Abstract
The class C β-lactamase from Enterobacter cloacae P99 confers resistance to a wide range of broad-spectrum β-lactams but not to the newer cephalosporin cefepime. Using PCR mutagenesis of the E. cloacae P99 ampC gene, we obtained a Leu-293-Pro mutant of the P99 β-lactamase conferring a higher MIC of cefepime (MIC, 8 μg/ml, compared with 0.5 μg/ml conferred by the wild-type enzyme). In addition, the mutant enzyme produced higher resistance to ceftazidime but not to the other β-lactams tested. Mutants with 15 other replacements of Leu-293 were prepared by site-directed random mutagenesis. None of these mutant enzymes conferred MICs of cefepime higher than that conferred by Leu-293-Pro. We determined the kinetic parameters of the purified E. cloacae P99 β-lactamase and the Leu-293-Pro mutant enzyme. The catalytic efficiencies (kcat/Km) of the Leu-293-Pro mutant β-lactamase for cefepime and ceftazidime were increased relative to the respective catalytic efficiencies of the wild-type P99 β-lactamase. These differences likely contribute to the higher MICs of cefepime and ceftazidime conferred by this mutant β-lactamase.
Class C β-lactamases are the second most common class of β-lactam-hydrolyzing enzymes (8). Most of the class C β-lactamases are chromosomally encoded; they are thus less prone to mutational alteration of structure than are plasmid-encoded enzymes, whose structural genes exist in greater numbers of copies. Moreover, the development of clinically important new resistance phenotypes from chromosomally encoded β-lactamases would require not only mutation of the structural gene, as in the case of constitutive plasmid-mediated enzymes such as TEM-1, but also the genetic derepression of the structural gene to produce large amounts of enzyme. Recently, several bacterial strains expressing plasmid-encoded class C β-lactamases have been identified among clinical isolates (1, 5, 11, 14, 30, 31). The discovery of plasmid-encoded class C β-lactamases bodes ill for the prospects of β-lactam therapy in the future and may be a prelude to a potential explosion among clinical isolates of new variants of class C enzymes with extended substrate profiles, such as we have witnessed for class A β-lactamases.
The principal mechanism of resistance to extended-spectrum β-lactams in nosocomial pathogens such as Enterobacter is the production of class C β-lactamases by derepression of chromosomal genes (16, 27). This results in high-level resistance to virtually all penicillins and cephalosporins and to the monobactam aztreonam. To overcome such resistance, novel β-lactams that are poor substrates for these enzymes have been developed. Two classes of such β-lactams are currently in clinical use: newer cephalosporins (10, 17, 28), such as cefepime and cefpirome, and carbapenems (13), such as imipenem and meropenem. Class A β-lactamases, such as TEM and SHV, can mutate easily in clinical isolates to confer resistance to newer cephalosporins but not to carbapenems (http://www.lahey.org/studies/webt.htm). In contrast, such mutations of class C enzymes have rarely been reported to occur in clinical isolates (2, 3, 25). From the class C β-lactamase of Enterobacter cloacae P99, chosen because its X-ray crystallographic structure is known (20), we have selected a mutant that confers resistance to cefepime as a result of a single amino acid substitution, Leu-293-Pro. Site-directed mutagenesis of the corresponding codon generated mutants with 14 additional replacements of Leu-293 that resulted in alterations of conferred resistance to β-lactams. The kinetic properties of the β-lactamase from the cefepime-resistant mutant with replacement of Leu-293 by proline have been compared to those of the parental enzyme in this report.
(This work was previously presented in part [S. B. Vakulenko, B. Geryk, and S. A. Lerner, Abstr. 37th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-202, 1997].)
MATERIALS AND METHODS
Bacterial strains.
Escherichia coli JM83 {F− ara Δ(lac-proAB) rpsL (Strr) [φ80 dlac Δ(lacZ)M15] thi} (New England Biolabs) was utilized as a recipient strain for plasmids and as a host for the determination of susceptibilities to antibiotics. E. coli BMH 71-18 mutS {thi supE Δ(lac-proAB) [mutS::Tn10] [F′ proAB lacIqZΔM15]} (Clontech) was used as the intermediate host in site-directed mutagenesis experiments. E. coli BL21(DE3) [F− ompT hsdSB(rB− mB−) gal dcm (DE3)] (Novagen) was the host for target gene expression.
Antibiotics.
Ampicillin, ceftazidime, ceftriaxone, cefoperazone, cephaloridine, and kanamycin were from Sigma, aztreonam and cefepime were from Bristol-Myers Squibb, and piperacillin was from Lederle (Wyeth-Ayerst).
