Abstract
Ten nonrepetitive Proteus mirabilis isolates, which were collected over 4 years (1996 to 1999) at the teaching hospital of Clermont-Ferrand, France, produced class D carbapenemase OXA-23. MICs of imipenem were 0.25 to 0.5 μg/ml for these clinical isolates. Molecular typing revealed that the 10 P. mirabilis isolates originated from the same clonal strain. Hybridization of I-CeuI-generated chromosome fragments with a blaOXA-23 probe showed that the gene was chromosome encoded in the P. mirabilis strain.
The β-lactamases are divided into four classes, designated A to D, on the basis of their amino acid contents (3). Class D β-lactamases include oxacillin-hydrolyzing, or OXA-type, enzymes, which are characterized by hydrolysis rates for cloxacillin and oxacillin higher than that for benzylpenicillin (9). Recently, oxacillinases OXA-23 to -27, which hydrolyze imipenem have been repeatedly associated with imipenem-resistant Acinetobacter baumannii (1, 2, 7, 11, 13).
During a survey of Proteus mirabilis β-lactamases performed in Clermont-Ferrand, France, we observed isolates that exhibited the β-lactam resistance phenotype of the IRT/OXA type and produced the same non-TEM β-lactamase of pI 6.9 (10). In this study, we report the characterization of this enzyme and its genetic support in 10 clinical P. mirabilis isolates. The 10 nonrepetitive P. mirabilis isolates were collected over 4 years (1996 to 1999) from different patients hospitalized in various wards of the teaching hospital of Clermont-Ferrand, France.
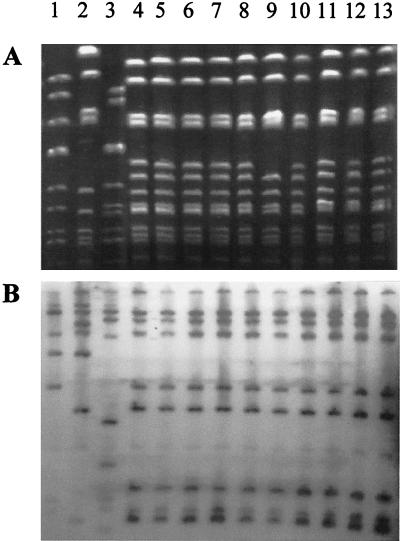
The 10 nonredundant P. mirabilis isolates were genotyped by ribotyping and pulsed-field gel electrophoresis (PFGE) (Fig. 1). Genomic DNAs were prepared from the 10 Proteus mirabilis isolates and 3 unrelated isolates. Ribotyping was performed as previously described (4) with endonuclease EcoRI (Boehringer). PFGE of restricted genomic DNA was performed with a CHEF-DR III apparatus (Bio-Rad Laboratories) as previously reported (12, 22), after DNA digestion by endonuclease NotI (New England Biolabs, Hertfordshire, United Kingdom). Lambda ladder (PFGE Marker 1; Boehringer Mannheim, Meylan, France) was used as a DNA molecular size marker. With both methods, the 10 OXA-23-producing isolates had indistinguishable (9 out of 10 strains) or closely related (1 strain displaying one genetic event of difference by PFGE only) patterns, which differed from those of non-OXA-23-producing P. mirabilis strains by more than seven DNA fragments in PFGE. The 10 OXA-23-producing P. mirabilis isolates therefore derived from a single clonal strain, designated CFO239, which had become established in our hospital over a period of at least 4 years.
FIG. 1.
Molecular typing of P. mirabilis isolates. (A) PFGE of NotI macrorestriction fragments. (B) Ribotyping. Lanes: 1 to 3, non-OXA-23-producing P. mirabilis strains; 4 to 13, OXA-23-producing P. mirabilis isolates.
The kinetic constants of the β-lactamase, carried out as previously described (14), showed that the β-lactamase of P. mirabilis CF0239 harbored the enzymatic features of a class D enzyme (data not shown). Probes specific to the major oxacillinase lineages (OXA-1, OXA-2, OXA-10, and OXA-9) did not hybridize with the total DNA of P. mirabilis. No amplification was obtained with primers located in the type I integrase gene and in the 5′ and 3′ conserved regions of type I integrons. The OXA-encoding gene was therefore cloned as follows. Genomic DNA was extracted as previously described (20) and partially cleaved by endonuclease Sau3A, and the resultant fragments were ligated in the BamHI site of pBK-CMV phagemid (Stratagene, La Jolla, Calif.). A transformant containing a >12-kb recombinant plasmid, pO239-8, was obtained on Mueller-Hinton agar supplemented with 32 mg of ticarcillin per ml. Subcloning was then carried out with plasmid pO239-8 by partial digestion with endonuclease Sau3A and purification of the resulting 1.5- to 3-kb DNA fragments by electroelution in a dialysis bag (20). Different recombinant plasmids were obtained, one of which, designated pO239-2, harbored a 2.2-kb insert. Sequencing of this insert by the procedure of Sanger et al. (21) showed the class D oxacillinase to be OXA-23, an enzyme previously observed in Acinetobacter baumannii 6B92 (11). Recombination sequence 59-BE, which is associated with gene cassettes inserted in integrons, was not observed, and the close genetic environment of blaOXA-23 in P. mirabilis was identical to that observed in A. baumannii.
