Abstract
The aminoglycosides paromomycin and neomycin were examined in Escherichia coli cells for an inhibitory effect on 30S ribosomal subunit assembly. Both compounds inhibited the growth rate, viable cell number, and protein synthesis rate with similar 50% inhibitory concentrations. Each drug also showed a concentration-dependent inhibition of 30S subunit formation. The inhibitory effect on 30S particle formation was approximately equivalent to the inhibitory effect on translation for these antibiotics.
Paromomycin and neomycin are aminoglycoside antibiotics, which are effective against a number of aerobic and facultative anaerobic gram-positive bacilli and staphylococci (9, 11). Paromomycin and neomycin differ in chemical structure by the functional group attached to the C′ 6 of ring 1. Paromomycin has a hydroxyl group at this position, while neomycin possesses an amino group (20).
Both antibiotics bind specifically to the 30S ribosomal subunit (20). They interact with the 16S rRNA of the ribosome within an internal loop of the decoding site (12, 16, 18). Binding to this region results in a conformational change of the conserved bases within the loop of the A-site, which facilitates high-affinity binding between the rRNA of the internal loop and rings I and II of the aminoglycoside antibiotic (7, 8, 15). The tightly bound antibiotic contributes to codon misreading and mistranslation of mRNA.
Previous work with a number of structurally different inhibitors of 50S subunit function has demonstrated that these antibiotics also inhibit 50S particle formation (reviewed by Champney [2]). These antibiotics halt 50S subunit assembly and cause accumulation of a precursor particle, which later becomes degraded by cellular ribonucleases (19). Inhibition of assembly is equivalent to inhibition of translation for most of these drugs (3-5). The present investigation examines the effects of paromomycin and neomycin on growing Escherichia coli cells. The results indicate that these aminoglycosides also have two inhibitory activities, preventing protein synthesis and 30S particle assembly in cells.
Studies were conducted with E. coli strain SK901 (1). Cells were grown at 37°C in tryptic soy broth (TSB) in the presence and absence of each antibiotic as described previously. Growth rates, cell viability, and 35S-labeled amino acid incorporation into proteins were determined as described previously (6). The previous methods for [3H]uridine labeling of cells and sucrose gradient sedimentation of ribosomal subunits were used, except the centrifugation time was 5.25 h (6).
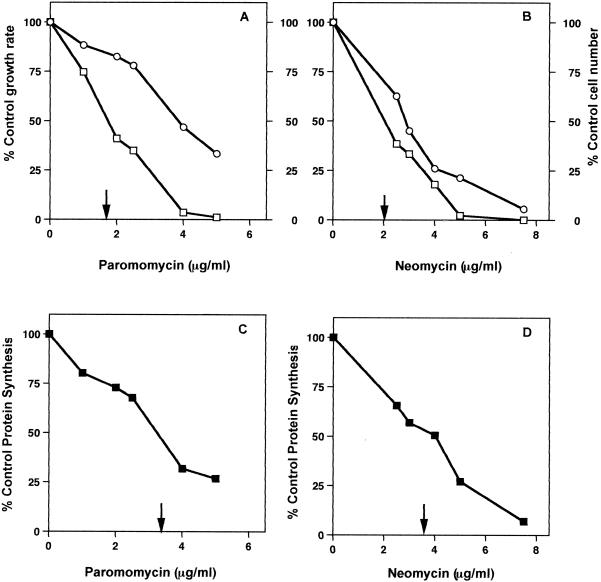
The MIC of each antibiotic was measured in E. coli cells growing in TSB. The MIC of paromomycin was 10 μg/ml, and that of neomycin was 15 μg/ml. The compounds were used at subinhibitory concentrations to investigate their effects on cell growth and protein synthesis. Each antibiotic increased the growth rate and reduced the cell viability in a similar fashion. Figure 1A and B show the concentration-dependent inhibition of the growth rate and cell number by these drugs. Paromomycin and neomycin had similar 50% inhibitory concentrations (IC50s) for effects on cell viability (1.6 and 2.0 μg/ml).
FIG. 1.
Inhibition of growth rates, viable cell numbers, and protein synthesis rates by increasing concentrations of paromomycin and neomycin in cells growing at 37°C. (A) Inhibitory effect of paromomycin on growth rate (○) and cell number (□). (B) Inhibitory effect of neomycin on growth rate (○) and cell number (□). (C) Concentration dependence of paromomycin inhibition of translation. (D) Concentration dependence of neomycin inhibition of translation. Results are the means of duplicate experiments with a standard error of ±13%. IC50s are indicated by arrows.
The rate of translation in growing E. coli cells was examined by measuring the incorporation of 35S-labeled amino acids into cellular proteins. Figure 1C and D show the effect of increasing concentrations of each drug on the rate of protein synthesis. Paromomycin and neomycin had IC50s of 3.2 and 3.6 μg/ml for inhibition of translation.