Plasmids for mutagenesis and DNA sequencing.
We constructed plasmid pUC19(ampC1) in the following manner. We replaced the DraI fragments of pUC19 containing the bla gene for the TEM β-lactamase with a HincII fragment of pUC4K (Pharmacia) containing the kanamycin resistance marker from Tn903 (construct I). The entire structural gene (plus the Shine-Dalgarno sequence) of the E. cloacae P99 β-lactamase was amplified by PCR from the genomic DNA of E. cloacae P99 (kindly provided by J.-M. Frère) with primers containing EcoRI sites near their 5′ ends. We used the resulting blunt-end product to replace the SspI-HindIII fragment in construct I after filling in the HindIII site. Expression of the E. cloacae P99 ampC β-lactamase gene was then driven by the TEM β-lactamase promoter, and therefore, a transformant containing this gene [pUC19(ampC1)] was selected by growth on ceftazidime-supplemented agar.
Mutagenesis and DNA sequencing.
We utilized Taq DNA polymerase and two primers containing EcoRI sites to reamplify the cloned ampC gene in order to introduce random mutations. After 30 cycles of PCR, the PCR product was digested with EcoRI, purified by using a QIAquick PCR purification kit (Qiagen), and substituted for the parental wild-type gene in pUC19(ampC1). The resulting construct was subsequently introduced into competent E. coli JM83 by high-efficiency electroporation, and selection was performed on the cefepime-supplemented agar. The entire sequences of the ampC genes from several cefepime-resistant strains were determined by the dideoxy chain termination method (29). Random site-directed mutagenesis of the E. cloacae P99 β-lactamase gene at the codon corresponding to Leu-293 of the enzyme was performed by using a transformer site-directed mutagenesis kit (Clontech) with the following mutagenic primer: GCGACAGTAAGGTAGCANNNGCGCCGTTGCCCGTGGCAG. After mutagenesis, transformants were selected by growth on cefepime or kanamycin, and the nucleotide sequence around and including the mutagenized codon was determined.
Expression and purification of parental and mutant E. cloacae P99 β-lactamases.
In order to facilitate β-lactamase secretion of both the wild-type and mutant β-lactamases into the growth medium, we replaced the DNA fragment corresponding to the original leader sequence of the AmpC β-lactamase with that of the leader sequence of the OmpA protein. To enhance the production of the parental and mutant enzymes, we cloned their genes fused to the OmpA leader sequence into the NdeI/HindIII sites of the pET-24a(+) vector (Novagen) under the control of the strong IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible bacteriophage T7 promoter. The resulting constructs were used to transform the recipient strain of E. coli BL21(DE3).
For enzyme purification, E. coli BL21(DE3) strains producing wild-type or mutant β-lactamase were inoculated in Luria-Bertani medium containing kanamycin at 20 μg/ml and grown overnight. Overnight cultures were diluted 100-fold in Terrific Broth (Difco) supplemented with 2 M sorbitol and 0.05 M betaine and incubated with shaking at 37°C until the cultures reached an optical density at 600 nm of 0.6. IPTG (0.4 mM) was added to each culture, and they were incubated overnight at 25°C. The cells were removed by centrifugation. The supernatant containing the enzyme was concentrated on an Amicon ultrafiltration device (membrane molecular size cutoff, 10,000 kDa). The concentrated protein was loaded onto a strong cation-exchange column (Macro-Prep high S support, 200 by 20 mm; Bio-Rad), equilibrated, and washed with 10 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] buffer (pH 6.8). The enzyme was eluted with a linear gradient of 0 to 1 M NaCl in 10 mM TES buffer (pH 6). The wild-type and mutant enzymes were purified by this procedure, and all appeared homogeneous according to the results of sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Determination of the amount of active enzymes in the periplasm.
Cells producing the wild-type and mutant P99 β-lactamases were grown at 37°C overnight in Luria-Bertani broth supplemented with 30 μg of kanamycin/ml. After 20-fold dilution into the same medium, the cells were regrown for another 2 h, and the cell densities of the two cultures were brought to the same value as determined spectrophotometrically at 600 nm. The cells were pelleted by centrifugation from 50 ml of culture, suspended in 1 ml of solution containing 27% (wt/vol) sucrose, 30 mM Tris, 5 mM EDTA, and 0.2 mg of lysozyme (pH 8.0), and incubated for 30 min at 4°C. Spheroplasts were removed by centrifugation for 5 min at 20,000 × g. Enzyme concentrations in the periplasm were determined from the hydrolysis rates of nitrocefin at the saturating concentration and turnover rate constants of both enzymes with nitrocefin.
