Abstract
Nucleotide-dependent unblocking of chain-terminated DNA by human immunodeficiency virus type 1 reverse transcriptase (RT) is enhanced by the presence of mutations associated with 3′-azido-3′-deoxythymidine (AZT) resistance. The increase in unblocking activity was greater for mutant combinations associated with higher levels of in vivo AZT resistance. The difference between mutant and wild-type activity was further enhanced by introduction of a methyl group into the nucleotide substrate and was decreased for a nonaromatic substrate, suggesting that π-π interactions between RT and an aromatic structure may be facilitated by these mutations.
Many nucleoside analogues, including 3′-azido-3′-deoxythymidine (AZT), inhibit human immunodeficiency virus type 1 (HIV-1) replication. The phosphorylated forms of these compounds are incorporated during DNA synthesis by the HIV-1 reverse transcriptase (RT), resulting in chain termination and inhibition of viral replication (7, 9, 12, 22, 31, 32). Mutations at codons 41, 67, 70, 210, 215, and 219 in the HIV-1 RT gene result in resistance of HIV-1 to AZT in cell culture assays (29). Substitutions of phenylalanine and tyrosine for threonine at position 215 (T215F and T215Y) are the predominant mutations observed in vivo and are considered the most important for the resistance phenotype (16, 17, 25).
The inhibitory effect of incorporating a chain-terminating nucleotide analogue can be partially relieved by a reaction catalyzed by RT in which the terminating nucleotide is removed from the 3′ end of a DNA chain by transfer to a nucleotide di- or triphosphate, producing an unblocked DNA chain and dinucleoside polyphosphate with the chain terminator linked to the nucleotide acceptor through a tri- or tetraphosphate chain (19, 21). HIV-1 RT can also transfer the chain-terminating residue to pyrophosphate (PPi), regenerating the triphosphate form of the chain terminator (1, 5, 10, 23). These observations have suggested that enhanced removal of 3′-azido-3′-deoxythymidine-5′-monophosphate (AZTMP) is a possible mechanism for AZT resistance, and an increase in the removal reaction has been reported for RT containing various AZT resistance mutations (1, 2, 18, 19). The biochemical contribution of each of these mutations in the removal reaction remains unclear. Boyer et al. (2) have modeled the amino acid substitutions associated with AZT resistance, as well as ATP or PPi, into the three-dimensional structure of HIV-1 RT and concluded that several of these amino acid substitutions could affect the binding of ATP but are unlikely to affect binding of PPi.
In this report we describe further investigation into the contributions made by specific mutations in HIV-1 RT to its removal activity and the effects of changes in the structure of the nucleotide substrate on the wild-type (WT) and mutant activities. The mutations associated with AZT resistance identify a region of HIV-1 RT that may interact with the nucleoside moiety of a transition intermediate to facilitate the formation of the dinucleoside tetraphosphate.
Removal of AZTMP or ddAMP from blocked primer-templates by WT and mutant RT.
Removal of AZTMP from a blocked primer-template was detected by the formation of unblocked primers that could be extended in a subsequent incubation as previously described (19-21). Removal of ddAMP was detected by the transfer of [32P]ddAMP from a labeled primer terminus to ATP to form [32P]Ap4ddA (19, 21). It has been shown that these assays give similar results for both AZTMP and ddAMP removal (19).