OXA-23 is the first member of a novel lineage of class D β-lactamases. This cluster of class D comprises five enzymes (OXA-23 to -27) that have 60% homology. The enzymes harbored a weak carbapenemase activity (relative Vmax, 1 to 3% of those for penicillin G) (1, 2). These class D carbapenemases have so far been associated with imipenem-resistant A. baumannii strains. P. mirabilis CFO239 is therefore the first instance of a clinical strain from the family Enterobacteriaceae producing a class D carbapenemase.
The MICs of β-lactam antibiotics for clinical P. mirabilis CFO239, recombinant OXA-23-producing P. mirabilis strain ATCC 29906(pO239-8), and recombinant OXA-23-producing Escherichia coli strain DH5α(pO239-8) (20) were determined by dilution in Mueller-Hinton agar (Sanofi Diagnostics Pasteur, Marnes la Coquette, France) with an inoculum of 104 CFU per spot (Table 1). The β-lactam resistance phenotype of OXA-23-producing strains was characterized by resistance to amoxicillin and ticarcillin alone and combined with clavulanate. MICs of cephalothin and cefpirome were higher than those of P. mirabilis ATCC 29906 and E. coli DH5α. MICs of cefoxitin, aztreonam, ceftazidime, and cefotaxime were unchanged. The MIC ranges of imipenem and meropenem for clinical P. mirabilis isolates were 0.25 to 0.5 and 2 to 4 μg/ml, respectively. The MICs of carbapenem for OXA-23-producing transformant E. coli DH5α(pO239-8) and P. mirabilis ATCC 29906(pO239-8) were fourfold higher than those for E. coli DH5α and P. mirabilis ATCC 29906. The weak carbapenemase activity of OXA-23 (1, 2, 11, 13) probably increases the MIC of imipenem, but our study provided no evidence that the enzyme confers imipenem resistance in P. mirabilis and E. coli. Therefore, the relative nonsusceptibility observed probably has no major clinical consequence, but could be involved in the persistence of the OXA-23-producing P. mirabilis strains in our hospital. P. mirabilis isolates exhibiting a higher level of resistance to imipenem (MIC, 8 μg/ml) have been previously reported in strains harboring alterations in penicillin-binding proteins or outer membrane proteins (17, 18, 23). Marier (16) suggests that the fairly high MIC of imipenem could explain why P. mirabilis is not eradicated from soft tissue. These results also suggest the presence of additional resistance factors that enhance resistance to imipenem in class D carbapenemase-producing A. baumannii, for which imipenem MICs were between 16 and 64 μg/ml.
TABLE 1.
MICs of β-lactam antibiotics for OXA-23-producing strains in comparison with those for P. mirabilis ATCC 29906 and E. coli DH5α
| Strains | MIC (μg/ml) for:
|
||||
|---|---|---|---|---|---|
|
P. mirabilis
|
E. coli
|
||||
| CFO239a | ATCC 29906 (pO239-8)b | ATCC 29906 | DH5α (pO239-8)b | DH5α | |
| Amoxicillin | 256 | >1,024 | 1 | >1,024 | 2 |
| Amoxicillin-CLAc | 256 | >1,024 | 1 | >1,024 | 2 |
| Ticarcillin | 64 | >1,024 | 0.5 | >1,024 | 2 |
| Ticarcillin-CLA | 64 | 1,024 | 0.5 | 1,024 | 1 |
| Piperacillin | 16 | 64 | 0.5 | 32 | 1 |
| Piperacillin-TZBd | 0.5 | 4 | 0.5 | 4 | 1 |
| Cephalothin | 4 | 64 | 8 | 16 | 4 |
| Cefoxitin | 2 | 2 | 4 | 4 | 4 |
| Cefotaxime | 0.06 | 0.25 | 0.06 | 0.06 | 0.06 |
| Cefpirome | 1 | 32 | 0.06 | 0.5 | 0.06 |
| Ceftazidime | 0.06 | 0.06 | 0.06 | 0.25 | 0.12 |
| Aztreonam | 0.06 | 0.06 | 0.06 | 0.12 | 0.12 |
| Meropenem | 2-4 | 4 | 1 | 0.5 | 0.12 |
| Imipenem | 0.25-0.5 | 1 | 0.25 | 0.5 | 0.12 |
MIC range for the 10 clinical P. mirabilis CFO239 isolates producing OXA-23.