Kinetic analysis of β-lactamases.
All kinetic measurements were performed on a Hewlett-Packard 8453 diode array spectrophotometer at 25°C in 100 mM sodium phosphate buffer (pH 7.0). Hydrolysis of each substrate was monitored at the corresponding wavelength: cephaloridine at 267 nm, Δɛ = 1,000 cm−1 M−1; ceftazidime at 280 nm, Δɛ = 3,725 cm−1 M−1; cefepime at 260 nm, Δɛ = 750 cm−1 M−1. The kinetic parameters for cephaloridine and ceftazidime turnover were evaluated from Hanes-Wolf plots, in which Km values were flanked by six points. The Km values of the wild-type enzyme for cefepime were determined as Ki by using this antibiotic as a competitive inhibitor versus nitrocefin, and the kcat/Km value was determined at substrate concentrations lower than Km with enzyme concentrations of 2 μM. The kinetic parameters for cefepime turnover by the mutant enzyme were evaluated from Hanes-Wolf plots, in which Km values were flanked by six points.
RESULTS
Cloning of the E. cloacae P99 ampC β-lactamase gene.
We constructed the pUC19(ampC1) vector by cloning the 1,172-bp fragment containing the entire structural gene for the E. cloacae P99 AmpC β-lactamase into construct I, a derivative of pUC19 bearing a kanamycin resistance marker, aph(3′)-Ia. The complete nucleotide sequence of the insert was confirmed, and restriction mapping revealed that the ampC and aph(3′)-Ia genes were transcribed from the same strand.
Generation and selection of cefepime-resistant mutants.
After PCR mutagenesis of the E. cloacae P99 ampC gene, the products were recloned into the pUC19(ampC1) vector. The resulting ligation mixture was used to electroporate E. coli JM83, and mutants were selected by growth on agar plates supplemented with 1, 2, 4, or 8 μg of cefepime/ml. Since there was no growth at the highest concentration, we chose four colonies from the plate with 4 μg of cefepime/ml for analysis of the β-lactamase gene. One mutant, with a single base change resulting in the replacement of Leu-293 with proline, was investigated further. The cefepime MIC for this mutant was 8 μg/ml. The β-lactamase structural gene from the mutant was recloned into the same vector and transformed into E. coli JM83; MICs of cefepime for transformants selected by growth in the presence of kanamycin were the same as those for the original mutant. Thus, the observed level of resistance to cefepime was due entirely to the replacement of Leu-293 by proline.
Mutagenic replacement of Leu-293.
Site-directed mutagenesis was carried out with a mixture of primers bearing all four nucleotides at the three positions of the codon corresponding to Leu-293. Initially, we sought mutants that conferred higher levels of resistance to cefepime by selection on cefepime plates, but all of the 20 mutants that grew in the presence of 4 μg of cefepime/ml had the same Leu-293-Pro mutation. Sequencing of ampC DNA from 70 of the transformants that grew on kanamycin revealed 15 different replacements of Leu-293, including those with proline.
Susceptibility of mutants to β-lactams.
Fifteen mutants, each with a different replacement of Leu-293 in the E. cloacae P99 β-lactamase, were tested for susceptibility to various β-lactams (Table 1). The cefepime MIC for the Leu-293-Pro mutant originally selected for growth on cefepime after random mutagenesis was the highest (8 μg/ml), 16-fold higher than that conferred by the wild-type enzyme. The same mutation increased the MIC of ceftazidime fourfold but reduced the MICs of ampicillin, aztreonam, ceftriaxone, and cephaloridine four- to eightfold. The cefepime resistance seen with the proline mutant was compromised by all of the other replacements. The highest residual MIC of cefepime (2 μg/ml) was conferred by residues with smaller, uncharged side chains (glycine, alanine, valine, and serine) but also by aspartate. The cefepime MICs for mutants with arginine, phenylalanine, histidine, tyrosine, and lysine were at or below that for the strain with the parental E. cloacae P99 β-lactamase. The profile of ceftazidime MICs for the various mutants followed closely that of cefepime MICs. In contrast to the profiles with cefepime and ceftazidime, there was no rise in resistance to ampicillin, piperacillin, ceftriaxone, cefoperazone, aztreonam, or cephaloridine. However, functional activity was retained against these β-lactams, since the MICs were in the range of 12.5 to 100% of those conferred by the wild-type E. cloacae P99 enzyme. The results with the Leu-293-Asp mutant were somewhat anomalous. Whereas the MICs of ampicillin, ceftazidime, cefoperazone, and cephaloridine conferred by this mutant were in line with those conferred by the other mutants, the MICs of piperacillin, ceftriaxone, and aztreonam were markedly (16- to 32-fold) reduced compared to those observed with the wild-type strain.
