Abstract
The synthetic peptide T-20 (enfuvirtide) represents the first of a new class of antiretroviral compounds to demonstrate in vivo potency by targeting a step in viral entry. T-20 inhibits a conformational change in the human immunodeficiency virus type 1 (HIV-1) transmembrane glycoprotein (gp41) that is required for fusion between HIV-1 and target cell membranes. The initial phase I clinical trial of T-20 treatment for HIV-infected patients thus provided a unique opportunity to evaluate the emergence of resistant virus in vivo to this novel class of antiretroviral agents. All four patients who received an intermediate dose of T-20 (30 mg twice daily) had an initial decline in plasma viral load over the first 10 days but a rising trend by day 14, suggestive of selection for resistant virus. Plasma virus derived from patients enrolled in all dosage groups of the phase I T-20 trial was analyzed by population sequencing before and after treatment. While no mutations were found within a highly conserved 3-amino-acid sequence (GIV) known to be critical for fusion at baseline, after 14 days of therapy, virus from one patient in the 30-mg dose group (30-1) developed a mutation in this motif, specifically an aspartic acid (D) substitution for glycine (G) at position 36. Multiple env clones were derived from the plasma virus of all four patients in the 30-mg dosage group. Sequence analysis of 49 clones derived from the plasma of patient 30-1 on day 14 revealed that 25 clones contained the G36D mutation, while 8 contained the V38A mutation. Dual mutations involving G36D and other residues within the HR1 domain were also identified. In 5 of the 49 env clones, other mutations involving residues 32 (Q32R or Q32H) and 39 (Q39R) were found in combination with G36D. Cloned env sequences derived from the plasma virus of subject 30-3 also had single mutations in the GIV sequence (V38M and I37V) detectable following therapy with T-20. The plasma virus from subjects 30-2 and 30-4 did not contain changes within the GIV sequence. To analyze the biological resistance properties of these mutations, we developed a novel single-cycle HIV-1 entry assay using JC53BL cells which express β-galactosidase and luciferase under control of the HIV-1 long terminal repeat. Full-length env clones were derived from the plasma virus of patients 30-1 and 30-3 and used to generate pseudotyped virus stocks. The mean 50% inhibition concentrations (IC50s) for mutants G36D and V38A (patient 30-1) were 2.3 μg/ml and 11.2 μg/ml, respectively, statistically significant increases of 9.1- and 45-fold, respectively, compared with those of wild-type Env. The IC50 for the V38 M mutation (patient 30-3) was 7.6 μg/ml, an 8-fold increase compared with that of the wild type. The I37V mutation resulted in an IC50 3.2-fold greater than that of the wild type. Envs with double mutations (Q32R plus G36D and Q32H plus G36D) exhibited a level of resistance similar to that of G36D alone. These findings provide the first evidence for the rapid emergence of clinical resistance to a novel class of HIV-1 entry inhibitors and may be relevant to future treatment strategies involving these agents.
The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein complex controls key processes of viral entry. This complex determines viral tropism and mediates membrane fusion and thus enables virus entry and infection of the host cell. The HIV-1 envelope protein is initially synthesized as a 160-kDa polyprotein precursor (gp160) that is extensively glycosylated. Proteolytic cleavage of gp160 produces the gp120 surface subunit and the gp41 transmembrane subunit, which form oligomers and associate with each other through noncovalent interactions on the surface of the virion. On the surface of target cells, the gp120 surface subunit binds to its receptor and coreceptors and the gp41 transmembrane subunit mediates the fusion of the viral and cellular membranes (for a review, see reference 8). Subsequent to receptor binding, conformational changes occur in gp41 that lead to membrane fusion (7, 13, 18, 19, 21). To attain a fusion-active conformation, specific regions of the gp41 ectodomain must interact. The ectodomain contains a hydrophobic fusion peptide (FP) sequence at the amino terminus, followed by two leucine zipper-like motifs (heptad repeat 1 [HR1] and HR2). HR1 and HR2 consist of a 4,3 hydrophobic repeat that is predictive of an alpha-helical secondary structure and characteristic of coiled coils. Ultimately, through coiled coil interactions a trimer of antiparallel dimers of HR1 and HR2 is predicted to form (12). The formation of this six-stranded helical bundle induces a hairpin structure that brings the viral and cell membranes into proximity for fusion (2, 23).
