4 cM distal to the Xic, in CK35Δ65 differentiated ES cell lines. A large and significant percentage of Xist domains lacked an associated Chic1 (Figure 5A–C) or Mecp2 (Figure 5D–F) signal. These data indicate that Xist RNA upregulation from the XΔ65 in differentiated ES cells results in the long-range silencing of X-linked genes. Silencing of the single XΔ65 chromosome is likely to be responsible for the cell mortality, which is most probably the cause of the decrease in the number of Xist domains observed at later differentiation time points (post 3 days) with the CK35Δ65 ES cell lines (CK35Δ65#3, Figure 4D; CK35Δ65#5, data not shown).
Figure 5.
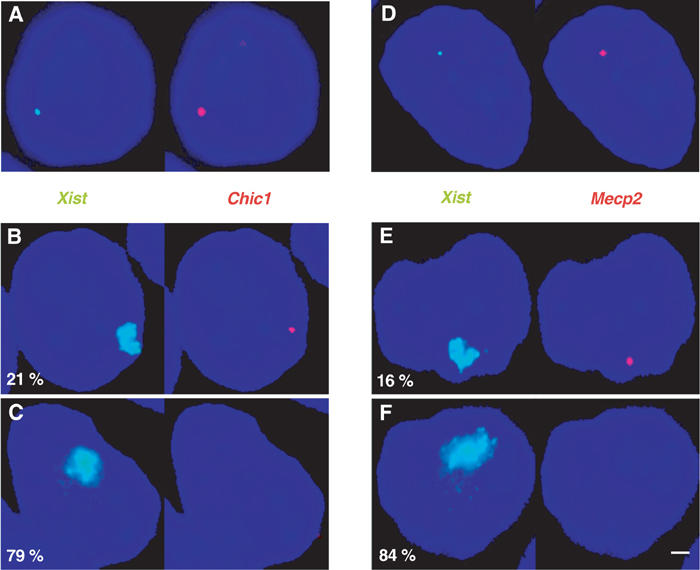
Inappropriate Xist accumulations in CK35Δ65 differentiated ES cells result in efficient X-linked genes silencing. (A–C) Silencing of the Chic1 gene. Two-colour RNA-FISH using probes for Chic1 (red) and Xist (green) in the CK35Δ65#3 ES cell line differentiated for 3 days in low-density cell culture. (A) shows an example of a nucleus where XCI has not yet occurred. In all, 93% of the cells presented a detectable Chic1 signal. To evaluate silencing, the number of nuclei (n=200) displaying a high-level Xist expression associated (B) or not (C) with a Chic1 signal was determined. Percentages are indicated. The bar equals 5 μm. (D–F) Silencing of the Mecp2 gene. A probe specific for the Mecp2 gene (red) was used to perform an analysis identical to that in (A–C). A 75% frequency of detection of Mecp2 was obtained in cells in which initiation of XCI had not yet occurred (D), whereas only 16% of the cells with an Xist RNA–FISH domain demonstrated a signal for Mecp2 (E).
Our data establish that the 65 kb deletion results in efficient initiation of XCI both in an XO ES cell line of 129/C3 H.Pgk1a background (the targeted Xic being of Pgk1a origin) and in a 129 XY ES cell line. This underlies the specificity of the 65 kb deletion phenotype, which is independent of both the genetic background and the XY/XX nature of the cell lines.
H3 Lys-9 methylation hotspot lying 5′ to Xist is unaffected by the 65 kb deletion 3′ to Xist in ES cells
In an attempt to characterize the function of the region 3′ to Xist and its interaction(s) with other parts of the Xic, we investigated whether the H3 Lys-9 methylation hotspot extending upstream of the Xist gene (Heard et al, 2001) would be affected by the 65 kb deletion 3′ to Xist. ChIP experiments were performed in parallel on the CK35 and CK35Δ65 ES cell lines, and explored a region encompassing 150 kb upstream of the Xist gene. Wild-type CK35 ES cells displayed high levels of H3 Lys-9 dimethylation over the hotspot region, similarly to what was previously described (Heard et al, 2001) (Figure 6A). Such high levels of H3 Lys-9 dimethylation were also observed 5′ to Xist in the deleted ES cell lines (Figure 6A). The slight differences seen at some positions in Figure 6A could not be reproduced in other ChIP experiments (data not shown). We conclude that the levels of H3 Lys-9 dimethylation in the hotspot region are unaffected by the 65 kb deletion and therefore are not controlled by Tsix, by Xite, or the element involved in counting that we have identified.
