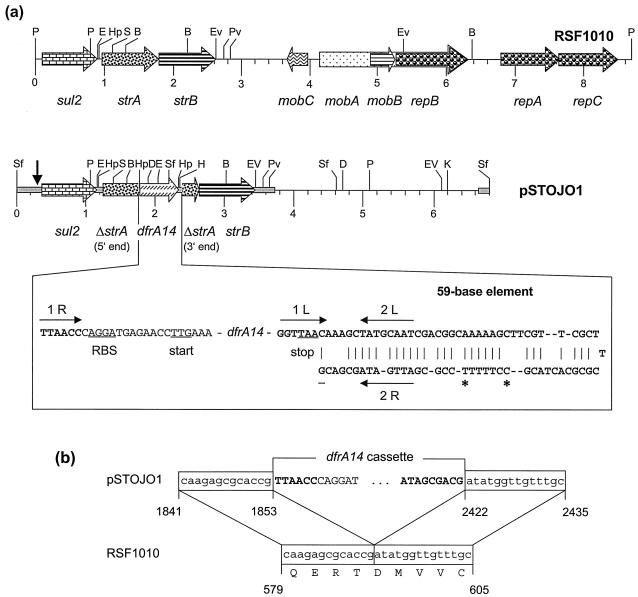
In a study of transferable antimicrobial resistance in uropathogenic Escherichia coli from humans in Nigeria, a small plasmid of 6.8 kb, designated pSTOJO1, was identified by transformation into E. coli JM107 to mediate resistance to sulfamethoxazole (Smz) and trimethoprim (Tmp). The MICs of Smz and the combination Tmp/Smz (4) were >1,024 μg/ml and >16/304 μg/ml, respectively, for both the original E. coli strain and E. coli JM107(pSTOJO1). Plasmid pSTOJO1 was mapped (Fig. 1a), and the two HindIII-KpnI fragments of 3.9 and 2.9 kb were cloned into pBluescript II SK+ (Stratagene, Amsterdam, The Netherlands). The sequence of a 3,839-bp segment of pSTOJO1 including the resistance genes was determined on both strands by primer walking starting at the HindIII cloning site of both fragments. It has been deposited with the EMBL database under accession no. AJ313522. Within this segment, a disrupted streptomycin resistance gene, strA, and three intact reading frames corresponding to the sulfonamide resistance gene sul2, the trimethoprim resistance gene dfrA14 (formerly known as dhfrIb), and the gene strB were detected (Fig. 1a).
FIG. 1.
(a) Restriction map and structural organization of plasmids RSF1010 (8) and pSTOJO1. Restriction endonucleases: B, BclI; D, DraI; E, EcoRI; EV, EcoRV; H, HindIII; Hp, HpaI; K, KpnI; P, PstI; Pv, PvuII; S, SacI; and Sf, SfuI. A distance scale in kilobases is presented below the map. The reading frames for the genes sul2, ΔstrA, strA, dfrA14, strB, mobA-C, and repA-C are shown as arrows, with the direction of transcription indicated by the arrowhead. The gray bar in pSTOJO1 indicates the sequenced part. The vertical arrow at the left end of the map of pSTOJO indicates the beginning of the RSF1010-related part of the pSTOJO1 sequence. Essential parts of the dfrA14 cassette are shown in more detail below the map of pSTOJO1. The ribosome binding site (RBS) and translational start and stop codons are underlined. In the 59-be, the putative IntI1 integrase binding domains 1L, 2L, 2R, and 1R (11) are indicated by arrows. The entire 59-be of the cassette is shown in boldface. The C marked with an asterisk is missing in the 59-be of the dfrA14 cassette of plasmid pUK1329 from E. coli (Z50805), and the T marked with an asterisk is missing in that of plasmid pHCM1 from S. enterica serovar Typhi (AL513383). (b) Comparison of the integration site of the dfrA14 cassette within strA in pSTOJO1 and the corresponding strA sequence of RSF1010 (8). The numbering refers to the positions in the database entries for pSTOJO1 (AJ313522) and RSF1010 (M28829).
The arrangement of the genes sul2-strA-strB closely resembled that known from plasmid RSF1010 (8), and sequence analysis confirmed 99% identity of the pSTOJO1 sequence from positions 353 to 1853 as well as 2422 to 3839 to the corresponding RSF1010 sequence. Except for these regions covering the genes sul2, strA, and strB, plasmids pSTOJO1 and RSF1010 appeared to be unrelated (Fig. 1a). The strA gene was disrupted by the insertion of a 568-bp element that carried a dfrA14 gene coding for a dihydrofolate reductase (2, 12). The dfrA14 gene was identical to the corresponding gene recently detected on a plasmid from Salmonella enterica serovar Typhimurium DT104 (AF393510) and exhibited 1-, 2-, and 5-bp differences from the sequences of the dfrA14 genes found in S. enterica serovar Typhi (AL513383) or E. coli (Z50805 and Z50804).