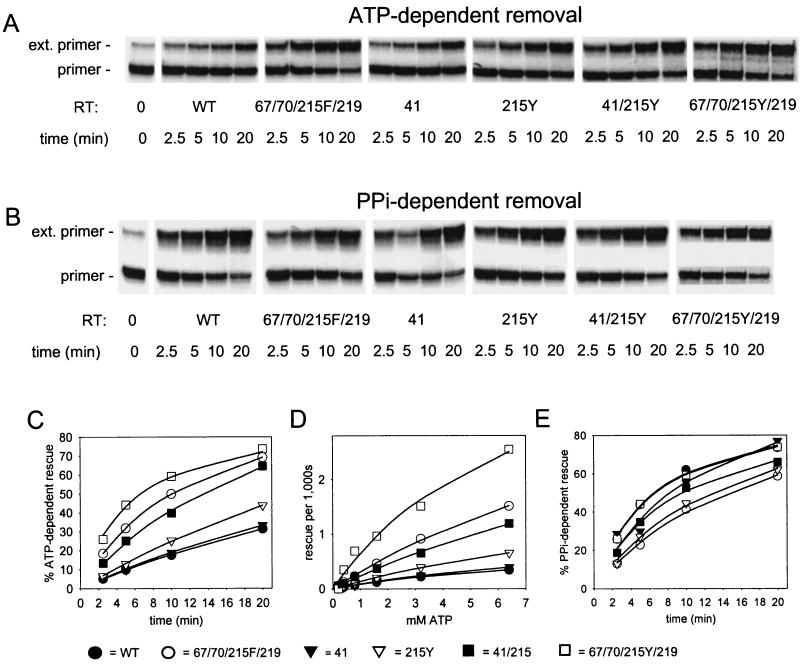
To measure AZTMP removal, DNA primer L33 (5′-CTACTAGTTTTCTCCATCTAGACGATACCAGAA-3′) was 5′ labeled with [γ-32P]ATP and T4 polynucleotide kinase, annealed with DNA template WL50 (5′-GAGTGCTGAGGTCTTCATTCTGGTATCGTCTAGATGGAGAAAACTAGTAG-3′) and chain terminated by extending with AZTTP as previously reported (19). AZTMP-terminated primer-template was reisolated and incubated with excess WT and mutant RT in the presence of ATP. RT was heat inactivated, and unblocked primers were extended by addition of exonuclease-free Klenow fragment of Escherichia coli DNA polymerase I and all four deoxynucleoside triphosphates (dNTPs) as previously described (19-21) (Fig. 1A and C). Formation of extendable primer was quantitated as a function of ATP concentration (Fig. 1D) and second-order rate constants (kcat/Km,ATP) were determined (Table 1). Figure 1 and Table 1 show that mutation at codon 41 alone had little effect on removal of AZTMP from AZTMP-terminated primers, mutation at codon 215 alone had a small but significant effect, and the combination 41L/215Y resulted in a 3.3-fold increase in AZTMP removal over removal by WT RT. The removal of AZTMP by 67N/70R/215F/219Q RT was 5-fold higher and that by 67N/70R/215Y/219Q RT was 10-fold higher than that by the WT. By contrast, with PPi as the substrate, the mutants had activities that were at most equal to that of WT RT (Fig. 1B and E).
FIG. 1.
Effects of the AZT resistance mutations on removal of AZTMP from blocked primer-template. (A, C, and D) ATP-dependent rescue of AZTMP-terminated primer-template by WT and mutant RTs (21). (A) AZTMP-terminated 5′-32P-L33 primer-WL50 template (5 nM) was incubated with the indicated RT (200 nM) in the presence of 3.2 mM ATP for the indicated times at 37°C. The RT was inactivated by heat treatment, and the unblocked primer was extended by incubation with exonuclease-free Klenow fragment of E. coli DNA polymerase I and all four dNTPs. Products were fractionated on a 20% denaturing polyacrylamide gel. Positions for unextended primer (primer) and for products formed after elongation to the end of the template (ext. primer) are shown on the left. (B and E) PPi-dependent rescue of AZTMP-terminated primer-template by WT and mutant RTs. (B) Rescue experiments were performed as for panel A, but with 50 μM PPi instead of ATP. (C) Radioactivity in products longer than 34 nucleotides (rescued primers) from experiments shown in panel A was quantitated by phosphorimaging, expressed as a percentage of total radioactivity in the lane, and plotted against time. In panels C, D, and E, the lines represent the best fit to the Michaelis-Menten equation using Sigmaplot 4.0. (D) Rate of rescue of AZTMP-terminated primer-template as a function of the ATP concentration. Rescue experiments were performed as described for panel A, except that the ATP concentration varied from 0.2 to 6.4 mM, and the incubation times varied from 2 to 90 min, depending on the RT, to allow a maximum of 35% rescue. (E) Radioactivity in products longer than 34 nucleotides (rescued primers) from experiments shown in panel B was quantitated by phosphorimaging, expressed as percentage of total radioactivity in the lane, and plotted against time. All dNTPs and ATP solutions had been pretreated with thermostable pyrophosphatase (100 μl of 64 mM nucleotide was incubated with 1 U of pyrophosphatase at 75°C for 10 min). Expression and purification of WT and mutant RT were described previously (21). Expression vectors for 41L, 215Y, 41L/215Y, and 67N/70R/215F/219Q RTs were derived from pKRT2 (8). Mutations were introduced by PCR using the megaprimer method (28). To construct an expression vector for 67N/70R/215Y/219Q RT, an RT sequence containing these mutations was transferred from the infectious proviral clone into pTRc99 (Pharmacia Biotech) using XbaI and XmaI sites introduced for that purpose (30). All constructs were modified to introduce an N-terminal hexahistidine extension on the expressed protein (21) and were confirmed by DNA sequence analysis.
