Abstract
The aim of the present study was to investigate the pharmacodynamics of moxifloxacin against strains of Streptococcus pyogenes with different susceptibilities to erythromycin by using an in vitro kinetic model simulating human pharmacokinetics of moxifloxacin at oral doses of 400 and 200 mg, respectively. When the different strains of S. pyogenes were exposed to the higher dose, the number of bacteria was reduced below the detection limit after 12 h and no regrowth was noted during the following 12 h. At the lower dose there was regrowth of the strains with constitutive and inducible erythromycin resistance of the MLSB phenotype. Replication assays of the regrowing bacteria indicated that the failure of moxifloxacin to kill the MLSB strains at the lower dose was likely caused by the emergence of preexisting resistant subpopulations. Thus, the present study indicates that the presently used 400-mg dose seems to have an advantage over the lower dose in that the risk for selection of resistant subpopulations is minimized.
During the past decade, pharmacodynamic parameters have become increasingly important for determining optimal dosing schedules of antibiotics (3-6, 12). It has been shown that the relationship between pharmacokinetic and pharmacodynamic parameters is different for different classes of antibiotics (3). The quinolones are characterized by a concentration-dependent bactericidal activity and the ability to induce a postantibiotic effect against both gram-positive and gram-negative bacteria (13-15). The ratio between the 24-h area under the concentration-time curve (AUC) and MIC (AUC/MIC) and the peak concentration/MIC ratio seem to be the parameters that correlate to efficacy both in animal studies and in human studies (3, 4, 5, 8, 12), and these ratios also have been correlated to higher survival rates in some animal studies (4). Several investigators have also studied the activity of quinolones in in vitro kinetic models and have shown that quinolones may differ in their pharmacodynamic properties (7, 10, 13, 15). Moxifloxacin is an 8-methoxyquinolone with enhanced antibacterial activity against gram-positive bacteria (1-2). The drug has a long terminal half-life of 12 h and protein binding of approximately 30%. The bioavailability is 90%, and the maximum concentration of drug in serum (Cmax) after a dose of 400 mg is approximately 3.8 mg/liter (16).
The aim of the present study was to investigate the pharmacodynamics of moxifloxacin against strains of Streptococcus pyogenes with different susceptibilities to erythromycin in an in vitro kinetic model simulating human pharmacokinetics. The strains used in the study included S. pyogenes NCTC (National Culture Type Collection) P1800 (erythromycin susceptible) and the following clinical strains: S. pyogenes 197 (erythromycin resistant, MLSB inducible, containing the gene ermTR), S. pyogenes 176 (erythromycin resistant, MLSB constitutive, containing the genes ermTR and ermB), and S. pyogenes 138 (erythromycin resistant by an efflux mechanism, containing the gene mefA), obtained from the Department of Bacteriology, Swedish Institute for Infectious Disease Control, Solna, Sweden. The MICs of moxifloxacin for the investigated strains were determined in triplicate in Todd-Hewitt broth on different occasions by a macrodilution technique with an inoculum of 5 × 105 CFU/ml, according to the guidelines of the National Committee for Clinical Laboratory Standards (11). The MICs were defined as the lowest concentrations of moxifloxacin that gave no visible growth. The MICs of benzylpenicillin and erythromycin for the same strains were determined with the Etest (Biodisk, Solna, Sweden).
(This material was presented in part at the 40th Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Ontario, Canada, 17 to 20 September, 2000.)