TABLE 1.
MICs of β-lactams conferred in E. coli JM83 by the E. cloacae P99 β-lactamase and its mutants with replacement of Leu-293
| Amino acid at res- idue 293 | MIC (μg/ml) of indicated β-lactama
|
|||||||
|---|---|---|---|---|---|---|---|---|
| AMP | CAZ | FEP | CRO | CFP | PIP | ATM | LOR | |
| Noneb | 4 | 0.125 | <0.015 | <0.03 | <0.06 | 0.5 | <0.03 | 4 |
| Leuc | 1,024 | 64 | 0.5 | 128 | 128 | 128 | 64 | 512 |
| Pro | 256 | 256 | 8 | 32 | 128 | 64 | 8 | 64 |
| Gly | 512 | 256 | 2 | 64 | 128 | 128 | 16 | 512 |
| Ala | 256 | 128 | 2 | 64 | 128 | 128 | 16 | 512 |
| Val | 512 | 128 | 2 | 64 | 128 | 256 | 32 | 256 |
| Ser | 512 | 128 | 2 | 64 | 128 | 128 | 16 | 256 |
| Cys | 512 | 128 | 1 | 64 | 64 | 128 | 32 | 256 |
| Asn | 512 | 128 | 1 | 32 | 64 | 64 | 32 | 512 |
| Glu | 512 | 64 | 1 | 32 | 128 | 128 | 16 | 256 |
| Met | 512 | 128 | 1 | 128 | 128 | 128 | 32 | 512 |
| Asp | 256 | 128 | 2 | 4 | 32 | 8 | 2 | 512 |
| Arg | 256 | 32 | 0.5 | 64 | 32 | 128 | 64 | 128 |
| Phe | 256 | 32 | 0.5 | 64 | 128 | 64 | 64 | 256 |
| His | 256 | 16 | 0.25 | 32 | 32 | 32 | 64 | 128 |
| Tyr | 256 | 32 | 0.25 | 32 | 128 | 32 | 64 | 256 |
| Lys | 256 | 32 | 0.25 | 64 | 32 | 64 | 64 | 128 |
Abbreviations: AMP, ampicillin; CAZ, ceftazidime; FEP, cefepime; CRO, ceftriaxone; CFP, cefoperazone; PIP, piperacillin; ATM, aztreonam; LOR, cephaloridine.
E. coli JM83 recipient strain.
Wild-type E. cloacae P99 β-lactamase.
Relative amount of active enzymes in the periplasm and their stability.
The experiments with enzymes extracted from the periplasm revealed that there was 2.4-fold less active Leu-293-Pro mutant enzyme than there was wild-type β-lactamase. Both enzymes were stable for at least 8 days at 4°C.
Kinetic studies.
We carried out kinetic studies with homogeneous preparations of the E. cloacae P99 β-lactamase and its mutant Leu-293-Pro variant (Table 2). The kinetic data that we obtained for the wild-type enzyme with cephaloridine, ceftazidime, and cefepime were within ranges reported previously (6, 9, 12, 24, 26). The catalytic efficiencies (kcat/Km) of the two enzymes with cephaloridine showed a 5.7-fold decline for the mutant β-lactamase, mostly resulting from the decline in kcat. On the other hand, the mutant enzyme was more efficient than the wild-type β-lactamase against ceftazidime and cefepime: the kcat/Km values were 11- and 27-fold higher, respectively. With ceftazidime, the change was mainly due to an eightfold increase in kcat, whereas with cefepime, the greater efficiency was due to the fourfold decrease in Km and the sixfold increase in kcat.
TABLE 2.
Kinetic constants for turnover of β-lactam substrates with the E. cloacae P99 β-lactamase and its Leu-293-Pro mutant derivativea
| β-Lactamase | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| LOR | |||
| Wild type | 68 ± 8 | 826 ± 62 | (12 ± 1) × 106 |
| Leu-293-Pro | 60 ± 6 | 126 ± 9 | (2.1 ± 0.2) × 106 |
| CAZ | |||
| Wild type | 15 ± 2 | 0.013 ± 0.001 | (9 ± 1) × 102 |
| Leu-293-Pro | 10 ± 3 | 0.10 ± 0.01 | (10 ± 3) × 103 |
| FEP | |||
| Wild type | 100 ± 16 | 0.5 ± 0.1 | (0.47 ± 0.03) × 104 |
| Leu-293-Pro | 24 ± 2 | 3.1 ± 0.2 | (13 ± 1) × 104 |
Abbreviations: LOR, cephaloridine; CAZ, ceftazidime; FEP, cefepime.