TABLE 1.
Apparent second-order rate constants for ATP-dependent removal of AZTMP and synthesis of Ap4ddA by HIV-1 WT and mutant RTs
| HIV-1 RT | ATP-dependent removal of AZTMPa, c
|
Formation of Ap4ddAb, c
|
||
|---|---|---|---|---|
| kcat/Km,ATP (M−1 s−1) | Fold increase | kcat/Km,ATP (M−1 s−1) | Fold increase | |
| WT | 0.066 ± 0.010 | 1.0 | 0.21 ± 0.04 | 1.0 |
| 67N/70R/215F/219Q | 0.33 ± 0.01 | 5.0d | 1.9 ± 0.4 | 9.0d |
| 41L | 0.11 ± 0.02 | 1.7e | 0.28 ± 0.01 | 1.3e |
| 215Y | 0.15 ± 0.01 | 2.3d | 0.83 ± 0.06 | 3.9d |
| 41L/215Y | 0.22 ± 0.03 | 3.3d | 1.9 ± 0.4 | 9.0d |
| 67N/70R/215Y/219Q | 0.66 ± 0.13 | 10d | 3.3 ± 0.6 | 16d |
Data obtained from experiments performed as described in the legend to Fig. 1D were fitted to hyperbolas to obtain kcat and Km,ATP.
Data from experiments performed as described in the legend to Fig. 2B were fitted to hyperbolas to obtain kcat and Km,ATP.
Values were derived from two to eight repeated determinations and are given as means ± standard deviation.
Significantly different from the WT value (P < 0.05) by Student's t test corrected for multiple comparisons.
Not significantly different from the WT value (P ≥ 0.05).
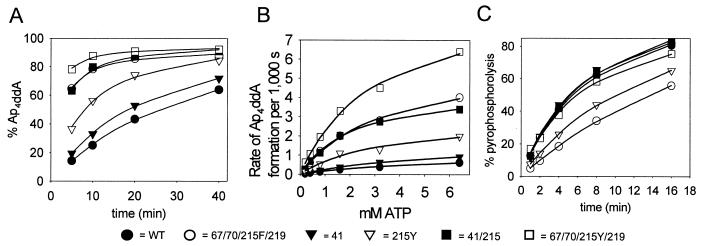
To measure removal of ddAMP, DNA primer L32 (5′-CTACTAGTTTTCTCCATCTAGACGATACCAGA-3′) was annealed with DNA template WL50 and chain terminated by extending with [32P]ddATP as previously reported (19, 21). The [32P]ddAMP-terminated L32 primer-WL50 template was reisolated and incubated with excess WT or mutant RT in the presence of ATP. Products were separated by polyacrylamide gel electrophoresis, and 32P-labeled dinucleoside tetraphosphate (Ap4ddA) was quantitated by phosphorimaging as a function of time of incubation with 3.2 mM ATP (Fig. 2A) and as a function of ATP concentration (Fig. 2B and Table 1). Synthesis of Ap4ddA was only slightly affected by the 41L mutation and more substantially increased by the 215Y mutation (1.3- and 3.9-fold increases over WT RT, respectively). Ap4ddA synthesis by 41L/215Y RT and 67N/70R/215F/219Q RT was increased 9-fold and that by 67N/70R/215Y/219Q RT was increased 16-fold over that by WT RT.
FIG. 2.
Effect of AZT resistance mutations on removal of [32P]ddAMP from blocked primer-template through transfer to ATP or PPi. (A) [32P]ddAMP-terminated L32 primer-WL50 template (5 nM) was incubated with the indicated RT and 3.2 mM ATP for 5, 10, 20, or 40 min at 37°C. Products were separated on a 20% denaturing polyacrylamide gel (data not shown). The radioactivity in primer and Ap4ddA was quantitated by phosphorimaging, and the percent Ap4ddA formed was plotted against time. The lines represent the best fit to the Michaelis-Menten equation using Sigmaplot 4.0. (B) Ap4ddA synthesis as a function of ATP concentration. Reactions were carried out as for panel A but with 0.2 to 6.4 mM ATP and for 1 to 10 min, depending on the enzyme, to allow a maximum of 35% Ap4ddA formation. The rate of Ap4ddA formation was calculated and plotted against time. (C) PPi-dependent formation of [32P]ddATP (pyrophosphorolysis). Experiments were performed as described for panel A but with 5 μM PPi instead of ATP. The amount of radioactivity in primer and ddATP was quantitated by phosphorimaging, and the percentage of label in ddATP was plotted against time.
