Abstract
Four Escherichia coli isolates harboring CTX-M-14, with a single Ala231→Val substitution compared to CTX-M-9, had three different ribotypes. Cefotaxime resistance was plasmid encoded and conjugatively transferable. Three isolates had the same plasmid restriction enzyme digestion profile, suggesting clonal spread of a resistant plasmid. A high kcat/Km value for cefotaxime (20.3 μM−1 s−1) but low values for ceftazidime and aztreonam (<0.02 μM−1 s−1) were observed in hydrolysis assays, indicating resistance to cefotaxime (MIC ⩾ 64 μg/ml) but susceptibility to ceftazidime (MIC ⩽ 2 μg/ml).
CTX-M type β-lactamases constitute a novel group of class A plasmid-encoded enzymes, and their carriers are highly resistant to cefotaxime but sometimes susceptible to ceftazidime. This family of enzymes is well inhibited by clavulanate and tazobactam (24). Recently, CTX-M type β-lactamases have been described in various members of the Enterobacteriaceae family, mostly in Salmonella enterica serovar Typhimurium and Escherichia coli; these include MEN-1, CTX-M-1 to CTX-M-12, Toho-1, and Toho-2. CTX-M type β-lactamases are endemic in Latin America and some northeastern European countries (24). Toho type β-lactamases have been found only in Japan (15, 19, 25). In Taiwan, CTX-M-3 has been previously described in E. coli (26).
According to GenBank, the CTX-M-14 sequence was submitted from China in 2000 (A. Chanawong, F. H. M'Zali, J. Heritage, J.-H. Xiong, and P. M. Hawkey, accession no. AF252622). However, the characteristics of this enzyme have not been described. In this study, we characterize this new CTX-M type β-lactamase obtained from E. coli in Taiwan. Four isolates were isolated from patients in three different hospitals in 1998 during an island-wide survey of antibiotic resistance (14).
To study the epidemiology, ribotyping, plasmid isolation, resistance transferal, and restriction enzyme digestion profiles of plasmids were performed. Ribotyping was performed with an automated Riboprinter microbial characterization system (Qualicon, Wilmington, Del.) according to the manufacturer's instructions. The results of ribotyping were analyzed as previously described (8). Plasmid isolation and plasmid profile analysis were performed by the alkaline extraction method (16). Resistance transferal was carried out by conjugation (23). For restriction enzyme digestion profiles of plasmids, plasmid DNA from the transconjugant was prepared as described previously (2). The restriction enzymes EcoRI and HincII (Gibco BRL, Gaithersburg, Md.) were used.
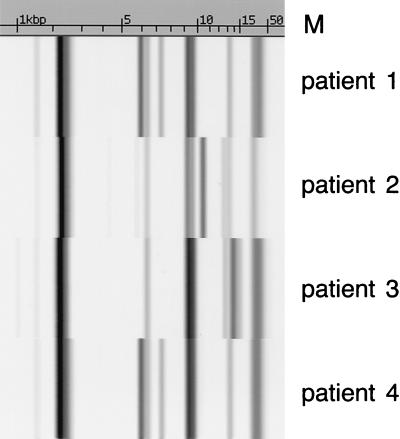
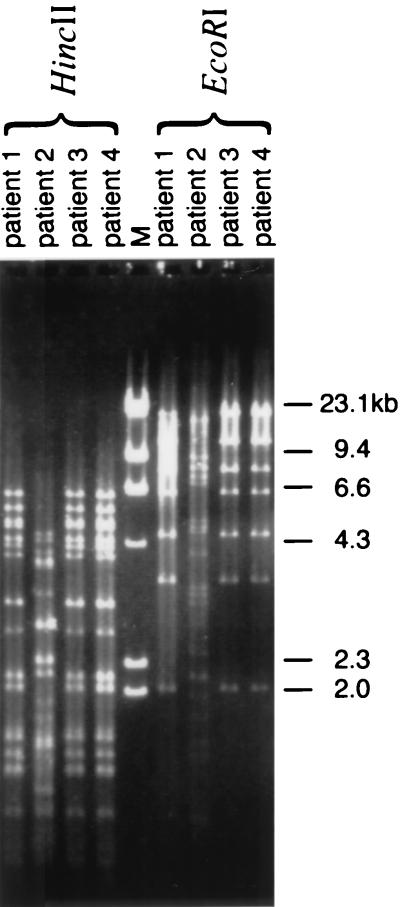
Among the four isolates, three different ribotypes were identified (Fig. 1). Two isolates with the same ribotype were obtained from two different hospitals and were geographically unrelated. The cefotaxime resistance of all isolates was found to be conjugatively transferable. Only one plasmid was transferred for each strain, and the resistant gene in each transconjugant was found to be located in a plasmid of >90 kb (data not shown). Plasmid typing of the transconjugants revealed that three isolates, including the two isolates with the same ribotype, had the same restriction enzyme digestion profile (Fig. 2).