DISCUSSION
The proliferation of extended-spectrum class A β-lactamases and the hyperproduction of AmpC β-lactamases has led to the potential compromise of the clinical utility of most of the β-lactam antibiotics, especially in nosocomial infections. Furthermore, in contrast to the class A β-lactamases, the intrinsic resistance of class C enzymes to clinically available β-lactamase inactivators limits the option of using such inactivators in combination with penicillins or cephalosporins against strains producing these enzymes. The carbapenems, such as imipenem and meropenem, fortunately retain activity against many strains that hyperproduce AmpC-type β-lactamases. Moreover, the zwitterionic cephalosporin, cefepime, may retain its activity against most AmpC-producing strains of Enterobacter, including those with reduced permeability of their outer membrane resulting from mutational alteration of porin proteins (21). In fact, cefepime was shown to retain much greater activity than carbapenems against an AmpC-producing Klebsiella pneumoniae strain that lacks one of the porin proteins (22).
Unlike the prototypical TEM-1 and SHV-1 class A β-lactamases, the class C enzymes confer resistance broadly to cephalosporins as well as to penicillins. The resulting lack of selective pressure on class C β-lactamases by most β-lactams and the requirement for derepression of these naturally inducible enzymes to produce resistance might explain in part why mutants of class C β-lactamases have apparently not emerged commonly in the clinical setting. Only three cases of mutant class C β-lactamases in clinical isolates have been reported. Two showed unusual types of mutations: a duplication of three amino acids (25) and a deletion of six consecutive amino acid residues in the P99 β-lactamase (2). Although the strain of E. cloacae from the first report was shown to be highly resistant to cefuroxime, ceftazidime, and aztreonam, the wild-type P99 β-lactamase itself is known to confer clinically significant, though lower, resistance to these β-lactams. Regrettably, the strain producing this mutant enzyme was not tested for susceptibility to cefepime and other β-lactams that retain activity against Enterobacter strains which hyperproduce an AmpC β-lactamase. The third report revealed that a point mutation in the plasmid-encoded CMY-1 β-lactamase had resulted in new resistance to ceftazidime (3).
The increased utilization of newer cephalosporins such as cefepime for the treatment of infections caused by AmpC-producing bacterial pathogens may now provide the setting for the selection of mutant class C enzymes that confer resistance to an even broader spectrum of β-lactams. With this in mind, novel cefepime resistance was previously selected in E. cloacae P99 in vitro; the mutant enzyme was shown to bear the replacements T147R and V298E (R. Gomez, S. B. Vakulenko, and S. A. Lerner, Abstr. 97th Gen. Meet. Am. Soc. Microbiol., abstr. A-1, 1997). The importance of the V298E mutation in producing cefepime resistance was subsequently confirmed (23).
We report here the selection of a mutant of the E. cloacae P99 β-lactamase conferring a 16-fold rise in the MIC of cefepime. At the same time, the MIC of ceftazidime was enhanced fourfold, and there was little compromise of activity for a variety of other β-lactams. Although the β-lactamase genes we describe were expressed in E. coli from the TEM-1 promoter in a multicopy plasmid, we should point out that the levels of resistance to the various β-lactams conferred by the cloned wild-type gene were similar to those for E. cloacae P99 (data not shown).
The significant increase in the MIC of cefepime resulted from a single base pair change that replaced Leu-293 with proline. Although the insertion of proline into the primary structure of an efficient enzyme may compromise its function, in this case it is noteworthy that the efficiency of the mutant enzyme for cefepime increased along with that for ceftazidime.