Similar methods were used to measure the pyrophosphorolysis reaction catalyzed by WT and mutant enzymes. The [32P]ddAMP-terminated primer-template was incubated with WT or mutant RTs in the presence of PPi, and 32P-labeled ddATP was quantitated by phosphorimaging (Fig. 2C). As with AZTMP removal, the mutant enzymes had pyrophosphorolysis activities that were at most equal to that of WT RT.
Specificity of the acceptor substrate used in dinucleoside polyphosphate synthesis.
To define the role of the AZT resistance mutations in the primer unblocking reaction, we tested the effects of various nucleotide acceptor substrates on the efficiency of removal of ddAMP (Table 2). For WT RT there was little difference in the kcat/Km (specificity constant) for dinucleoside polyphosphate synthesis using ATP, UTP, UDP, or methylated ATP or UTP as the substrate. In contrast, for the three mutant enzymes, the specificity constants for ATP were greater than for UTP, and the specificity constants for the methylated forms of ATP and UTP were greater than for the unmethylated nucleotides. Each enzyme used 5,6-dihydro-UTP less efficiently than UTP, and the differences between mutant and WT RT were smaller with dihydro-UTP. For example, kcat/Km for dinucleoside polyphosphate synthesis by 67N/70R/215Y/219Q RT was 7.4 times that of WT RT with UTP as the substrate but only 3.4 times that of WT RT with dihydro-UTP as the acceptor substrate. In agreement with a previous report (19), the mutant enzymes were similar to WT RT when UDP was used as the substrate.
TABLE 2.
Ability of WT and AZT-resistant RTs to use various nucleotides as substrate acceptors for removal of ddAMP from blocked primer-templates through dinucleoside polyphosphate synthesisa
| HIV-1 RT |
kcat/Km (M−1 s−1)
|
|||||
|---|---|---|---|---|---|---|
| ATP | Me-ATP | UTP | Me-UTP | Dihydro-UTP | UDP | |
| WT | 0.21 ± 0.04 (1.0) | 0.22 ± 0.02 (1.0) | 0.19 ± 0.01 (1.0) | 0.17 ± 0.08 (1.0) | 0.056 ± 0.008 (1.0) | 0.32 ± 0.04 (1.0) |
| 215Y | 0.82 ± 0.06 (3.9)b | 1.3 ± 0.3 (5.9)b | 0.42 ± 0.10 (2.2)c | 0.59 ± 0.08 (3.5)b | 0.056 ± 0.003 (1.0)c | 0.23 ± 0.001 (0.7)c |
| 41L/215Y | 1.9 ± 0.4 (9.0)b | 3.0 ± 1.1 (14)b | 0.55 ± 0.03 (2.9)b | 0.98 ± 0.26 (5.8)b | 0.10 ± 0.002 (1.8)b | 0.29 ± 0.01 (0.9)c |
| 67N/70R/215Y/219Q | 3.3 ± 0.6 (16)b | 4.6 ± 2.3 (21)c | 1.4 ± 0.1 (7.4)b | 1.6 ± 0.6 (9.4)b | 0.19 ± 0.01 (3.4)b | 0.51 ± 0.01 (1.6)c |
Experiments were performed as described in the legend to Fig. 2B with the indicated RT and nucleotide substrate. The rate of dinucleoside polyphosphate synthesis was plotted against nucleotide concentration and fitted to a hyperbola using Sigmaplot 4.0 to obtain the kcat and Km. Data are the averages of two to eight experiments ± standard deviations. Numbers in parenthesis are increase (fold) versus the WT value. Significance of difference from WT was estimated by Student's t test corrected for multiple comparisons. N6-methyladenosine-5′-triphosphate (Me-ATP), 5-methyluridine-5′-triphosphate (Me-UTP), and dihydro-UTP were purchased from TriLink Biotechnologies, Inc. and treated with thermostable pyrophosphatase as described in the legend to Fig. 1, except dihydro-UTP, which was treated with inorganic pyrophosphatase (Roche Molecular Biochemicals) at 37°C to minimize breakdown to the diphosphate.
P < 0.05.
Not significantly different from WT value.
Inhibition of primer rescue and dinucleoside polyphosphate synthesis by the next complementary dNTP.
It was demonstrated previously (19, 20) that removal of chain terminators by HIV-1 RT is determined by at least two factors, the catalytic efficiency of mutant or WT RT to carry out the removal reaction and the sensitivity of that reaction to inhibition by the next complementary dNTP. Removal of ddAMP, d4TMP, and ddTMP is inhibited by low concentrations of the next complementary dNTP, whereas removal of AZTMP is much less sensitive to this inhibition. Therefore, it was important to assess the sensitivity to dNTP inhibition for each mutant enzyme.