Figure 6.
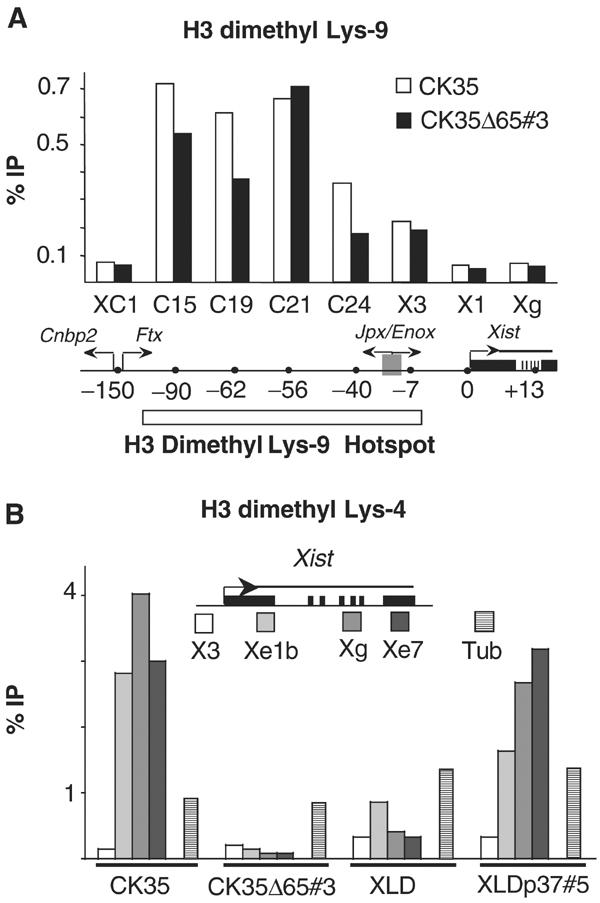
Effects of mutations 3′ to Xist on H3 Lys-9 and Lys-4 dimethylation 5′ and within the Xist gene in ES cells. (A) ChIP analysis of H3 Lys-9 dimethylation in the hotspot region and in the Xist locus in the CK35 and CK35Δ65 ES cell lines. The immunoprecipitated chromatin was analysed by real-time quantitative PCR using assays spanning over 150 kb of the H3 Lys-9 dimethyl hotspot region 5′ to Xist (Heard et al, 2001). Two primer pairs located within the Xist gene were also used (X1 and Xg). Coordinates of the assays relative to the Xist P1 promoter are indicated in kilobases. High levels of H3 lys-9 methylation over a 150 kb region 5′ to Xist were observed in both the CK35 and the CK35Δ65#3 ES cell lines. No significant differences between the CK35 and CK35Δ65#3 ES cell lines in the levels of H3 Lys-9 methylation at the positions tested could be reproducibly observed in the course of repeat experiments. Results from a single representative experiment are shown. (B) ChIP analysis of H3 Lys-4 dimethylation within the Xist gene in wild-type and mutated ES cells. PCR assay X3 is located 7 kb upstream of the Xist promoter P1. PCR assays Xe1b, Xg and Xe7 are located, respectively, within Xist exon 1, exon 5–intron 5 junction and exon 7. The ChIP efficiencies are controlled using PCR assays located within the α-tubulin (stripped columns), myc and G6pd genes (not shown). In the CK35 wild-type ES cell line, high levels of H3 Lys-4 methylation were observed within the Xist gene (assays Xe1b, Xg and Xe7), but not upstream of it (assay X3). A drastic loss of H3 Lys-4 dimethylation is observed in the Xist gene of the 65-kb-deleted X in the CK35Δ65#3 ES cell line. The levels of H3 Lys-4 dimethylation are similarly low in the Xist gene in the XLD ES cell line, and increased in the complemented XLDp37#5 ES cell line. Three independent experiments provided results essentially identical to the example shown here.
The region 3′ to Xist controls H3 Lys-4 dimethylation within the Xist gene without affecting H3 Lys-9 methylation in ES cells
To further characterize the role of the region lying 3′ to Xist, we examined the levels of H3 Lys-4 and Lys-9 dimethylation within the Xist gene in the CK35, CK35Δ65, XLD and XLDp37 ES cell lines.