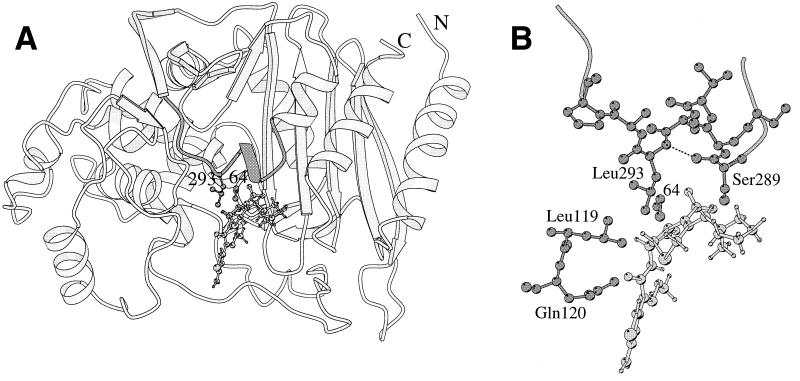
The crystal structure of the E. cloacae P99 β-lactamase (20) shows that the side chain of Leu-293 is located rather far (9.5 Å) from the reactive Ser-64, yet near the expected position of C3 substituents of cephalosporins (Fig. 1A). The side chain is rather exposed and not intimately involved in stabilizing the folded protein. Therefore, one might expect that the enzyme could tolerate many changes at this position, even though it is fairly conserved among class C enzymes. In fact, the modest effect on resistance to various β-lactams resulting from other substitutions at position 293 in our set of mutants supports this expectation.
FIG. 1.
(A) Ribbon representation of the crystallographic structure of the E. cloacae P99 β-lactamase (20). Cefepime is modeled near the reactive Ser-64. (B) A closer view of the environment of Leu-293. The hydrogen bond from the 293-NH to the carbonyl oxygen atom of Ser-289 is drawn as a dashed line. Hydrogen atoms are omitted from the protein structure. The figure was drawn by using MOLSCRIPT (18).
Since the relatively large side chain of Leu-293 in the wild-type P99 β-lactamase may possibly contact the C3 substituent of cefepime, substitution with a smaller side chain, as in proline, glycine, and alanine, apparently provides more space or a better fit for cefepime. Conversely, bulky residues such as phenylalanine and tyrosine are associated with the lowest cefepime MICs conferred. In addition, one may consider the electrostatic interaction between the positively charged C3 substituent of cefepime and residues in proximity such as that at position 293. Thus, positively charged residues at that site (arginine, histidine, and lysine) are also associated with the lowest levels of resistance to cefepime, perhaps because of mutual repulsion. On the other hand, negatively charged residues are associated with somewhat higher levels of conferred cefepime resistance.
The crystal structure shows that the replacement of Leu-293 by proline will have at least two structural consequences. An existing hydrogen bond (2.9 Å) from the backbone NH of Leu-293 to the CO of Ser-289 will be lost, as will a stabilizing hydrophobic interaction with the side chain of a neighboring Leu-119 (Fig. 1B), which lies in a conserved sequence: GLPL (residues 116 to 119). The first change is possible only with the substitution by proline. Combined with the narrowness of backbone conformations available to proline, both changes are highly likely to produce a conformational change somewhere in the 288 to 296 loop above the binding site. While details of the change are, of course, unknown, it is quite possible that a more flexible loop, or a more open one, results from the mutation, such that more space is available to an approaching substrate, with a reduction of Km. It is noteworthy that a recently described mutant E. cloacae β-lactamase that was responsible in part for resistance to cefepime in a clinical isolate lacked residues 289 to 294 (2).
The MIC of a β-lactam for β-lactamase-producing bacteria is a complex observation that includes not only the catalytic efficiency of the enzyme but also its concentration in the periplasm and the penetration of the drug though the outer membrane (6, 15, 19, 24). It should be noted (Tables 1 and 2) that the differences in catalytic efficiency between the wild-type and mutant enzymes for each substrate parallel the differences in the MICs of the respective β-lactam conferred by the two enzymes in the intact cells. Thus, the mutant enzyme conferred an MIC of cephaloridine eight times lower than that conferred by the P99 β-lactamase, and its catalytic efficiency (kcat/Km) was six times lower. The kcat/Km values of the mutant enzyme for ceftazidime and cefepime were 11 and 27 times higher, respectively, than those of the wild-type enzyme, and the MICs of these β-lactams conferred by the mutant enzyme were also higher, 4- and 16-fold, respectively. If one takes into account the fact that the concentration of the mutant enzyme in the periplasm is 2.4-fold lower than that of the wild type, the correlations between the MICs and kinetic parameters, especially for ceftazidime and cefepime, become even closer.
We have shown that the development of resistance to cefepime by mutational alteration of the E. cloacae P99 β-lactamase can occur readily by a single mutation. Recent reports have described AmpC-type β-lactamases encoded by plasmid genes that confer broad β-lactam resistance but not to cefepime (4, 7). Concern about the evolution of class C β-lactamases to confer resistance to cefepime is further heightened by the possibility of the plasmid-mediated spread of such mutant enzymes.