Rescue of AZTMP-terminated primer-template was measured as described above and inhibition by the next complementary dNTP (dGTP) was determined. The percent inhibition was plotted against dGTP concentration and fitted to a hyperbola to obtain the 50% inhibitory concentration (IC50) for each enzyme (Table 3). Rescue by WT, 41L, 41L/215Y, and 67N/70R/215Y/219Q RTs was slightly more sensitive to this inhibition than was rescue by either 215Y or 67N/70R/215F/219Q RT, but the differences were not significant.
TABLE 3.
Ability of the next complementary dNTP to inhibit ATP-dependent primer rescue by WT and mutant RTs
| HIV-1 RT | IC50 (μM) for next dNTPa
|
|
|---|---|---|
| AZTMP terminated primer-templateb | ddAMP terminated except as indicatedc | |
| WT | 80 ± 13 | 4.0 ± 0.7 |
| 67N/70R/215F/219Q | 149 ± 3 | 10.8 ± 1.9d |
| 41L | 53 ± 3 | 3.2 ± 0.1 |
| 215Y | 135 ± 25 | 8.3 ± 0.6d |
| 41L/215Y | 85 ± 16 | 4.9 ± 2.3 |
| 67N/70R/215Y/219Q | 62 ± 2 | 6.6 ± 2.2 |
The numbers were obtained from two to six experiments and are given as means ± standard deviations. By Student's t test, the difference from WT values was not significant for any of the mutant enzymes tested (P ≥ 0.05) except as indicated.
Primer rescue experiments were performed as described in the legend to Fig. 1A in the presence of dGTP. The percent of inhibition of primer rescue was plotted against [dGTP], and the data were fitted to a hyperbola to obtain the IC50s.
Ap4ddA synthesis experiments were performed as described in the legend to Fig. 2A in the presence of TTP. The percent of inhibition of Ap4ddA synthesis was plotted against [TTP], and the data were fitted to a hyperbola to obtain the IC50s.
P < 0.05.
We also measured inhibition of removal of ddAMP residues by TTP, the dNTP complementary to the next position on the [32P]ddAMP-terminated primer-template, by quantitating the formation of [32P]Ap4ddA in the presence of various concentrations of TTP for each of the mutant enzymes (Table 3). As previously reported (19), removal of AZTMP was much less sensitive to inhibition by the next complementary dNTP than removal of ddAMP for all of the enzymes studied. Effects of the mutations were parallel for AZTMP removal and Ap4ddA synthesis. In summary, the 215Y and 67N/70R/215F/219Q enzymes show slightly reduced ability to interact with the next complementary dNTP, but the effect is too small to be convincing in these experiments.
Conclusions.
The present studies show that the in vitro removal of AZTMP from the 3′ termini of DNA chains by HIV-1 RT containing AZT resistance mutations is roughly proportional to the level of AZT resistance observed in cell culture assays for recombinant viruses containing the same mutations. To illustrate, the rate of in vitro removal of AZTMP at a single termination site by 67N/70R/215Y/219Q RT and 67N/70R/215F/219Q RT was 5 to 10 times that by the WT enzyme, whereas in vivo, in plaque reduction assays, HIV-1 containing these mutations was 120- to 150-fold less sensitive to AZT than WT HIV-1 (14). 41L/215Y RT catalyzed AZTMP removal at 3.3 times the rate of WT enzyme, compared with 64-fold AZT resistance of virus encoding 41L/215Y RT in cell culture (14). Mutant RT containing only the 215Y mutation removed AZTMP at a rate 2.3 times that of WT RT, compared with a 16-fold AZT resistance for virus containing this mutation (14). RT containing the M41L mutation alone exhibited a minimal increase in AZTMP removal, and the M41L mutant virus showed only fourfold AZT resistance (14). Since many factors contribute to the quantitative measurement of drug sensitivity in cells, the phenotypic assay results cannot be interpreted in terms of the number of AZTMP residues incorporated and removed in vivo; however, we have shown that the difference between mutant and WT enzyme is greater when multiple unblocking and retermination events occur (19), and it is reasonable that an increase in removal of AZTMP at each site where termination occurs will be further amplified when replication of the entire viral genome is considered. Our results demonstrate a strong correlation between the in vitro and the in vivo effects of these mutations.