Recombinant strains containing OXA-23-encoding plasmid pO239-8.
CLA, clavulanate at a fixed concentration of 2 μg/ml.
TZB, tazobactam at a fixed concentration of 4 μg/ml.
In A.baumannii 6B92, blaOXA-23 is located on a 40-kb plasmid. As in previous reports on A. baumannii 6B92, blaOXA-23 was not transferred in E. coli C600 (20) at 37 or 30°C in solid or liquid Mueller-Hinton medium. No plasmid DNA was observed in the extracts, which were prepared by the Qiagen method (Diagen, Hilden, Germany) and the method of Birnboim and Doly (5). Electroporation of DNA extracts did not yield transformants on Mueller-Hinton agar containing ticarcillin (8 μg/ml).
To search for a possible chromosomal location of OXA-23 genetic support, we used the endonuclease I-CeuI (New England Biolabs, Hertfordshire, United Kingdom), which digests a 26-bp sequence in rrn genes for the 23S large-subunit rRNA (15). After digestion, the separation of the resulting fragments was performed on the CHEF-DRIII apparatus as previously reported (15). The sizes of I-CeuI-generated fragments were determined by comparison with Saccharomyces cerevisiae chromosomal DNA (Bio-Rad Laboratories). As with other bacterial species of the family Enterobacteriaceae (15), seven fragments were generated from DNA of OXA-23-producing P. mirabilis CF0239 and clinical P. mirabilis strains without the blaOXA-23 gene (>2.2 Mb, 680 kb, 600 kb, 365 kb, 325 kb, 285 kb, and 42 kb), as well as from DNA of an E. coli strain.
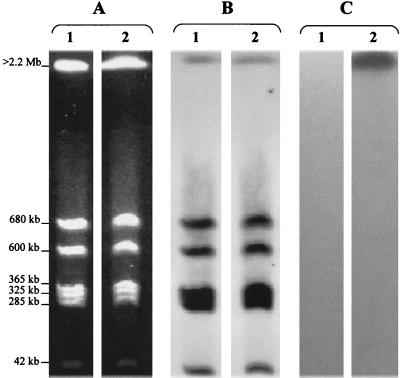
After immobilization on Nytran filters (Schleicher and Schuell), the I-CeuI-generated fragments were hybridized with three different probes: the 16S and 23S rRNA probe (Roche Diagnostics, Meylan, France), a blaOXA-23 probe, and a probe specific to the ampC gene of E. coli. Labeling, hybridization, and revelation were performed as previously reported (6). The rRNA probe hybridized with the seven I-CeuI-generated fragments. The blaOXA-23 probe hybridized only with the >2.2-Mb fragment of strain CF0239 (Fig. 2). ampC probes hybridized only with the DNA of E. coli, according to the nucleic sequence of the E. coli K12 chromosome (data not shown). These data indicate the chromosomal location of the blaOXA-23 gene in P. mirabilis CFO239 and that the overall organization of the genome observed in P. mirabilis CFO239 (as in Salmonella spp., Morganella morganii, and Proteus rettgeri) (15) is closely related to that of E. coli.
FIG. 2.
Localization of blaOXA-23 in I-Ceu I-generated chromosome fragments of P. mirabilis CFO239 separated by PFGE. (A) Chromosome restriction patterns. (B) Hybridization of restricted patterns with a probe specific to 23S rRNA genes. (C) Hybridization of restricted patterns with a probe specific to the blaOXA-23 gene. Lanes: 1, clinical P. mirabilis without the blaOXA-23 gene; 2, OXA-23-producing P. mirabilis CFO239.
Initially, P. mirabilis CFO239 must have acquired an OXA-23-encoding plasmid. The plasmid may have been unable to replicate in P. mirabilis. For the blaOXA-23 gene to be maintained in the strain, it must therefore have been inserted in the chromosome probably by genetic events of recombination, cointegration, or transposition. To our knowledge, this is the first report of a chromosome-encoded class D enzyme in the family Enterobacteriaceae. Class A and C enzymes (GN-79 and CMY-3), which are generally plasmid encoded, have been observed in the chromosome of P. mirabilis strains (8, 19). The putative insert sequence ISEcp-1 was observed upstream of blaCMY-3 (data not shown), but was not detected in P. mirabilis CFO239.