FIG. 1.
Ribotyping of CTX-M-14 carriers isolated from four different patients. The scale at the top is measured in kilobase pairs.
FIG. 2.
HincII- or EcoRI-digested plasmid profiles of transconjugants of CTX-M-14 carriers isolated from four different patients.
The CTX-M-14 resistance gene was amplified by PCR with the primers CTX-F (5′-AAAAATGATTGAAAGGTGGTTGT-3′) and CTX-R (5′-TTACAGCCCTTCGGCGATGA-3′) and cloned into a vector (PCR-ScriptCamSK) according to the instructions for a PCR-ScriptCam cloning kit (Stratagene, La Jolla, Calif.). Our sequence data indicated an open reading frame of 876 bp, corresponding to 291 amino acids. A comparison of the nucleotide and deduced amino acid sequences of the CTX-M-14 β-lactamase with known β-lactamase sequences revealed the consensus sequences STSK, SDN, and KTG, which are conserved motifs characteristic of class A β-lactamases. Amino acid alignments with other blaCTX types revealed 99, 87, and 87% similarity with CTX-M-9, Toho-2, and CTX-M-1, respectively. CTX-M-9 and CTX-M-14 β-lactamases differ by a single amino acid at position 231 (Ala→Val). For other CTX-M type extended-spectrum β-lactamases (ESBLs), homology of not more than 85% was observed. The amino acid relationships of CTX-M type proteins can be found at the following website: http://www.lahey.org/studies/webt.htm.
Antimicrobial susceptibility was determined by a broth microdilution test (TREK Diagnostic Systems Ltd., West Sussex, United Kingdom) in serial twofold concentrations from 0.025 to 64 μg/ml, and the results were interpreted according to the method of the NCCLS (21). The CTX-M-14 carriers, transconjugants, and cloned strains were found to be resistant to ampicillin, cephalothin, and cefotaxime. They were susceptible to ceftazidime, aztreonam, ciprofloxacin, amikacin, and imipenem. When clavulanic acid at a fixed concentration of 4 μg/ml was combined with cefotaxime, ceftriaxone, ceftazidime, or cefpodoxime, >4-fold reductions in the MIC were observed, a characteristic of ESBL. The CTX-M-14 cloned strain was also susceptible to cefoxitin (Table 1).
TABLE 1.
MICs of various antibiotics for strains producing CTX-M-14 β-lactamases
| Antibiotica | MIC (μg/ml) against:
|
|||
|---|---|---|---|---|
| Clinical isolates (KTC984167) | Trans- conjugant | Cloned strains | JP-995 | |
| AMP | ≥32 | ≥32 | ≥32 | 2 |
| LOT | ≥32 | ≥32 | ≥32 | 4 |
| CFX | 8 | 2 | 8 | 4 |
| CTX | ≥64 | ≥64 | ≥64 | ≤0.25 |
| CTX plus CAL | ≤0.25 | ≤0.25 | ≤0.25 | ≤0.25 |
| CTR | ≥64 | ≥64 | ≥64 | ≤0.25 |
| CTR plus CAL | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
| CPD | ≥64 | ≥64 | ≥64 | ≤0.5 |
| CPD plus CAL | ≤0.5 | ≤0.5 | 4 | ≤0.5 |
| CAZ | 2 | 1 | 16 | ≤0.25 |
| CAZ plus CAL | ≤0.25 | ≤0.25 | 0.5 | ≤0.25 |
| ATM | 8 | 4 | ≥64 | ≤0.25 |
| IMP | ≤0.25 | 0.5 | 0.5 | 0.5 |
| GEN | >8 | ≤0.5 | ≤0.5 | ≤0.5 |
| AMK | 4 | ≤2 | ≤2 | ≤2 |
| CIP | ≤0.06 | ≤0.06 | 0.25 | ≤0.06 |
Abbreviations: AMP, ampicillin; LOT, cephalothin; CFX, cefoxitin; CTX, cefotaxime; CAL, clavulanic acid; CTR, ceftriaxone; CPD, cefpodoxime; CAZ, ceftazidime; ATM, aztreonam; IMP, imipenem; GEN, gentamicin; AMK, amikacin; CIP, ciprofloxacin.