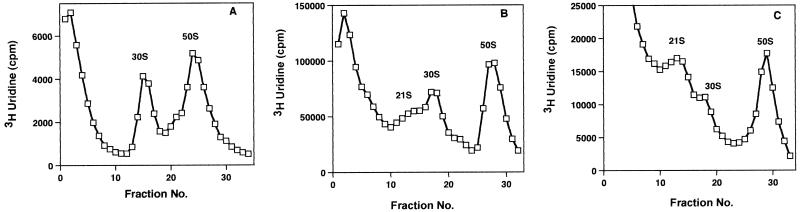
Both antibiotics were also examined to see if they had an effect upon 30S subunit formation. Figure 2 shows a comparison of the sucrose gradient profiles of lysates from [3H]uridine-labeled E. coli cells grown in the absence or presence of the two aminoglycosides. Growth in the presence of each antibiotic led to a reduction in 30S subunit amounts and to the accumulation of a 21S particle, suggestive of the presence of a precursor to the 30S subunit (17).
FIG. 2.
Sucrose gradient profiles for cells grown at 27°C with paromomycin or neomycin. (A) Sucrose gradient profile of [3H]uridine-labeled control cell lysate. (B) Sucrose gradient profiles from cells grown with paromomycin at 5 μg/ml. (C) Sucrose gradient profiles from cells grown with neomycin at 5 μg/ml.
The kinetics of ribosomal subunit assembly were examined by a [3H]uridine pulse- and chase-labeling procedure performed on cells growing at 27°C. We have shown previously that subunit assembly kinetics can be seen most clearly in cells growing at a reduced rate at 27°C (19), since ribosome assembly times are proportional to the growth rate of cells (10, 17).
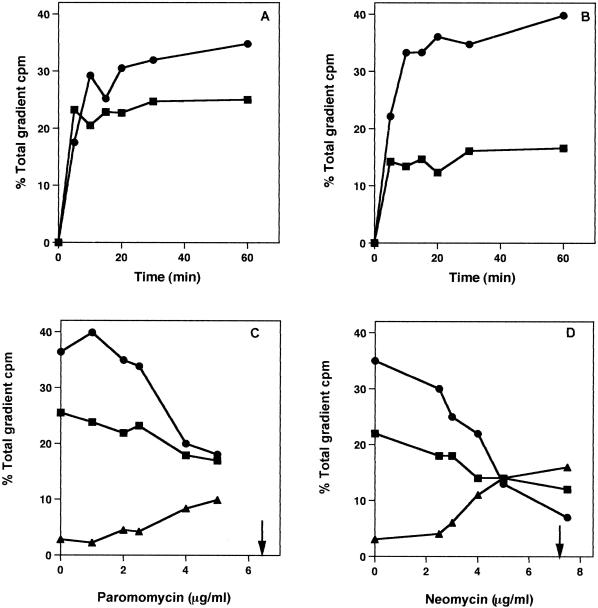
Figure 3 shows the kinetics of subunit assembly for the control (Fig. 3A) and for paromomycin (Fig. 3B)-treated cells. In the absence of the antibiotics, 30S subunit formation was completed in 15 min, and 50S particle synthesis was finished in 30 min (Fig. 3A). When the cells were treated with paromomycin, 30S subunit formation was halted at approximately 50% of the control amount, while 50S ribosomal subunit assembly was little affected. Similar results were found with neomycin-treated cells.
FIG. 3.
Ribosomal subunit synthesis rates and concentration dependence of ribosomal subunit assembly inhibition by paromomycin and neomycin. (A) Pulse-chase-labeling kinetics of 30S (▪) and 50S (•) subunits in control cells. (B). Pulse-chase-labeling kinetics of 30S (▪) and 50S (•) subunits in cells grown with paromomycin. (C) Inhibition of 30S (▪) and 50S (•) subunit formation by increasing concentrations of paromomycin. Increase in 21S particle amounts (▴). (D) Inhibition of 30S (▪) and 50S (•) subunit formation by increasing concentrations of neomycin. Increase in 21S particle amounts (▴). Results are the means of duplicate experiments with a standard error of ±3.5%. IC50s for 30S synthesis are indicated by arrows.
The antibiotic concentration dependence of subunit formation was tested as well. Figure 3C and D show the inhibitory effects of increasing concentrations of each antibiotic on 30S ribosomal subunit assembly. With increasing drug concentration, there was a decrease in 30S subunit amounts and an increased accumulation of a 21S particle. This relationship exists for both paromomycin (Fig. 3C) and neomycin (Fig. 3D). IC50s of 6.4 and 7.2 μg/ml were found for the two compounds. A decline in 50S subunit amounts was apparent at higher concentrations of the antibiotics.
The IC50s for the inhibition of translation and 30S particle formation in these cells differ by a factor of 2 for both drugs. This is the expected outcome for antibiotics with equivalent inhibitory effects on the two processes, since at the IC50 for 30S subunit formation, the cell contains only 50% of the number of functional subunits of control cells. Of these, 50% are inhibited in their translational function. As documented with a number of other antibiotics, inhibition of translation and 50S formation are equivalent effects of these compounds (3-5).