The analysis of H3 Lys-4 dimethylation at the Xist locus gave striking results. In wild-type CK35 ES cells, the body of the Xist gene (positions Xe1b, Xg and Xe7, located respectively within Xist exon 1, exon 5–intron 5 junction and exon 7, Figure 6B) was associated with high levels of H3 dimethyl Lys-4 as compared to a position 7 kb upstream of the Xist promoter (position X3; Figure 6B). The 65 kb deletion 3′ to Xist affected dramatically H3 Lys-4 dimethylation levels within the Xist gene, as a near-complete loss of methylation was detected in the CK35Δ65#3 cell line (positions Xe1, Xg and Xe7; Figure 6B). Low levels of H3 Lys-4 dimethylation were also observed in the Xist gene in the 65-kb-deleted XLD ES cell line and, interestingly, the H3 Lys-4 dimethylation levels were significantly increased by the 37 kb add back present in XLDp37 ES cells (Figure 6B). Our results clearly establish that the 37 kb region downstream to Xist controls the H3 Lys-4 dimethylation in the Xist gene. This effect seems to be restricted to within the Xist gene itself, as no difference could be observed at a position 7 kb 5′ to Xist. In addition, we found comparable low levels of H3 Lys-9 methylation at the Xist locus (positions X1 and Xg; Figure 6A) in the CK35 wild-type and CK35Δ65 ES cell lines, suggesting that the region 3′ to Xist has no impact on H3 Lys-9 methylation within the Xist gene.
Altogether, our results establish that the region downstream to Xist specifically controls the levels of H3 Lys-4 dimethylation within the Xist gene without affecting the levels of H3 Lys-9 methylation.
Discussion
Our study explores the temporal and mechanistical aspects of the counting process. We report on the phenotypic rescue of counting through site-specific transgenesis in an XO ES cell line deleted for a 65 kb region downstream to Xist and defective for normal counting. The fact that normal counting could be rescued has important implications for the timing of the counting process during development and restricts the location for the elements of counting to 20 kb of genomic DNA. In parallel, we have generated a 65 kb deletion 3′ to Xist in an XY ES cell line and described its capacity to efficiently undergo inappropriate silencing upon differentiation. Importantly, we show that the 37 kb region closest to the 3′ end of the Xist gene specifically controls H3 Lys-4 dimethylation within the Xist gene, but not H3 Lys-9 methylation either at the H3 Lys-9 hotspot lying 5′ to Xist or within the Xist gene itself.
Three mutations targeting Tsix (Lee et al, 1999b; Luikenhuis et al, 2001; Sado et al, 2001) have previously demonstrated only minor or no effects on the initiation of XCI in differentiated male ES cells. These results differ strikingly from our observation of efficient XCI in an XO ES cell line bearing a 65 kb deletion encompassing Tsix, which was derived from an XX ES cell line (this work and Clerc and Avner (1998)). This has led Lee et al (1999b) to suggest that a ‘competence factor' strictly required for the initiation of XCI might be produced in XX cells. According to this hypothesis, XO ES cells would possess this factor and be competent to initiate XCI due to their XX origin. In contrast, XY ES cells would lack this factor and should therefore be unable to undergo XCI even in the presence of the mutation. Our observation that the 65 kb deletion induces XCI with high efficiency in XY differentiating ES cells excludes the involvement of such a ‘competence factor' in the differential programming of XCI in males and females. The similar phenotype of the 65 kb deletion in XO and XY differentiated ES cells also suggests that the differential chromatin marks present on the X chromosomes in male and female ES cells (O'Neill et al, 1999, 2003) can have, if any, only a minor influence on XY/XX XCI differential programming. Taken together, our results indicate that the phenotypic differences between the various mutations can likely be attributed solely to the nature of the mutations and not to parameters specific to the ES cell lines like the sex of the embryo of origin or its genetic background. Our data strongly support a role for the region 3′ to Xist in the counting pathway.