The pharmacodynamics of moxifloxacin for the investigated strains was studied in an in vitro kinetic model, consisting of a spinner flask with a 0.45-μm-pore-size filter membrane and a prefilter fitted in between the upper part and the bottom part in order to prevent bacterial dilution (9). A magnetic stirrer ensures homogenous mixing of the culture and prevents membrane pore blockage. In one of the sidearms of the culture vessel, a silicon membrane is inserted to enable repeated sampling. The other arm is connected by thin plastic tubing to a vessel containing fresh medium. The medium is removed from the culture flask through the filter at a constant rate with a pump. Fresh sterile medium is sucked into the flask at the same rate by the negative pressure built up inside the culture vessel. The antibiotic was added to the vessel and eliminated at a constant rate according to the first-order kinetics C = Co × e−kt, where Co is the initial antibiotic level, C is the antibiotic level at time t, k is the rate of elimination, and t is the time elapsed since the addition of antibiotic. The apparatus was placed in a thermostatic room at 37°C during the experiments. Before the experiments, the culture vessel was filled with Todd-Hewitt broth and bacteria were added at a starting inoculum of 5 × 105 CFU/ml. Two dose levels of moxifloxacin were used, which were under clinical evaluation when this study was initiated. The antibiotic was added at concentrations of 1.3 and 2.7 mg/liter, corresponding to the free (non-protein bound) fraction obtained after doses of 200 and 400 mg, respectively, and the flow rate was adjusted to give a half-life of 12 h. One sample was withdrawn at each of various times (0, 2, 4, 6, 8, 13, and 24 h), and if necessary the sample was diluted in phosphate-buffered saline. At least three dilutions of each sample were spread onto blood agar plates (Colombia agar base with 5% horse blood) and incubated at 37°C in 5% CO2, and the colonies were counted after 24 h. The limit of detection of the method was 5 × 101 CFU/ml. The initial experiments were performed in duplicate for each bacterial strain and concentration. Due to regrowth at the lower concentration, additional experiments were later performed for strains 197 and 176. To investigate if the regrowth was explained by the selection of resistant subpopulations, a replication assay was performed at 24 h for strains 197 and 176, respectively. In short, colonies grown from the plate at 24 h were replicated on moxifloxacin-containing blood agar plates (2 times, 5 times, and 20 times the MIC) and incubated for another 24 h, and the colonies were counted. Mutation frequency analysis was also done with the four original strains. Thirty individual cultures were inoculated with 1,000 bacteria, diluted from a fresh broth culture, and incubated overnight to obtain approximately 5 × 108 CFU/ml. The cultures were concentrated by centrifugation, and the pellets were suspended in phosphate-buffered saline. Plates containing moxifloxacin (2 times, 5 times, and 20 times the MIC) were seeded with 60 μl of the bacterial suspensions, giving approximately 5 × 107 bacterial cells. The plates were incubated overnight, and colonies were counted.
The concentrations of moxifloxacin during the in vitro kinetic experiments were determined by a microbiological agar diffusion method, with Escherichia coli MB 3804 as the test organism. A standardized inoculum of the bacterial suspension was mixed with Iso-Sensitest agar (Oxoid Ltd., Basingstoke, Hampshire, England) and poured onto plates. After the plates were dried, 0.03-ml volumes of all samples and standards diluted in Todd-Hewitt broth were applied to agar wells. The assays were made in triplicate, and the plates were incubated overnight at 37°C.
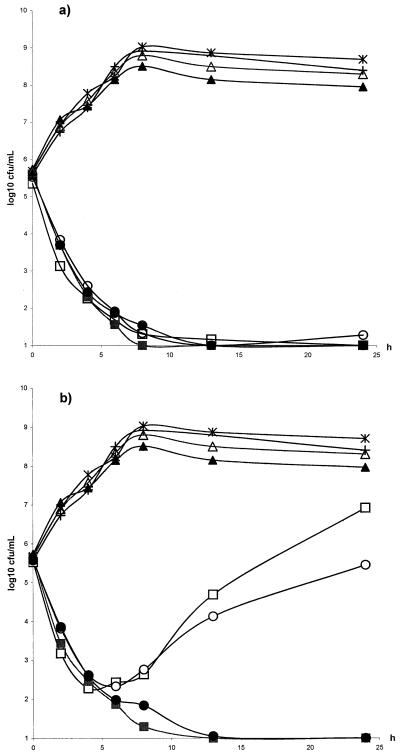
The strains investigated in the study were fully susceptible to penicillin (MIC = 0.016 mg/liter). Only strain NCTC P1800 was susceptible to erythromycin (MIC = 0.06 mg/liter). For all other strains, the MIC of erythromycin was ≥256 mg/liter. The MICs of moxifloxacin for the various strains are shown in Table 1. The MICs of moxifloxacin for the investigated strains of S. pyogenes did not differ more than one dilution step and were thus not correlated to erythromycin resistance, which is in accordance with the findings of other authors (1). The concentrations of moxifloxacin in the in vitro kinetic model corresponding to the free concentration obtained after a dose of 400 mg at 0, 12, and 24 h were the following (mean ± standard deviation): 2.7 ± 0.2 mg/liter, 1.49 ± 0.2 mg/liter, and 0.64 ± 0.13 mg/liter. The corresponding figures for the dose of 200 mg were 1.32 ± 0.15 mg/liter, 0.64 ± 0.08 mg/liter, and 0.28 ± 0.06 mg/liter. The ratios of AUC/MIC and peak concentration/MIC are presented in Table 1. At both doses, the 24-h time above MIC (T > MIC) was >100% for all strains. When the different strains of S. pyogenes were exposed to the