Acknowledgments
We thank Mathieu Longeon, Rolande Perroux, Marlène Jan, and Dominique Rubio for technical assistance. We are also grateful to Jeffrey Watts for help with the English in the manuscript.
This work was supported in part by a grant from Ministère de l'Education Nationale, de la Recherche et de la Technologie.
REFERENCES
- 1.Afzal-Shah, M., H. E. Villar, and D. M. Livermore. 1999. Biochemical characteristics of a carbapenemase from an Acinetobacter baumannii isolate collected in Buenos Aires, Argentina. J. Antimicrob. Chemother. 43:127-131. [DOI] [PubMed] [Google Scholar]
- 2.Afzal-Shah, M., N. Woodford, and D. M. Livermore. 2001. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D β-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 45:583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambler, R. P., A. F. W. Coulson, J.-M. Frère, J.-M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276:269-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bingen, E., C. Boissinot, P. Desjardins, H. Cave, N. Brahimi, N. Lambert-Zechovsky, E. Denamur, P. Blot, and J. Elion. 1993. Arbitrarily primed polymerase chain reaction provides rapid differentiation of Proteus mirabilis isolates from a pediatric hospital. J. Clin. Microbiol. 31:1055-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnet, R., J. L. M. Sampaio, C. Chanal, D. Sirot, C. De Champs, J. L. Viallard, R. Labia, and J. Sirot. 2000. A novel class A extended-spectrum β-lactamase (BES-1) in Serratia marcescens isolated in Brazil. Antimicrob. Agents Chemother. 44:3061-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bou, G., G. Cerveró, M. A. Dominguez, C. Quereda, and J. Martínez-Beltrán. 2000. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of β-lactamases. J. Clin. Microbiol. 38:3299-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bret, L., C. Chanal-Claris, D. Sirot, E. B. Chaibi, R. Labia, and J. Sirot. 1998. Chromosomally encoded AmpC-type β-lactamase in a clinical isolate of Proteus mirabilis. Antimicrob. Agents Chemother. 42:1110-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chanal, C., R. Bonnet, C. De Champs, D. Sirot, R. Labia, and J. Sirot. 2000. Prevalence of β-lactamases among 1,072 clinical strains of Proteus mirabilis: a 2-year survey in a French hospital. Antimicrob. Agents Chemother. 44:1930-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donald, H. M., W. Scaife, S. G. Amyes, and H.-K. Young. 2000. Sequence analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob. Agents Chemother. 44:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gouby, A., C. Neuwirth, G. Bourg, N. Bouzigues, M. J. Carles-Nurit, E. Despaux, and M. Ramuz. 1994. Epidemiological study by pulsed-field gel electrophoresis of an outbreak of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in a geriatric hospital. J. Clin. Microbiol. 32:301-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hornstein, M., C. Sautjeau-Rostoker, J. Péduzzi, A. Vessiéres, L. T. Hong, M. Barthélémy, M. Scavizzi, and R. Labia. 1997. Oxacillin-hydrolyzing beta-lactamase involved in resistance to imipenem in Acinetobacter baumannii. FEMS Microbiol. Lett. 153:333-339. [DOI] [PubMed] [Google Scholar]
- 14.Labia, R., J. Andrillon, and F. Le Goffic. 1973. Computerized microacidimetric determination of β-lactamase Michaelis-Menten constants. FEBS Lett. 33:42-44. [DOI] [PubMed] [Google Scholar]
- 15.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marier, R. L. 1985. Role of imipenem/cilastatin in the treatment of soft tissue infections. Am. J. Med. 78:140-144. [DOI] [PubMed] [Google Scholar]
- 17.Mehtar, S., A. Tsakris, and T. L. Pitt. 1991. Imipenem resistance in Proteus mirabilis. J. Antimicrob. Chemother. 28:612-615. [DOI] [PubMed] [Google Scholar]
- 18.Neuwirth, C., E. Siebor, J. M. Duez, A. Pechinot, and A. Kazmierczak. 1995. Imipenem resistance in clinical isolates of Proteus mirabilis associated with alterations in penicillin-binding proteins. J. Antimicrob. Chemother. 36:335-342. [DOI] [PubMed] [Google Scholar]
- 19.Sakurai, Y., K. Tsukamoto, and T. Sawai. 1991. Nucleotide sequence and characterization of a carbenicillin-hydrolyzing penicillinase gene from Proteus mirabilis. J. Bacteriol. 173:7038-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villar, H. E., F. Danel, and D. M. Livermore. 1997. Permeability to carbapenems of Proteus mirabilis mutants selected for resistance to imipenem or other beta-lactams. J. Antimicrob. Chemother. 40:365-370. [DOI] [PubMed] [Google Scholar]