The re-establishment of normal counting obtained on reinsertion of cloned DNA into ES cells implies that ES cells possess all the factors necessary for functional reconstitution. Our data demonstrate clearly that X chromosomes cannot have been definitively precounted at embryonic stages preceding that of the ICM from which ES cells are derived, and that at least part of the counting process operates at the time of cell differentiation. This finding is of major importance for subsequent experimentation aimed at identifying precisely the ‘counting element' that will exploit the ES cell system. Furthermore, our site-specific transgenesis approach locates the elements of counting to within the 37 kb region 3′ to Xist. Both Tsix and Xite, which lie within this interval, have been shown respectively by a 3.7 kb and by a 12.5 kb deletion not to be directly involved in the counting mechanism (Lee et al, 1999a; Ogawa and Lee, 2003). We, therefore, propose that the element important for counting lies within a 20 kb bipartite region depicted in Figure 7. Interestingly, comparative sequence analysis has identified a 2 kb region of mouse/human conservation (80610–82632 of Genbank no. X99946), located proximal to DXPas34 (Chureau et al, 2002), which, based on our results, must constitute an interesting candidate for a function in the counting pathway. The precise nature of this function has yet to be determined. Counting seems to involve both a sensing of the X chromosome number relative to cell ploidy and the output of this information into the control of the initiation of XCI. As the degree of integration of these two steps is still unclear, the element located in the region that we have identified might be the ‘counting element' that is postulated to interact with a ‘blocking factor', or an effector acting downstream, which would repress in cis the initiation of XCI. It is intriguing that Tsix, a well-characterized repressor in cis of the initiation of XCI (Luikenhuis et al, 2001; Morey et al, 2001; Stavropoulos et al, 2001), has been excluded for a function in counting (Lee et al, 1999b). We must therefore postulate the existence of yet another repressor element or of the ‘counting element' itself within the 20 kb region that we have identified.
Figure 7.
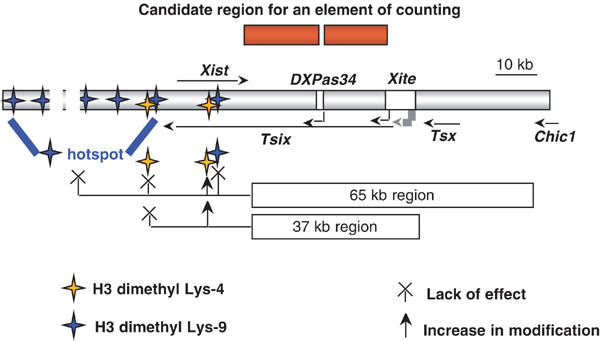
Schematic representation of the roles of the region 3′ to Xist in counting and in control of H3 Lys-9 and Lys-4 dimethylation at the Xist locus. The schema depicts the Xist locus, as well as the DXPas34/Tsix and Xite elements, which have been previously excluded from having a function in counting. The location of the bipartite 20 kb region identified in this study as containing an element of counting is shown above the central schema. The positions tested for histone H3 Lys-9 and Lys-4 dimethylation are, respectively, indicated by blue and yellow stars. The role of the region lying 3′ to Xist in histone H3 modification is illustrated in the lower part of the diagram. In ES cells, the 65 kb region has no effect on H3 Lys-9 dimethylation in either the hotspot region or the Xist locus. In contrast, the region 3′ to Xist targets the deposition of the Lys-4 dimethyl form of H3 to within the body of the Xist gene. This regulation is specifically supported by the proximal 37 kb subregion of the 65 kb span.
Our observation of a differential effect of the region 3′ to Xist on H3 Lys-4 and Lys-9 methylation within the Xist gene itself emphasizes the complexity of the regulatory process involving the Xist locus, which is summarized in Figure 7. We observed no significant effect of the region 3′ to Xist on H3 Lys-9 methylation either within Xist or within a region extending 5′ to Xist, which is part of an H3 Lys-9 methylation hotspot proposed to act as a nucleation centre for the spreading of XCI. This indicates that the H3 Lys-9 methylation hotspot is not acting downstream of the element of counting present in the region 3′ to Xist and is likely independent of Xist RNA steady-state levels, which are elevated in the CK35Δ65 ES cell lines. Conversely, the 65 kb deletion induced a dramatic loss of H3 Lys-4 dimethylation restricted to the Xist gene itself. H3 Lys-4 and Lys-9 methylations have frequently been described as mirrors of each other (Turner, 2002), and a dissociation of this inverse correlation has been reported only rarely (Yan et al, 2003). Such a lack of correlation between H3 Lys-4 and Lys-9 dimethylations may therefore be specific to particular loci, including the Xist gene.