Acknowledgments
This work was supported in part by grants from Bristol-Myers Squibb and Dura/Elan Pharmaceuticals.
REFERENCES
- 1.Barnaud, G., G. Arlet, C. Verdet, O. Gaillot, P. H. Lagrange, and A. Philippon. 1998. Salmonella enteritidis: AmpC plasmid-mediated inducible β-lactamase (DHA-1) with an ampR gene from Morganella morganii. Antimicrob. Agents Chemother. 42:2352-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnaud, G., R. Labia, L. Raskine, M. J. Sanson-Le Pors, A. Philippon, and G. Arlet. 2001. Extension of resistance to cefepime and cefpirome associated to a six amino acid deletion in the H-10 helix of the cephalosporinase of an Enterobacter cloacae clinical isolate. FEMS Microbiol. Lett. 195:185-190. [DOI] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., Y. Chong, and K. Lee. 1998. Plasmid-encoded AmpC β-lactamases: how far have we gone 10 years after the discovery? Yonsei Med. J. 39:520-525. [DOI] [PubMed] [Google Scholar]
- 4.Bauernfeind, A., I. Schneider, R. Jungwirth, H. Sahly, and U. Ullmann. 1999. A novel type of AmpC β-lactamase, ACC-1, produced by a Klebsiella pneumoniae strain causing nosocomial pneumonia. Antimicrob. Agents Chemother. 43:1924-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauernfeind, A., I. Stemplinger, and R. Jungwirth. 1996. Characterization of the plasmidic β-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob. Agents Chemother. 40:221-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellido, F., J.-C. Pechère, and R. E. W. Hancock. 1991. Reevaluation of the factors involved in the efficacy of new β-lactams against Enterobacter cloacae. Antimicrob. Agents Chemother. 35:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bou, G., A. Oliver, M. Ojeda, C. Monzón, and J. Martínez-Beltran. 2000. Molecular characterization of FOX-4, a new AmpC-type plasmid-mediated β-lactamase from an Escherichia coli strain isolated in Spain. Antimicrob. Agents Chemother. 44:2549-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bush, K., and S. Mobashery. 1998. How β-lactamases have driven pharmaceutical drug discovery: from mechanistic knowledge to clinical circumvention, p. 71-98. In B. P. Rosen and S. Mobashery (ed.), Resolving the antibiotic paradox: progress in understanding drug resistance and development of new antibiotics. Plenum Press, New York, N.Y.
- 9.Bush, K., S. K. Tanaka, D. P. Bonner, and R. B. Sykes. 1985. Resistance caused by decreased penetration of β-lactam antibiotics into Enterobacter cloacae. Antimicrob. Agents Chemother. 27:555-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunha, B. A., and M. V. Gill. 1995. Cefepime. Med. Clin. N. Am. 79:721-732. [DOI] [PubMed] [Google Scholar]
- 11.Fosberry, A. P., D. J. Payne, E. J. Lawlor, and J. E. Hodgson. 1994. Cloning and sequence analysis of blaBIL-1, a plasmid-mediated class C β-lactamase gene in Escherichia coli BS. Antimicrob. Agents Chemother. 38:1182-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galleni, M., G. Amicosante, and J.-M. Frere. 1988. A survey of the kinetic parameters of class C β-lactamases. Cephalosporins and other β-lactam compounds. Biochem. J. 255:123-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garau, J., J. Blanquer, L. Cobo, S. Corcia, M. Daguerre, F. J. de Latorre, C. Leon, F. Del Nogal, A. Net, and J. Rello. 1997. Prospective, randomised, multicentre study of meropenem versus imipenem/cilastatin as empiric monotherapy in severe nosocomial infections. Eur. J. Clin. Microbiol. Infect. Dis. 16:789-796. [DOI] [PubMed] [Google Scholar]
- 14.Gazouli, M., L. S. Tzouvelekis, A. C. Vatopoulos, and E. Tzelepi. 1998. Transferable class C beta-lactamases in Escherichia coli strains isolated in Greek hospitals and characterization of two enzyme variants (LAT-3 and LAT-4) closely related to Citrobacter freundii AmpC beta-lactamase. J. Antimicrob. Chemother. 42:419-425. [DOI] [PubMed] [Google Scholar]
- 15.Hancock, R. E. W., and F. Bellido. 1992. Factors involved in the enhanced efficacy against Gram-negative bacteria of fourth generation cephalosporins. J. Antimicrob. Chemother. 29(Suppl. A):1-6. [DOI] [PubMed] [Google Scholar]