higher dose, the number of bacteria was reduced below the detection limit after 12 h and no regrowth was noted during the following 12 h (Fig. 1a). At the lower dose there was regrowth of the strains with erythromycin resistance of the MLSB phenotype, both constitutive and inducible (Fig. 1b). The replication assay showed heavy growth for these two strains on plates containing two times the MIC. The frequency of the regrowing population of strains 197 and 176 on the plates containing five times the MIC was 10−4 and 2 × 10−3, respectively. Also, a few colonies of both strains grew on the plates containing 20 times the MIC. The mutation frequency analysis at twice the MIC showed at least a 1,000-fold increase in frequency for strains 197 and 176 compared to those for NCTC P1800 and strain 138. To our knowledge, there are no earlier pharmacokinetic/pharmacodynamic studies of quinolones and S. pyogenes. However, there are data concerning other pathogens. Earlier in our in vitro kinetic model we studied grepafloxacin and trovafloxacin against both penicillin-susceptible and penicillin-resistant strains of Streptococcus pneumoniae. When human pharmacokinetics were simulated, AUC24/MIC ratios with high values (>100) were obtained. No regrowth was noted in any of the experiments, but a 99.9% reduction in bacterial counts was obtained earlier with trovafloxacin compared with that obtained with grepafloxacin (2 h versus 6 h) (15). Lister and Sanders described similar results in an in vitro kinetic model examining trovafloxacin, ofloxacin, and ciprofloxacin. They noted a 99.9% reduction in bacterial counts for trovafloxacin against S. pneumoniae after 1 to 3 h, which was faster compared to that of the other two agents. In their experiments, the AUC/MIC for trovafloxacin was 300 to 600 (7). Craig has found in a thigh infection model that the 24-h AUC/MIC ratio is the parameter that correlates best with the efficacy of quinolones. They found that a ratio of ≥35 was necessary to produce a bacteriostatic effect. However, an AUC/MIC ratio of ≥100 was associated with almost zero mortality (3). These studies included both gram-positive and gram-negative strains. Forrest and coworkers found a similar ratio correlating to clinical and microbiological cure in critically ill patients with nosocomial pneumonia (5). However, it seems that the AUC/MIC needed to treat community-acquired infections and infections caused by gram-positive bacteria might be lower (12).
TABLE 1.
The MICs, AUC24/MICs, and peak concentration/MICs for moxifloxacin against the investigated strains
| Strain | MIC | AUC24/MIC at:
|
Peak concentration/MIC at:
|
||
|---|---|---|---|---|---|
| 200 mg | 400 mg | 200 mg | 400 mg | ||
| S. pyogenes NCTC P1800 | 0.125 | 138 | 286 | 10.6 | 21.6 |
| S. pyogenes 197 | 0.125 | 138 | 286 | 10.6 | 21.6 |
| S. pyogenes 176 | 0.25 | 69 | 143 | 5.3 | 10.8 |
| S. pyogenes 138 | 0.25 | 69 | 143 | 5.3 | 10.8 |
FIG. 1.
(a) Killing curves of moxifloxacin in the in vitro kinetic model. Cmax = 2.7 mg/liter, T1/2 = 12 h. The strains tested were S. pyogenes 197 (open squares), S. pyogenes 138 (filled squares), S. pyogenes 176 (open circles), and S. pyogenes NCTC P1800 (filled circles). The control strains were S. pyogenes 197 (open triangles), S. pyogenes 138 (filled triangles), S. pyogenes 176 (multiplication signs), and S. pyogenes NCTC 1800 (plus signs). (b) Killing curves of moxifloxacin in the in vitro kinetic model. Cmax = 1.3 mg/liter, T1/2 = 11.5 h. The strains tested were S. pyogenes 197 (open squares), S. pyogenes 138 (filled squares), S. pyogenes 176 (open circles), and S. pyogenes NCTC P1800 (filled circles). The control strains were S. pyogenes 197 (open triangles), S. pyogenes 138 (filled triangles), S. pyogenes 176 (multiplication signs), and S. pyogenes NCTC 1800 (plus signs).
In order to prevent selection of resistant mutants, Madaras-Kelly et al. studied ciprofloxacin and ofloxacin against Pseudomonas aeruginosa in an in vitro pharmacodynamic model, and their results suggested a value for AUC24/MIC of 100 as a breakpoint (10). In an animal model Drusano et al. could show that peak values of 10:1 and greater for lomefloxacin against P. aeruginosa were associated with better outcome, but when peak values of <10:1 were not reached the AUC/MIC ratio was a better marker to predict outcome (4). In the present study there was a consistent regrowth of the MLSB strains at the lower dose. The peak concentration/MIC ratios for the original population of strains 197 and 176 were 10.6 and 5.3, respectively, and the AUC24/MIC ratios were 138 and 69 at this dose. Our results indicate that the failure of moxifloxacin to kill the MLSB constitutive and inducible strains at the lower dose was likely caused by the emergence of preexisting resistant subpopulations. Thus, the present study indicates that the presently used 400-mg dose seems to have an advantage over the lower dose in that the emergence of resistant subpopulations is minimized.