As shown by our site-specific transgenesis approach, the control of H3 Lys-4 methylation is primarily exerted by the 37 kb domain, which is responsible both for antisense transcription within Xist and for normal counting. It remains for the moment an open and important question as to whether H3 Lys-4 methylation within the Xist gene is linked to antisense transcription, or counting, or both. In an attempt to explore this question, we have looked at the Xist H3 Lys-4 methylation profile in our previously described D102 and c16 XX ES cell lines (Clerc and Avner, 1998; Morey et al, 2001). These cell lines both carry a wild-type X chromosome accompanied, respectively, by a 65-kb-deleted X or a 65-kb-deleted X chromosome complemented for the proximal 16 kb consisting in the Tsix antisense transcription unit. Our preliminary results suggest that the H3 Lys-4 methylation within the Xist gene, lost on deletion of the 65 kb, is partially recovered after the Tsix restoration (data not shown). Although further experimental validations would be necessary, this leads us to favour the hypothesis of an association between antisense transcription and the chromatin remodelling we have detected. Indeed, transcriptionally active chromatin domains generally show elevated levels of H3 Lys-4 dimethylation and, in yeast, this mark results at least in part from the association of the Set1 H3 methyl transferase with the elongating RNA polII (Krogan et al, 2003; Ng et al, 2003). Our results show that H3 Lys-4 dimethylation is lost in the 65-kb-deleted allele, despite ongoing Xist sense transcription. This suggests that the Tsix, but not the Xist, transcriptional machinery might recruit an H3 lys-4 methyl transferase. A more speculative possibility is that an H3 Lys-4 methyl transferase could be recruited to the Xist gene as a result of the sense/antisense transcriptional activity within the locus, using a putative nuclear RNAi pathway reminiscent of the one existing in Schizosaccharomyces pombe (Volpe et al, 2002). Finally, the link between H3 Lys-4 dimethylation within Xist and the counting process leads us to postulate, for the first time, a role of genomic elements within the Xist gene itself in counting. Our results indicate that Lys-4 methylation within Xist is associated with an inhibition of the initiation of XCI and a functional counting process. It is conceivable that Lys-4 dimethylation within the Xist gene, by creating an open chromatin conformation, could potentiate the interaction of this domain with regulatory factors involved in preventing the initiation of XCI, and thereby mediate counting. Indeed, a function ensured by H3 Lys-4 dimethylation may well be to regulate the accessibility of cis-acting DNA elements, as suggested by recent findings in the immunoglobulin heavy-chain locus (Morshead et al, 2003). Further work will be required to explore these exciting avenues and for a complete understanding of the counting process of X chromosome inactivation.
Materials and methods
ES cell site-specific re-insertion
The XLD ES cell line was electroporated using a mixture of the cre-expressing plasmid pOG231 (Clerc and Avner, 1998) and the pGln37 plasmid (see below). The complemented clones were selected for the reconstitution of a functional neomycin cassette, and neomycin-resistant colonies were PCR screened as previously described (Morey et al, 2001). After preliminary FISH analysis for X aneuploidy, two positive clones were extended and characterized. The pGln37 plasmid was obtained by inserting the loxP-neomycin ORF from the pC3Nleo (Clerc and Avner, 1998) plasmid and the 37 kb ClaI fragment from a BAC clone encompassing DXPas34 (Chureau et al, 2002) into a plasmid bearing the low-copy replication origin of the pLG339 vector (Stoker et al, 1982) and the ampicillin resistance gene.
Targeted deletions
The genomic targeting for the 65 kb deletion was realized on the CK35 male ES cell line originating from the 129 mouse strain. ES cell transfections and selections of recombinants were performed as previously described for the HP3.10 ES cell line (Clerc and Avner, 1998). Both proximal and distal homology regions were generated as previously described (Clerc and Avner, 1998). The CK35Δ65Pur ES cell clones were selected for the reconstitution of the puromycin gene (1 μg/ml) and were PCR screened (primer pair D651Up-Del1Lo; see Figure 3A). The puromycin excision was carried out by lipofecting two independent CK35Δ65Pur ES cell clones with the pCAGGS-FLPe supercoiled flp-expressing plasmid (Schaft et al, 2001). Two CK35Δ65 ES cell clones derived from each of the two CK35Δ65Pur clones were selected on the basis of their sensitivity to puromycin and using the PCR screening assay, which gives a smaller product after the loss of the puromycin gene.