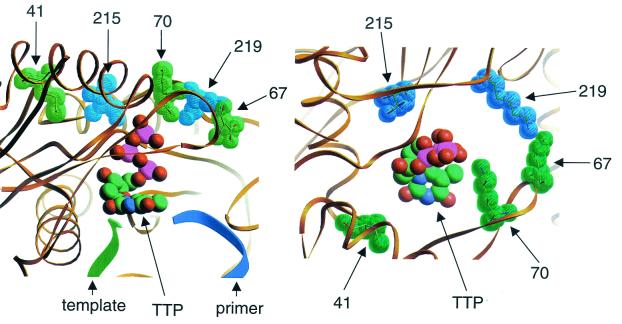
An indication of the role of these amino acid substitutions in the removal of chain terminators can be obtained from the molecular structures that have been reported for HIV-1 RT (11, 13, 15, 24, 26). Figure 3 shows the location of residues 41, 67, 70, 215, and 219 in WT RT in the structure of a covalently trapped catalytic complex reported by Huang et al. (11), which contains primer-template as well as the next complementary dNTP, TTP. The complex is identical whether the bound TTP is considered the substrate for polymerization or the product of PPi-dependent removal of TMP from the primer terminus. An analogous structure can be envisioned after ATP-dependent removal of TMP. However, instead of TTP, the product would be Ap4T, where the TTP portion, consisting of the TMP from the primer terminus and the γ- and β-phosphates from ATP, would occupy the position of TTP in the Huang et al. structure, and the α-phosphate and the adenosine moieties of ATP (not depicted in Fig. 3) would extend beyond the three phosphates of TTP in this structure.
FIG. 3.
Location of residues M41, D67, K70, T215, and K219 in the ternary complex of WT HIV-1 RT (11). The five residues that were substituted in the AZT resistance mutants in this study are shown as space-filling models. The rest of the protein is depicted as thin ribbons. DNA primer and template are shown as thick ribbons, and bound TTP is shown as a ball-and-stick model. The figure was produced by using RIBBONS (6) with the structure 1RTD coordinates retrieved from the Brookhaven Protein Data Bank. (Left) Side view; (right) top view looking down the phosphate chain of TTP. The primer and template are omitted from the right panel for simplicity. The shortest distances between the side chains of the residues and the γ-phosphate of TTP are as follows: M41, 11 Å; D67, 10 Å; K70, 7 Å; T215, 9 Å; K219, 8 Å. The corresponding distances to the 3′ OH of TTP are as follows: M41, 10 Å; D67, 15 Å; K70, 13 Å; T215, 10 Å; K219, 15 Å.
The amino acids corresponding to codons 41, 67, 70, 215, and 219 in the WT enzyme are shown by space-filling models in Fig. 3. These residues identify a region on the surface of the protein just distal to the γ-phosphate of the bound TTP (Fig. 3, left panel). When the structure is viewed by looking down the phosphate chain of TTP (Fig. 3, right panel), these amino acids surround a position that would be occupied by an extension of the phosphate chain described in the previous paragraph. Their locations suggest that they may facilitate the ATP-dependent removal reaction through interactions with the adenosine and phosphate moieties in reaction intermediates. Introduction of either phenylalanine or tyrosine at codon 215 is crucial for AZT resistance (16, 17, 25), suggesting that resistance is facilitated by the introduction of an aromatic group in a position that could allow formation of π-π interactions with the adenine base derived from ATP. This hypothesis is strongly supported by the data in Table 2. ATP is a more efficient substrate than UTP when tested with the mutant enzymes, in agreement with the expectation that a purine base will form stronger π-π interactions than a pyrimidine base (27). Introduction of a methyl group on the aromatic base of either ATP or UTP increased the catalytic efficiency of the reaction by the mutant enzymes, and methylation has been shown to enhance π-π interactions (3, 27). Finally, reduction of a double bond in UTP to form dihydro-UTP preferentially reduced the catalytic efficiency of the mutant enzymes, in agreement with the loss of aromatic structure resulting from this modification, which would disfavor π-π interactions. The M41L and T215Y mutations interact to enhance resistance in vivo (14) and our results are also consistent with a cooperative interaction between these mutations in vitro. These residues are not close enough to each other for direct interaction, and simultaneous interaction with the adenosine moiety of the reaction intermediate also seems unlikely. The molecular basis of their functional interaction remains to be determined.