Isoelectric focusing (IEF) was performed in ampholine gel (pH 3.0 to 10.0; Pharmacia). Preparations from standard strains known to harbor CTX-M-3, SHV-1, and SHV-5 were used as standards (20). IEF revealed that CTX-M-14 had a pI of 8.0, which was similar to those of CTX-M-9 and CTX-M-2 but different from those of other CTX-M type β-lactamases.
The β-lactamase from each cloned bacterial strain was purified as described previously (19). The purity of enzyme was >95%. The rate of hydrolysis of antibiotics by CTX-M-14 was determined by monitoring the variation in the absorbance of the β-lactam in 50 mM phosphate buffer (pH 7.0). The peak wavelength for each antibiotic used for the measurement was set according to those described in previous reports (15). The steady-state kinetic parameters (Km or Ki and kcat) were determined by analyzing the complete hydrolysis time courses as described by De Meester et al. (9) and Galleni et al. (10). Lower values of Km were determined as Ki with the help of a substrate reporter. Km and kcat values were obtained at different substrate concentrations ranging from 10 to 100 μM. Kinetic parameters for poor substrates (k2, k3, K, or k2/K) were determined by using nitrocefin as a reporter substrate (10).
The kcat and Km values were determined for a representative set of β-lactam antibiotics (Table 2). The results showed that the β-lactamase exhibited a broad-spectrum activity profile, although with notable differences for different substrates. Penicillin G, cephalothin, cephaloridine, cefotaxime, and nitrocefin were good substrates for CTX-M-14, with kcat values ranging from 150 to 6,900 s−1. The catalytic efficiency of CTX-M-14 against these drugs was greater than 20 μM−1 s−1. On the other hand, ceftazidime and aztreonam were poorly hydrolyzed (kcat/Km ⩽ 0.02 μM−1 s−1). For the reporter substrate of nitrocefin, Km and kcat were 1.12 μM and 150 s−1, respectively (Table 2). Cefoxitin and imipenem were also poor substrates for CTX-M-14. The k2/K values of cefoxitin and imipenem were 0.059 and 0.072 μM−1 s−1, respectively. However, the k3 values of the deacylation constant for these two substrates were 0.01 and 0.0007 s−1, respectively.
TABLE 2.
Kinetic parameters (kcat and Km) and physiologic efficiency (kcat/Km) of CTX-M-14 enzyme against various β-lactam antibiotics
| Antibiotic | kcat (s−1) | Km (μM) | kcat/Km | Relative kcat/Km |
|---|---|---|---|---|
| Penicillin G | 1,200 ± 60 | 52 ± 4.3 | 22.9 | 100 |
| Cephalothin | 6,900 ± 320 | 98 ± 8.0 | 70.5 | 308 |
| Cephaloridine | 6,500 ± 430 | 168 ± 4.0 | 38.7 | 169 |
| Cefotaxime | 1,100 ± 40 | 54 ± 3.2 | 20.3 | 87 |
| Ceftazidime | <0.01 | 440 ± 26 | <0.01 | <0.04 |
| Aztreonam | <0.01 | 0.48 ± 0.06 | <0.02 | <0.04 |
| Nitrocefin | 150 ± 1.9 | 1.12 ± 0.02 | 133 | 581 |
The CTX-M type β-lactamase is considered an enzyme with a potent hydrolytic activity against cefotaxime. Substitution of Ser-237 (1), which is known to enhance hydrolysis of cefotaxime, is observed in CTX-M-14 (3). Comparison with other CTX-M type enzymes revealed 99 and 87% similarity with CTX-M-9 and Toho-2, respectively, and less similarity with other CTX-M type β-lactamases. The amino acid substitutions at positions 104, 164, 179, 238, and 240 (1), which are associated with expansion of the spectrum of activity towards oximino-cephalosporins and aztreonam in TEM and SHV type ESBLs (18), were not observed in CTX-M-14, for which the MICs of aztreonam and ceftazidime were low. IEF revealed a pI value similar to those of CTX-M-9 and CTX-M-2. Substrate profiles showed that cefotaxime is a good substrate with a high kcat/Km value, such that hydrolysis by CTX-M-14 resulted in resistance of the organism to cefotaxime. The reverse was the case with ceftazidime.