RNA-FISH
The preparation of nuclei, hybridization and washes were performed as described (Clerc and Avner, 1998) using DNA probes labelled by nick translation with Spectrum Red or Green dUTP (Vysis, Downer Grove, IL, USA). A Zeiss Axioplan fluorescence microscope with a Quantix CCD camera (Photometrix) and the SmartCapture 2 software (Digital Scientific) were used for image acquisition. All the probes used in this study have been previously described (Clerc and Avner, 1998; Debrand et al, 1999; Heard et al, 1999).
Real-time quantitative RT–PCR
Quantitative real-time PCR measurements of Xist random primed cDNA were performed using TaqMan fluorescent probes and TaqMan Universal PCR mix (Perkin-Elmer Applied Biosystem) on ABI Prism 7700 (Perkin-Elmer Applied Biosystem). Xist cDNA quantitations were internally standardized against the endogenous 18S cDNA or against the Rrm2 cDNA. The PCR assays for Xist and 18S genes were the same as previously described (Morey et al, 2001). For Rrm2, a new system was set (primer pair Rrm1F-Rrm1R, TaqMan probe Rrm2), which spans an exonic junction to prevent any contaminating amplification of genomic DNA. The lack of interference between the amplification and detection systems for Xist and Rrm2 was verified. The specificity of each PCR was checked by running in parallel a PCR on an RT reaction lacking the enzyme.
ES cell culture and in vitro differentiation
ES cells were grown in DMEM (GIBCO), 15% foetal calf serum (FCS; ES cell grade, GIBCO) and 1000 U/ml leukemia inhibitory factor (LIF; Chemicon) on male embryonic feeders for two passages prior to analysis. For the ChIP analysis, ES cells were cultured on gelatin-coated flasks in the absence of feeder cells (Heard et al, 2001). ES cell differentiation into embryoid bodies was performed as previously described (Morey et al, 2001). For ES cell differentiation under low-density cell culture conditions, ES cells were plated at 7000 cells/cm2 on gelatin-coated plates for RNA extraction or on gelatinized slide flasks (Nunc) for RNA-FISH analysis without feeders, and grown for 4 days in a medium with 10% of FCS without LIF. The medium was changed daily throughout differentiation. All ES cell lines exhibited morphological features of differentiated cells after 2 days of culture under these conditions and only low levels of cell mortality before 4 days of differentiation.
Chromatin immunoprecipitation (ChIP)
Immunoprecipitations were performed using a dimethylated H3 Lys-4 antibody or a dimethylated H3 Lys-9 antibody (Upstate Biotechnology, Lake Placid, NY, USA), as described previously (Heard et al, 2001). Cells were crosslinked with 1% formaldehyde for 15 min at RT. Isolated nuclei were sonicated to an average length of 300–1000 bp. Immunoprecipitated DNA and input DNA were analysed by real-time PCR using SYBR Green Universal Mix and an ABI Prism 7700 (Perkin-Elmer Applied Biosystem). Each PCR was run in triplicate. For standardization, the results are represented as a percentage of immunoprecipitation, calculated by dividing the average value of the IP by the average value of the corresponding input, both values being first normalized by the dilution factor. Each experiment was repeated on independent chromatin extracts a total of three times.
Supplementary Material
Oligonucleotide sequences
Acknowledgments
CM is a doctoral fellow supported by the French Ministry for Scientific Research and by the Association pour la Recherche contre le Cancer (ARC). This work was supported by grants to PA from the ARC.