The gene dfrA14 has been reported to be part of a gene cassette, the size of which and the structure and length of the 59-base element (59-be) of which have been unknown (5). Analysis of the pSTOJO1 sequence identified the dfrA14 cassette to be 568 bp. The 59-be of the dfrA14 cassette consists of 87 bp and shows a central axis of symmetry (Fig. 1a). Since the submission of the pSTOJO1 sequence to the EMBL database, another two database entries for complete dfrA14 cassettes have become available (AL513383 and Z50805). Both 59-be sequences comprised 86 bp; the differences from the 59-be of the dfrA14 cassette from pSTOJO1 are indicated in Fig. 1a. In pSTOJO1, the dfrA14 cassette was found to be integrated at a secondary site within the strA gene. A similar situation was also seen in the E. coli plasmid pUK1329 (Z50805). No base pairs were lost or gained at the integration site (Fig. 1b), suggesting precise integration of the cassette. As a result, the strA gene was inactivated. Integration of a gene cassette at a secondary site was assumed to be an IntI-catalyzed recombination event, which involves a secondary recombination site (5, 6). The strA sequence at the integration site, GATAT, corresponded to the consensus sequences for secondary sites: Gt/aT (7) or Ga/tTa/ca/t (1). Precise integration of a complete aadB cassette at a secondary site between the genes repB and repA of RSF1010 (6), as well as in plasmid pRAY of a clinical isolate of Acinetobacter (9), has previously been reported. Moreover, truncation of an RSF1010-like strA gene by the insertion of the non-cassette-borne Tmp resistance gene, dfrA9, has also been reported (10) and is believed to have occurred as a consequence of the high selective pressure imposed by the frequent use of Tmp (3). A similar condition can be assumed for the development of the Smz/Tmp resistance plasmid pSTOJO1, since sulfonamides and Tmp are among the most frequently used antimicrobial drugs in Nigeria. Since integrase genes as well as plasmids carrying sul2-strA-strB genes are widespread among gram-negative bacteria, it is impossible to determine in retrospect where or when the recombination event between the dfrA14 cassette and the strA gene occurred.
REFERENCES
- 1.Francia, M. V., F. de la Cruz, and J. M. Garcia Lobo. 1993. Secondary sites for integration mediated by the Tn21 integrase. Mol. Microbiol. 10:823-828. [DOI] [PubMed] [Google Scholar]
- 2.Huovinen, P., L. Sundström, G. Swedberg, and O. Sköld. 1995. Trimethoprim and sulfonamide resistance. Antimicrob. Agents Chemother. 39:279-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jansson, C., A. Franklin, and O. Sköld. 1992. Spread of a newly found trimethoprim resistance gene, dhfrIX, among porcine isolates and human pathogens. Antimicrob. Agents Chemother. 36:2704-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; Approved standard, 5th ed. NCCLS document M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 5.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 6.Recchia, G. D., and R. M. Hall. 1995. Plasmid evolution by acquisition of mobile gene cassettes: plasmid pIE723 contains the aadB gene cassette precisely inserted at a secondary site in the IncQ plasmid RSF1010. Mol. Microbiol. 15:179-187. [DOI] [PubMed] [Google Scholar]
- 7.Recchia, G. D., H. W. Stokes, and R. M. Hall. 1994. Characterisation of specific and secondary recombination sites recognised by the integron DNA integrase. Nucleic Acids Res. 22:2071-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scholz, P., V. Haring, B. Wittmann-Liebold, K. Ashman, M. Bagdasarian, and E. Scherzinger. 1989. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75:271-288. [DOI] [PubMed] [Google Scholar]
- 9.Segal, H., and B. G. Elisha. 1997. Identification and characterization of an aadB gene cassette at a secondary site in a plasmid from Acinetobacter. FEMS Microbiol. Lett. 153:321-326. [DOI] [PubMed] [Google Scholar]
- 10.Sköld, O. 2001. Resistance to trimethoprim and sulfonamides. Vet. Res. 32:261-273. [DOI] [PubMed] [Google Scholar]
- 11.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 12.Young, H. K., M. J. Qumsieh, and M. L. McIntosh. 1994. Nucleotide sequence and genetic analysis of the type Ib trimethoprim resistant, Tn4132-encoded dihydrofolate reductase. J. Antimicrob. Chemother. 34:715-725. [DOI] [PubMed] [Google Scholar]