From the structural considerations described above, we would predict that the rate of removal of chain-terminators would not be increased in the mutant enzyme when PPi is used as the acceptor substrate, and our results agree with this prediction. By contrast, Arion et al. (1) reported that the rate of pyrophosphorolysis of AZTMP-terminated primers was increased for 67N/70R/215F/219Q and 67N/70R mutant RTs. Canard et al. (4) have reported that the 67N/70R/215F/219Q RT has enhanced affinity for AZTMP-terminated primer-template, and it is possible that increased pyrophosphorolysis reported by Arion et al. (1) reflects this difference, since limiting enzyme concentrations were used in their assays. Our experiments were carried out with a large excess of WT or mutant enzyme, which should minimize the effects of differences in affinity for primer-template.
Ren et al. (24) have shown that the 215F/219Q mutations can induce long-range conformational changes in the enzyme and these structural perturbations may affect the dNTP-binding site. Our results and those in reference 19 suggest that three mutant enzymes (215Y, 215F/219Q, and 67N/70R/215F/219Q RTs) have decreased sensitivity to inhibition of removal of ddAMP by the next complementary dNTP, implying that the mutations facilitate the removal reaction both directly and indirectly. While the data suggest that dNTP binding is slightly impaired in these mutant enzymes, more direct assays may be needed to confirm this observation. Additional studies are also necessary to investigate the possible roles of other mutations associated with AZT resistance (including the 210W mutation and insertions at codon 69) in the removal reaction; however, the results in this report extend the previous conclusion (19) that enhanced nucleotide-dependent removal of AZTMP by AZT resistant RT plays an important role in the biochemical mechanism of AZT resistance.
Acknowledgments
We thank Hengemeh Bazmi at the University of Pittsburgh for providing us with the expression vector for 67N/70R/215Y/219Q RT and Arun Malhotra of the University of Miami for help with Fig. 3.
This work was supported by NIH grants AI-39973 (W.A.S.) and DK-26206 (A.G.S.).
REFERENCES
- 1.Arion, D., N. Kaushik, S. McCormick, G. Borkow, and M. A. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908-15917. [DOI] [PubMed] [Google Scholar]
- 2.Boyer, P. L., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broom, A. D., M. P. Schweizer, and P. O. P. Ts'o. 1967. Interaction and association of bases and nucleosides in aqueous solution. V. Studies of the association of purine nucleosides by vapor pressure osmometry and by proton magnetic resonance. J. Am. Chem. Soc. 89:3612-3622. [DOI] [PubMed] [Google Scholar]
- 4.Canard, B., S. R. Sarfati, and C. C. Richardson. 1998. Enhanced binding of azidothymidine-resistant human immunodeficiency virus 1 reverse transcriptase to the 3′-azido-3′-deoxythymidine 5′-monophosphate-terminated primer. J. Biol. Chem. 273:14596-14604. [DOI] [PubMed] [Google Scholar]
- 5.Carroll, S. S., J. Geib, D. B. Olsen, M. Stahlhut, J. A. Shafer, and L. C. Kuo. 1994. Sensitivity of HIV-1 reverse transcriptase and its mutants to inhibition by azidothymidine triphosphate. Biochemistry 33:2113-2120. [DOI] [PubMed] [Google Scholar]
- 6.Carson, M. 1997. Ribbons. Methods Enzymol. 277:493-505. [PubMed] [Google Scholar]
- 7.Cheng, Y.-C., G. E. Dutschman, K. F. Bastow, M. G. Sarngadharan, and R. Y. C. Ting. 1987. Human immunodeficiency virus reverse transcriptase. General properties and its interactions with nucleoside triphosphate analogs. J. Biol. Chem. 262:2187-2189. [PubMed] [Google Scholar]
- 8.D'Aquila, R. T., and W. C. Summers. 1989. HIV-1 reverse transcriptase/ribonuclease H: high level expression in Escherichia coli from a plasmid constructed using the polymerase chain reaction. J. Acquir. Immune Defic. Syndr. 2:579-587. [PubMed] [Google Scholar]
- 9.Furman, P. A., J. A. Fyfe, M. H. St. Clair, K. Weinhold, J. L. Rideout, G. A. Freeman, S. Nusinoff-Lehrman, D. P. Bolognesi, S. Broder, H. Mitsuya, and D. W. Barry. 1986. Phosphorylation of 3′-azido-3′-deoxythymidine and selective interaction of the 5′-triphosphate with human immunodeficiency virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 83:8333-8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh, J.-C., S. Zinnen, and P. Modrich. 1993. Kinetic mechanism of the DNA-dependent DNA polymerase activity of human immunodeficiency virus reverse transcriptase. J. Biol. Chem. 268:24607-24613. [PubMed] [Google Scholar]