Inhibition of 50S particle assembly by 50S translational inhibitors does not affect 30S particle synthesis (2). The opposite may not be true. At higher concentrations of both drugs, an inhibition of 50S particle formation was also observed. This effect may be due to downstream inhibition of 50S synthesis. Transcription of 16S rRNA precedes transcription of 23S and 5S rRNAs, and concomitant 30S assembly precedes 50S particle formation (13, 14). Stalling of 30S synthesis could have a nonspecific downstream effect of slowing 50S synthesis without a direct effect of the antibiotic. Both aminoglycosides bind with specificity only to the 30S particle and stimulate misreading on this subunit (9, 20).
Paromomycin has been shown to stimulate mistranslation by locking a particular conformation of the 30S particle so that base pair mismatches are stimulated in the presence of the antibiotic (7, 8, 15). This observation suggests that these antibiotics could lock a precursor particle into a unique conformation and prevent rearrangements needed for further particle assembly and maturation. Conformational changes are required during particle assembly both in vivo and in vitro (14). Accumulation and turnover of a stalled 30S precursor particle could account for the bactericidal activity of these antibiotics in some cells (11, 20).
These results extend observations made with 50S subunit inhibitors to include antibiotics affecting 30S particle function. This expands the generality of the effect of ribosomal antibiotics as having two modes of inhibitory activity in growing cells.
Acknowledgments
We are pleased to acknowledge the assistance of Craig Tober with some of this work.
This research was supported in part by a grant from Pfizer Pharmaceuticals.
REFERENCES
- 1.Champney, W. S. 1979. Localized mutagenesis for the isolation of temperature-sensitive mutants of Escherichia coli affected in protein synthesis. Genetics 91:215-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Champney, W. S. 2001. Bacterial ribosomal subunit synthesis: a novel antibiotic target. Curr. Drug Targets-Infect. Disorders 1:19-36. [DOI] [PubMed] [Google Scholar]
- 3.Champney, W. S., and R. Burdine. 1996. 50S ribosomal subunit synthesis and translation are equivalent targets for erythromycin inhibition in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1301-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Champney, W. S., and R. Burdine. 1998. Azithromycin and clarithromycin inhibition of 50S ribosomal subunit formation in Staphylococcus aureus cells. Curr. Microbiol. 36:119-123. [DOI] [PubMed] [Google Scholar]
- 5.Champney, W. S., and C. L. Tober. 2001. Structure-activity relationships for six ketolide antibiotics. Curr. Microbiol. 42:203-210. [DOI] [PubMed] [Google Scholar]
- 6.Champney, W. S., C. L. Tober, and R. Burdine. 1998. A comparison of the inhibition of translation and 50S ribosomal subunit formation in Staphylococcus aureus cells by nine different macrolide antibiotics. Curr. Microbiol. 37:412-417. [DOI] [PubMed] [Google Scholar]
- 7.Fourmy, D., M. I. Recht, S. C. Blanchard, and J. D. Puglisi. 1996. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science 274:1367-1371. [DOI] [PubMed] [Google Scholar]
- 8.Fourmy, D., S. Yoshizawa, and J. D. Puglisi. 1998. Paromomycin binding induces a local conformational change in the A-site of 16 S rRNA. J. Mol. Biol. 277:333-345. [DOI] [PubMed] [Google Scholar]
- 9.Kotra, L. P., J. Haddad, and S. Mobashery. 2000. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 44:3249-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michaels, G. A. 1971. Variation in the proportion of 50 S and 30 S ribosomal subunits at different growth rates. J. Bacteriol. 107:385-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mingeot-Leclercq, M.-P., Y. Glupczynski, and P. M. Tulkens. 1999. Aminoglycosides: activity and resistance. Antimicrob. Agents Chemother. 43:727-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389-394. [DOI] [PubMed] [Google Scholar]
- 13.Nierhaus, K. 1982. Structure, assembly and function of ribosomes. Curr. Top. Microbiol. Immunol. 97:81-155. [DOI] [PubMed] [Google Scholar]
- 14.Nomura, M. 1973. Assembly of bacterial ribosomes. Science 179:863-873. [DOI] [PubMed] [Google Scholar]
- 15.Ogle, J. M., D. E. Brodersen, W. M. Clemons, M. J. Tarry, A. P. Carter, and V. Ramakrishnan. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897-902. [DOI] [PubMed] [Google Scholar]
- 16.Recht, M. I., S. Douthwaite, K. D. Dahlquist, and J. D. Puglisi. 1999. Effect of mutations in the A site of 16S RNA on aminoglycoside antibiotic-ribosome interactions. J. Mol. Biol. 286:33-43. [DOI] [PubMed] [Google Scholar]
- 17.Schlessinger, D. 1974. Ribosome formation in Escherichia coli, p. 393-416. In M. Nomura, A. Tissieres, and P. Lengyel (ed.), Ribosomes. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Schroeder, R., C. Waldsich, and H. Wank. 2000. Modulation of RNA function by aminoglycoside antibiotics. EMBO J. 19:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Usary, J., and W. S. Champney. 2001. Erythromycin inhibition of 50S ribosomal subunit formation in Escherichia coli cells. Mol. Microbiol. 40:951-962. [DOI] [PubMed] [Google Scholar]
- 20.Vazquez, D. 1979. Inhibitors of protein biosynthesis. Springer-Verlag, Berlin, Germany.