Cefoxitin and imipenem were poorly hydrolyzed by CTX-M-14. In the case of cefoxitin, we determined the K, k2, k3, and k2/K values. These values explained why these were poor substrates for CTX-M-14. The k2 value, a constant for the acylation speed from the Michaelis-Menten complex to the acyl intermediate, was four times greater than k3, a constant for the deacylation speed. Accordingly, the acyl enzyme accumulated in the reaction mixture. The k2/K and k3 values of imipenem were close to those of cefoxitin. These values also indicated that the deacylation speed of imipenem was slower than the acylation speed.
Bacterial strains harboring CTX-M type β-lactamases have been identified for over 10 years as isolated incidents or outbreaks in various geographic regions. Since the first reports of CTX-M-1/MEN-1 in 1989 in Germany and France (3, 4), 13 CTX-M type β-lactamases have been reported: CTX-M-2 in Argentina (5), CTX-M-3 in Poland (13) and Taiwan (26), CTX-M-4 in Russia (12), CTX-M-5 in Latvia (7), CTX-M-6 in Greece (11), CTX-M-8 in Brazil (6), CTX-M-9 in Spain (22), CTX-M-12 in Kenya (17), and Toho-1 and Toho-2 in Japan (15, 19). In 2000, one report of CTX-M-3 was documented in southern Taiwan (26). In our laboratory, apart from CTX-M-3, CTX-M-14 is the most frequent CTX-M type ESBL detected in E. coli (unpublished data), and it has also been isolated in central and northern Taiwan. In the present study, the CTX-M-14 in all isolates was encoded on a transferable plasmid of >90 kb. Molecular typing revealed that two isolates had the same ribotype and that three had the same plasmid type, suggesting clonal and plasmid spread. To our knowledge, this is the first report of CTX-M-14 in Taiwan, and our results raise the important question of whether this enzyme has disseminated to other geographic areas. Unfortunately, the isolates in the present study were retrieved from a nonselective antibiotic surveillance study (14) and we cannot estimate the magnitude of this clonal or plasmid spread. Thus, collection of isolates with specific selection criteria should be undertaken to delineate the epidemiology of CTX-M-14 in Taiwan.
Acknowledgments
This work was supported by a grant from the National Health Research Institutes.
We also thank Moreno Galleni, Centre d'Ingénierie des Protéines, Université de Liège, Liège, Belgium, for his critical review of the kinetics studies.
REFERENCES
- 1.Ambler, R. P., A. F. Coulson, J. M. Frere, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A beta-lactamases. Biochem. J. 276:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and J. A. Smith (ed.). 1995. Miniprep of bacterial genomic DNA, p. 2.4.2. In Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Barthelemy, M., J. Peduzzi, H. Bernard, C. Tancrede, and R. Labia. 1992. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum beta-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim. Biophys. Acta 1122:15-22. [DOI] [PubMed] [Google Scholar]
- 4.Bauernfeind, A., J. M. Casellas, M. Goldberg, M. Holley, R. Jungwirth, P. Mangold, T. Rohnisch, S. Schweighart, and R. Wilhelm. 1992. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection 20:158-163. [DOI] [PubMed] [Google Scholar]
- 5.Bauernfeind, A., I. Stemplinger, R. Jungwirth, S. Ernst, and J. M. Casellas. 1996. Sequences of beta-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other beta-lactamases. Antimicrob. Agents Chemother. 40:509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnet, R., J. L. Sampaio, R. Labia, C. De Champs, D. Sirot, C. Chanal, and J. Sirot. 2000. A novel CTX-M beta-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 44:1936-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, P. A., Y. Yang, D. Sahm, I. Grope, D. Gardovska, and G. Storch. 1998. CTX-M-5, a novel cefotaxime-hydrolyzing beta-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob. Agents Chemother. 4:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brisse, S., D. Milatovic, A. C. Fluit, K. Kusters, A. Toelstra, J. Verhoef, and F. J. Schmitz. 2000. Molecular surveillance of European quinolone-resistant clinical isolates of Pseudomonas aeruginosa and Acinetobacter spp. using automated ribotyping. J. Clin. Microbiol. 38:3636-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Meester, F., B. Joris, G. Reckinger, C. Bellefroid-Bourguignon, J. M. Frere, and S. G. Waley. 1987. Automated analysis of enzyme inactivation phenomena. Application to beta-lactamases and DD-peptidases. Biochem. Pharmacol. 36:2393-2403. [DOI] [PubMed] [Google Scholar]