- 11.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 12.Huang, P., D. Farquhar, and W. Plunkett. 1990. Selective action of 3′-azido-3′-deoxythymidine 5′-triphosphate on viral reverse transcriptases and human DNA polymerases. J. Biol. Chem. 265:11914-11918. [PubMed] [Google Scholar]
- 13.Jacobo-Molina, A., J. Ding, R. G. Nanni, A. D. Clark, Jr., X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, P. Clark, A. Hizi, S. H. Hughes, and E. Arnold. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 90:6320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellam, P., C. A. B. Boucher, J. M. G. H. Tijnagel, and B. A. Larder. 1994. Zidovudine treatment results in the selection of human immunodeficiency virus type 1 variants whose genotypes confer increasing levels of drug resistance. J. Gen. Virol. 75:341-351. [DOI] [PubMed] [Google Scholar]
- 15.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 16.Lacey, S. F., and B. Larder. 1994. Mutagenic study of codons 74 and 215 of the human immunodeficiency virus type 1 reverse transcriptase, which are significant in nucleoside analog resistance. J. Virol. 68:3421-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larder, B. 1994. Interactions between drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase. J. Gen. Virol. 75:951-957. [DOI] [PubMed] [Google Scholar]
- 18.Lennerstrand, J., K. Hertogs, D. K. Stammers, and B. A. Larder. 2001. Correlation between viral resistance to zidovudine and resistance at the reverse transcriptase level for a panel of human immunodeficiency virus type 1 mutants. J. Virol. 75:7202-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer, P. R., S. E. Matsuura, A. M. Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35-43. [DOI] [PubMed] [Google Scholar]
- 20.Meyer, P. R., S. E. Matsuura, R. Schinazi, A. G. So, and W. A. Scott. 2000. Differential removal of thymidine nucleotide analogues from blocked DNA chains by human immunodeficiency virus reverse transcriptase in the presence of physiological concentrations of 2′-deoxynucleoside triphosphates. Antimicrob. Agents Chemother. 44:3465-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer, P. R., S. E. Matsuura, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 95:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitsuya, H., R. Yarchoan, and S. Broder. 1990. Molecular targets for AIDS therapy. Science 249:1533-1544. [DOI] [PubMed] [Google Scholar]
- 23.Reardon, J. E. 1993. Human immunodeficiency virus reverse transcriptase. A kinetic analysis of RNA-dependent and DNA-dependent DNA polymerization. J. Biol. Chem. 268:8743-8751. [PubMed] [Google Scholar]
- 24.Ren, J., R. M. Esnouf, A. L. Hopkins, E. Y. Jones, I. Kirby, J. Keeling, C. K. Ross, B. A. Larder, D. I. Stuart, and D. K. Stammers. 1998. 3′-Azido-3′-deoxythymidine drug-resistance mutations in HIV-1 reverse transcriptase can induce long range conformational changes. Proc. Natl. Acad. Sci. USA 95:9518-9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richman, D. D., J. C. Guatelli, J. Grimes, A. Tsiatis, and T. Gingeras. 1991. Detection of mutations associated with zidovudine resistance in human immunodeficiency virus by use of polymerase chain reaction. J. Infect. Dis. 164:1075-1081. [DOI] [PubMed] [Google Scholar]
- 26.Rodgers, D. W., S. J. Gamblin, B. A. Harris, S. Ray, J. S. Culp, B. Hellmig, D. J. Woolf, C. Debouck, and S. C. Harrison. 1995. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 92:1222-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saenger, W. 1984. Principles of nucleic acid structure, p. 134-137. Springer-Verlag, New York, N.Y.
- 28.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 29.Schinazi, R. F., B. Larder, and J. W. Mellors. 2000. Mutations in retroviral genes associated with drug resistance: 2000-2001 update. Int. Antivir. News 8:65-91. [Google Scholar]
- 30.Shi, C., and J. W. Mellors. 1997. A recombinant retroviral system for rapid in vivo analysis of HIV-1 susceptibility to reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 41:2781-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.St. Clair, M. H., C. A. Richards, T. Spector, K. J. Weinhold, W. H. Miller, A. J. Langlois, and P. A. Furman. 1987. 3′-azido-3′-deoxythymidine triphosphate as an inhibitor and substrate of purified human immunodeficiency virus reverse transcriptase. Antimicrob. Agents Chemother. 31:1972-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueno, T., and H. Mitsuya. 1997. Comparative enzymatic study of HIV-1 reverse transcriptase resistant to 2′-3′-dideoxynucleotide analogs using the single-nucleotide incorporation assay. Biochemistry 36:1092-1099. [DOI] [PubMed] [Google Scholar]