- 16.Itokazu, G. S., J. P. Quinn, C. Bell-Dixon, F. M. Kahan, and R. A. Weinstein. 1996. Antimicrobial resistance rates among aerobic gram-negative bacilli recovered from patients in intensive care units: evaluation of a national postmarketing surveillance program. Clin. Infect. Dis. 23:779-784. [DOI] [PubMed] [Google Scholar]
- 17.Jones, R. N., and S. A. Marshall. 1994. Antimicrobial activity of cefepime tested against Bush group I beta-lactamase-producing strains resistant to ceftazidime. Diagn. Microbiol. Infect. Dis. 19:33-38. [DOI] [PubMed] [Google Scholar]
- 18.Kraulis, P. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 19.Lakaye, B., A. Dubus, S. Lepage, S. Groslambert, and J.-M. Frère. 1999. When drug inactivation renders the target irrelevant to antibiotic resistance: a case story with β-lactams. Mol. Microbiol. 31:89-101. [DOI] [PubMed] [Google Scholar]
- 20.Lobkovsky, E., P. C. Moews, H. Liu, H. Zhao, J.-M. Frere, and J. R. Knox. 1993. Evolution of an enzyme activity: crystallographic structure at 2 Å resolution of the cephalosporinase from the ampC gene of Enterobacter cloacae P99 and comparison with a class A penicillinase. Proc. Natl. Acad. Sci. USA 90:11257-11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez-Martínez, L., M. C. Conejo, A. Pascual, S. Hernández-Allés, S. Ballesta, E. Ramírez de Arellano-Ramos, V. J. Benedí, and E. J. Perea. 2000. Activities of imipenem and cephalosporins against clonally related strains of Escherichia coli hyperproducing chromosomal β-lactamase and showing altered porin profiles. Antimicrob. Agents Chemother. 44:2534-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martínez-Martínez, L., A. Pascual, S. Hernández-Allés, D. Alvarez-Díaz, A. I. Suárez, J. Tran, V. J. Benedí, and G. A. Jacoby. 1999. Roles of β-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob. Agents Chemother. 43:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morosini, M.-I., M.-C. Negri, B. Schoichet, M.-R. Baquero, F. Baquero, and J. Blázquez. 1998. An extended-spectrum AmpC-type β-lactamase obtained by in vitro antibiotic selection. FEMS Microbiol. Lett. 165:85-90. [DOI] [PubMed] [Google Scholar]
- 24.Nikaido, H., and E. Y. Rosenberg. 1990. Outer membrane permeability and beta-lactamase stability of dipolar ionic cephalosporins containing methoxyimino substituents. Antimicrob. Agents Chemother. 34:337-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nukaga, M., S. Haruta, K. Tanimoto, K. Kogure, K. Taniguchi, M. Tamaki, and T. Sawai. 1995. Molecular evolution of a class C beta-lactamase extending its substrate specificity. J. Biol. Chem. 270:5729-5735. [DOI] [PubMed] [Google Scholar]
- 26.Nukaga, M., K. Taniguchi, Y. Washio, and T. Sawai. 1998. Effect of an amino acid insertion into the omega loop region of a class C β-lactamase on its substrate specificity. Biochemistry 37:10461-10468. [DOI] [PubMed] [Google Scholar]
- 27.Pitout, J. D. D., E. S. Moland, C. C. Sanders, K. S. Thomson, and S. R. Fitzsimmons. 1997. β-Lactamases and detection of β-lactam resistance in Enterobacter spp. Antimicrob. Agents Chemother. 41:35-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanders, C. C. 1993. Cefepime: the next generation? Clin. Infect. Dis. 17: 369-379. [PubMed] [Google Scholar]
- 29.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verdet, C., G. Arlet, S. Ben Redjeb, A. Ben Hassen, P. H. Lagrange, and A. Philippon. 1998. Characterization of CMY-4, an Amp C-type plasmid-mediated β-lactamase in a Tunisian clinical isolate of Proteus mirabilis. FEMS Microbiol. Lett. 169:235-240. [DOI] [PubMed] [Google Scholar]
- 31.Wu, S. W., K. Dornbusch, G. Kronvall, and M. Norgren. 1999. Characterization and nucleotide sequence of a Klebsiella oxytoca cryptic plasmid encoding a CMY-type β-lactamase: confirmation that the plasmid-mediated cephamycinase originated from the Citrobacter freundii AmpC β-lactamase. Antimicrob. Agents Chemother. 43:1350-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]