References
- Chureau C, Prissette M, Bourdet A, Barbe V, Cattolico L, Jones L, Eggen A, Avner P, Duret L (2002) Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res 12: 894–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc P, Avner P (1998) Role of the region 3′ to Xist exon 6 in the counting process of X-chromosome inactivation. Nat Genet 19: 249–253 [DOI] [PubMed] [Google Scholar]
- Debrand E, Chureau C, Arnaud D, Avner P, Heard E (1999) Functional analysis of the DXPas34 locus, a 3′ regulator of Xist expression. Mol Cell Biol 19: 8513–8525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartler SM, Riggs AD (1983) Mammalian X-chromosome inactivation. Annu Rev Genet 17: 155–190 [DOI] [PubMed] [Google Scholar]
- Heard E, Mongelard F, Arnaud D, Avner P (1999) Xist yeast artificial chromosome transgenes function as X-inactivation centers only in multicopy arrays and not as single copies. Mol Cell Biol 19: 3156–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL (2001) Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107: 727–738 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A (2003) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11: 721–729 [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D (1999a) Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 21: 400–404 [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N, Keohane AM, O'Neill LP, Belyaev ND, Lavender JS, Turner BM (1999b) Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99: 47–57 [DOI] [PubMed] [Google Scholar]
- Luikenhuis S, Wutz A, Jaenisch R (2001) Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol Cell Biol 21: 8512–8520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermoud JE, Popova B, Peters AHFM, Jenuwein T, Brockdorff N (2002) Histone H3 lysine 9 methylation occurs rapidly at the onset of random X chromosome inactivation. Curr Biol 12: 247–251 [DOI] [PubMed] [Google Scholar]
- Morey C, Arnaud D, Avner P, Clerc P (2001) Tsix-mediated repression of Xist accumulation is not sufficient for normal random X inactivation. Hum Mol Genet 10: 1403–1411 [DOI] [PubMed] [Google Scholar]
- Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA (2003) Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc Natl Acad Sci USA 100: 11577–11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- O'Neill LP, Keohane AM, Lavender JS, McCabe V, Heard E, Avner P, Brockdorff N, Turner BM (1999) A developmental switch in H4 acetylation upstream of Xist plays a role in X chromosome inactivation. EMBO J 18: 2897–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LP, Randall TE, Lavender J, Spotswood HT, Lee JT, Turner BM (2003) X-linked genes in female embryonic stem cells carry an epigenetic mark prior to the onset of X inactivation. Hum Mol Genet 12: 1783–1790 [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Lee JT (2003) Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol Cell 11: 731–743 [DOI] [PubMed] [Google Scholar]
- Panning B, Dausman J, Jaenisch R, Avner P, Brockdorff N, Turner BM (1997) X chromosome inactivation is mediated by Xist RNA stabilization. Cell 90: 907–916 [DOI] [PubMed] [Google Scholar]
- Panning B, Jaenisch R (1996) DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev 10: 1991–2002 [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N (1996) Requirement for Xist in X chromosome inactivation. Nature 379: 131–137 [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y (2003) Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135 [DOI] [PubMed] [Google Scholar]
- Rastan S, Robertson EJ, Rasmussen TP, Mastrangelo MA, Eden A, Pehrson JR, Jaenisch R (1985) X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J Embryol Exp Morphol 90: 379–388 [PubMed] [Google Scholar]
- Rice JC, Allis CD (2001) Code of silence. Nature 414: 258–261 [DOI] [PubMed] [Google Scholar]
- Sado T, Li E, Sasaki H (2002) Effect of TSIX disruption on XIST expression in male ES cells. Cytogenet Genome Res 99: 115–118 [DOI] [PubMed] [Google Scholar]
- Sado T, Wang Z, Sasaki H, Li E (2001) Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development (Cambridge, England) 128: 1275–1286 [DOI] [PubMed] [Google Scholar]
- Schaft J, Ashery-Padan R, van der Hoeven F, Gruss P, Stewart AF (2001) Efficient FLP recombination in mouse ES cells and oocytes. Genesis 31: 6–10 [DOI] [PubMed] [Google Scholar]
- Sheardown SA, Duthie SM, Johnston CM, Newall AE, Formstone EJ, Arkell RM, Nesterova TB, Alghisi GC, Rastan S, Brockdorff N (1997) Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell 91: 99–107 [DOI] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N (2003) Establishment of histone h3 methylation on the inactive x chromosome requires transient recruitment of eed-enx1 polycomb group complexes. Dev Cell 4: 481–495 [DOI] [PubMed] [Google Scholar]
- Stavropoulos N, Lu N, Lee JT (2001) A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc Natl Acad Sci USA 98: 10232–10237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoker NG, Fairweather NF, Spratt BG (1982) Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene 18: 335–341 [DOI] [PubMed] [Google Scholar]
- Turner BM (2002) Cellular memory and the histone code. Cell 111: 285–291 [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Yan Q, Huang J, Fan T, Zhu H, Muegge K (2003) Lsh, a modulator of CpG methylation, is crucial for normal histone methylation. EMBO J 22: 5154–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotide sequences