Acknowledgments
This study was supported by a grant from Bayer AB.
We thank André Bryskier for identification of the resistant genes.
REFERENCES
- 1.Betriu, C., M. Redondo, M. L. Palau, A. Sanchez, M. Gómez, E. Culebras, A. Boloix, and J. J. Picazo. 2000. Comparative in vitro activities of linezolid, quinopristin-dalfopristin, moxifloxacin, and trovafloxacin against erythromycin-susceptible and -resistant streptococci. Antimicrob. Agents Chemother. 44:1838-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blondeau, J. M. 1999. A review of the comparative in-vitro activities of 12 antimicrobial agents, with a focus on the five new respiratory quinolones. J. Antimicrob. Chemother. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 3.Craig, W. A. 1998. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing in mice and men. Clin. Infect. Dis. 26:1-12. [DOI] [PubMed] [Google Scholar]
- 4.Drusano, G. L., D. E. Johnson, M. Rosen, and H. C. Standiford. 1993. Pharmacodynamics of a fluoroquinolone antimicrobial agent in a neutropenic rat model of pseudomonas sepsis. Antimicrob. Agents Chemother. 37:483-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forrest, A., D. E. Nix, C. H. Ballow, T. F. Goss, M. C. Birmingham, and J. J. Schentag. 1993. Pharmacodynamics of intravenous ciprofloxacin in seriously ill patients. Antimicrob. Agents Chemother. 37:1073-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leggett, J. E., B. Fantin, S. Ebert, K. Totsuka, B. Vogelman, W. Calame, et al. 1989. Comparative antibiotic dose-effect relations at several dosing intervals in murine pneumonitis and thigh-infection models. J. Infect. Dis. 159:281-292. [DOI] [PubMed] [Google Scholar]
- 7.Lister, P. D., and C. C. Sanders. 1999. Pharmacodynamics of trovafloxacin, ofloxacin and ciprofloxacin against Streptococcus pneumoniae in an in vitro pharmacokinetic model. Antimicrob. Agents Chemother. 43:1118-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lode, H., K. Borner, and P. Koeppe. 1998. Pharmacodynamics of fluoroquinolones. Clin. Infect. Dis. 27:33-39. [DOI] [PubMed] [Google Scholar]
- 9.Löwdin, E., I. Odenholt, and O. Cars. 1996. Pharmacodynamic effects of sub-MICs of benzylpenicillin against Streptococcus pyogenes in a newly developed in vitro kinetic model. Antimicrob. Agents Chemother. 40:2478-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madaras-Kelly, K. J., B. E. Ostergaard, L. Baeker Hovde, and J. C. Rotschafer. 1996. Twenty-four hour area under the concentration-time curve/MIC ratio as a generic predictor of fluoroquinolone antimicrobial effect by using three strains of Pseudomonas aeruginosa and an in vitro pharmacodynamic model. Antimicrob. Agents Chemother. 40:627-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. 1992. Methods for determining bactericidal activity of antimicrobial agents. Tentative Guideline M26-T. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 12.Preston, S. L., G. L. Drusano, A. L. Berman, C. L. Fowler, A. T. Chow, B. Dornseif, V. Reichl, J. Natarajan, and M. Corrado. 1998. Pharmacodynamics of levofloxacin; a new paradigm for early clinical trials. JAMA 279:125-129. [DOI] [PubMed] [Google Scholar]
- 13.Odenholt, I., E. Löwdin, and O. Cars. 1998. Bactericidal effects of levofloxacin in comparison with those of ciprofloxacin and sparfloxacin. Clin. Microbiol. Infect. 4:264-270. [DOI] [PubMed] [Google Scholar]
- 14.Odenholt, I., and S. Bengtsson. 1994. Postantibiotic-effect and postantibiotic effect of subinhibitory concentrations of sparfloxacin on gram-negative bacteria. Chemotherapy 40:30-36. [DOI] [PubMed] [Google Scholar]
- 15.Odenholt, I., T. Cars, and, E. Löwdin. 2000. Pharmacodynamic studies of trovafloxacin and grepafloxacin in vitro against gram-positive and gram-negative bacteria. J. Antimicrob. Chemother. 46:35-43. [DOI] [PubMed] [Google Scholar]
- 16.Stass, H., and D. Kubitza. 1999. Pharmacokinetics and elimination of moxifloxacin after oral and intravenous administration in man. J. Antimicrob. Chemother. 43:83-90. [DOI] [PubMed] [Google Scholar]