- 10.Galleni, M., G. Amicosante, and J. M. Frere. 1988. A survey of the kinetic parameters of class C beta-lactamases. Cephalosporins and other beta-lactam compounds. Biochem. J. 255:123-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gazouli, M., E. Tzelepi, A. Markogiannakis, N. J. Legakis, and L. S. Tzouvelekis. 1998. Two novel plasmid-mediated cefotaxime-hydrolyzing beta-lactamases (CTX-M-5 and CTX-M-6) from Salmonella typhimurium. FEMS Microbiol. Lett. 165:289-293. [DOI] [PubMed] [Google Scholar]
- 12.Gazouli, M., E. Tzelepi, S. V. Sidorenko, and L. S. Tzouvelekis. 1998. Sequence of the gene encoding a plasmid-mediated cefotaxime-hydrolyzing class A beta-lactamase (CTX-M-4): involvement of serine 237 in cephalosporin hydrolysis. Antimicrob. Agents Chemother. 42:1259-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gniadkowski, M., I. Schneider, A. Palucha, R. Jungwirth, B. Mikiewicz, and A. Bauernfeind. 1998. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing beta-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 42:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho, M., L. C. McDonald, T. L. Lauderdale, L. L. Yeh, P. C. Chen, and Y. R. Shiau. 1999. Surveillance of antibiotic resistance in Taiwan, 1998. J. Microbiol. Immunol. Infect. 32:239-249. [PubMed] [Google Scholar]
- 15.Ishii, Y., A. Ohno, H. Taguchi, S. Imajo, M. Ishiguro, and H. Matsuzawa. 1995. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A beta-lactamase isolated from Escherichia coli. Antimicrob. Agents Chemother. 39:2269-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kariuki, S., J. E. Corkill, G. Revathi, R. Musoke, and C. A. Hart. 2001. Molecular characterization of a novel plasmid-encoded cefotaximase (CTX-M-12) found in clinical Klebsiella pneumoniae isolates from Kenya. Antimicrob. Agents Chemother. 45:2141-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knox, J. R. 1995. Extended-spectrum and inhibitor-resistant TEM-type beta-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob. Agents Chemother. 39:2593-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma, L., Y. Ishii, M. Ishiguro, H. Matsuzawa, and K. Yamaguchi. 1998. Cloning and sequencing of the gene encoding Toho-2, a class A beta-lactamase preferentially inhibited by tazobactam. Antimicrob. Agents Chemother. 42:1181-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthew, M., and A. M. Harris. 1976. Identification of beta-lactamases by analytical isoelectric focusing: correlation with bacterial taxonomy. J. Gen. Microbiol. 94:55-67. [DOI] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. 1999. Performance standards for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 22.Sabate, M., R. Tarrago, F. Navarro, E. Miro, C. Verges, J. Barbe, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing beta-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siu, L. K., P. L. Lu, P. R. Hsueh, F. M. Lin, S. C. Chang, K. T. Luh, M. Ho, and C. Y. Lee. 1999. Bacteremia due to extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a pediatric oncology ward: clinical features and identification of different plasmids carrying both SHV-5 and TEM-1 genes. J. Clin. Microbiol. 37:4020-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tzouvelekis, L. S., E. Tzelepi, P. T. Tassios, and N. J. Legakis. 2000. CTX-M-type beta-lactamases: an emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents 14:137-142. [DOI] [PubMed] [Google Scholar]
- 25.Yagi, T., H. Kurokawa, K. Senda, S. Ichiyama, H. Ito, S. Ohsuka, K. Shibayama, K. Shimokata, N. Kato, M. Ohta, and Y. Arakawa. 1997. Nosocomial spread of cephem-resistant Escherichia coli strains carrying multiple Toho-1-like beta-lactamase genes. Antimicrob. Agents Chemother. 41:2606-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan, J. J., W. C. Ko, S. H. Tsai, H. M. Wu, Y. T. Jin, and J. J. Wu. 2000. Dissemination of CTX-M-3 and CMY-2 beta-lactamases among clinical isolates of Escherichia coli in southern Taiwan. J. Clin. Microbiol. 38:4320-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]