Previous studies reported that synthetic peptides based on the HR2 sequence effectively block HIV-induced membrane fusion and infection by cell-free virus (9, 12, 24-26). T-20, previously known as DP178 and now as enfuvirtide, is a 36-amino-acid synthetic peptide homologous to the last 36 amino acids of HR2 (3, 24). By competitively binding HR1, T-20 blocks formation of the hairpin structure necessary for fusion (3, 26). Studies in vitro have shown that T-20 inhibits cell-free HIV-1 infection and virus-mediated cell-cell fusion (24, 26). After in vitro passage for 6 weeks in the presence of increasing concentrations of T-20, resistant variants of HIV-1 evolved (16). Sequence analysis indicated that resistant virus contained mutations within a sequence of three amino acid residues (GIV, positions 36 to 38) that are highly conserved in the HR1 domain (16). Mutations at both position 36 (G to D or S) and 38 (V to M) caused a marked decrease in susceptibility to T-20 inhibition (16). An intermediate level of sensitivity was observed with single mutations at position 36 (G36S).
Kilby et al. reported results from the first phase I clinical trial of T-20, which provided proof of concept for potent, dose-related virologic suppression by inhibiting a step in viral entry and/or fusion (10). All four of the subjects who received a 100-mg dose twice daily experienced a marked decline (mean, −1.96 log10) in plasma HIV-1 RNA (vRNA), while those in the 3- and 10-mg BID dose groups had minimal or no changes in viral load after the 14-day course of therapy. Subjects treated with the 30-mg BID dose were particularly intriguing from the standpoint of this investigation, because incomplete suppression of virus replication (median decline of −0.62 log10 among these four patients) suggested the possibility of selection for T-20-resistant HIV-1. Here we report, for the first time, the in vivo emergence of HIV-1 resistance to a viral entry inhibitor.
MATERIALS AND METHODS
Population nucleotide sequencing.
Viral RNA was prepared from the plasma of 12 of the 16 patients enrolled in a phase I clinical trial of T-20 (10) by using the QIAamp viral RNA purification protocol (Qiagen). A 692-bp fragment of gp41 that includes the FP, HR1, and HR2 was amplified by nested PCR as described earlier (22) by using the following primers: outer sense primer 5′-GAGGGACAATTGGAGAAGTGAATT-3′ (nt 7663 to 7686), outer antisense primer 5′-GTGAGTATCCCTGCCTAACTCTAT-3′ (nt 8355 to 8378), inner sense primer 5′-GGAGAAGTGAATTATATAAATATAAAG-3′ (nt 7674 to 7700), and inner antisense primer 5′-AGCTGGATCCGTCTCGAGATACTGCTCCCACCC-3′ (nt 8896 to 8918). Nucleotide numbers indicate the primer locations, using pHXB2 proviral DNA as the reference sequence. After gel purification, the PCR products were population sequenced using an automated ABI 377 Sequenator with dye terminator cycle sequencing kits (Applied Biosystems, Inc.). Sequences were analyzed using Sequencher (Gene Codes Corp.) software.
Cloning and subcloning env.
Full-length env genes were amplified from plasma vRNA by nested PCR using the following primers: outer sense primer 5′-TAGAGCCCTGGAAGCATCCAGGAAG-3′ (nt 5852 to 5876), outer antisense primer 5′-TTGCTACTTGTGATTGCTCCATGT-3′ (nt 8927 to 8950), inner sense primer 5′-GATCAAGCTTTAGGCATCTCCTATGGCAGGAAGAAG-3′ (nt 5957 to 5982), and inner antisense primer 5′-AGCTGGATCCGTCTCGAGATACTGCTCCCACCC-3′ (nt 8896 to 8918). The PCR products of the full-length env genes were cloned into pCR-XL-TOPO (Invitrogen) and the individual clones were sequenced and analyzed as described above. Some of clones were chosen for phenotypic analysis and were subcloned into pCDNA3.1 (Invitrogen).
JC53BL reporter cells.
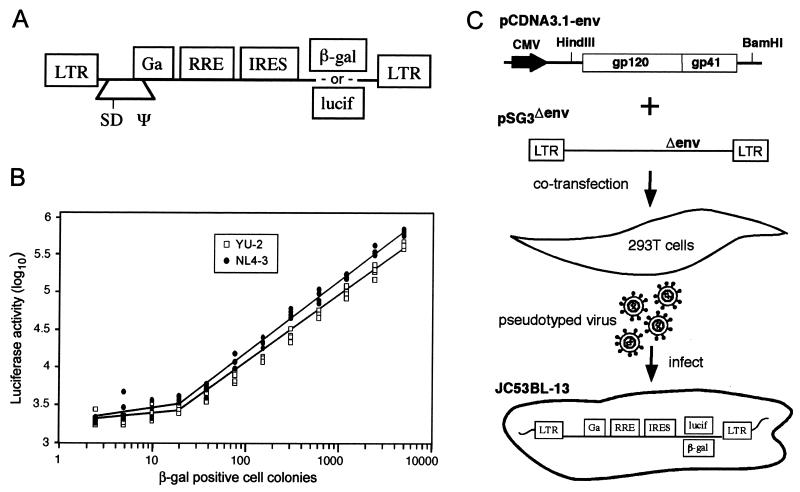
The HeLa-CD4/CCR5 (JC53) cell line (15) expresses relatively high surface levels of both CD4 and CCR5 and is susceptible to infection by both R5 and X4 HIV-1 isolates. We used an HIV-based vector (Fig. 1A) to stably introduce genes that encode the Escherichia coli β-galactosidase (β-Gal) and firefly luciferase (lucif) coding sequences into the JC53 cell line. The vector was similar to that described previously (14) except the encephalomyocarditis virus internal ribosomal entry site was inserted 5′ of the β-Gal or lucif coding regions. The β-Gal and lucif genes were introduced into JC53BL cells separately, each under transcriptional control of the HIV-1 long terminal repeat. The β-Gal reporter was included to allow direct enumeration of infectious viral units by counting β-Gal expression-positive infected cell colonies under a microscope. The lucif reporter enables automated quantitation of HIV infection. The transduced cells were biologically cloned by limiting dilution, and from 36 expanded cultures characterized for high-level expression of CD4 and CCR5 and the absence of constitutive expression of β-Gal and lucif, one clone (JC53BL-13) was selected for this study (Fig. 1C). The JC53BL-13 cell line did not nonspecifically stain positive for β-Gal expression, exhibited a low basal level of lucif, and was strongly responsive for both β-Gal and lucif expression upon HIV infection. By infecting JC53BL-13 cells with a wide range of virus concentrations, we demonstrated a near-linear relationship between the numbers of infectious viral particles (β-Gal-positive cell colonies) and lucif activity. The linear range of detection using lucif was approximately 3 orders of magnitude, and as few as 20 to 30 infected cells out of approximately 100,000 were sufficient to generate bona fide virus-positive results (Fig. 1B). JC53BL cells were analyzed for their susceptibility to infection with primary HIV-1 isolates. Compared with phytohemagglutinin (PHA)-stimulated cultures of peripheral blood mononuclear cells (PBMC), the JC53BL cells were at least two- to fivefold more sensitive to infection with primary HIV-1 isolates (Table 1).
FIG. 1.
HIV-1 entry assay. (A) Illustration of lentivirus vector used to transduce JC53 cells (LTR, long terminal repeat; SD, splice donor; Ψ, RNA packaging signal; RRE, rev-responsive element; IRES, internal ribosomal entry site). Infectious stocks of vector containing either β-Gal or lucif were prepared by transfection of 293T cells as described previously (14) and used to transduce JC53 cells. (B) Analysis of viral infectious units and lucif activity in JC53BL-13 cells. Serial twofold dilutions of the T-tropic NL4-3 and M-tropic YU-2 viruses (1, 11), beginning at 5,120 infectious units, as determined by the β-Gal assay, were used to infect (in quadruplicate) JC53BL-13 cells plated in a 96-well tissue culture plate. Forty-eight hours after infection, the replica wells were lysed, pooled, and analyzed for lucif activity by using an automated Lumistar XL system (BMG Lab Technologies). lucif activity was plotted against the number of infectious units. (C) Phenotypic analysis of the HIV-1 env clones for susceptibility to inhibition by T-20. Full-length env genes were cloned into the pCDNA3.1 eukaryotic expression plasmid. Individual pCDNA3.1-env clones were cotransfected with pSG3Δenv into 293T cells. Pseudotyped progeny virions collected from supernatants of the transfected cells were used to infect JC53BL-13 cells in either the absence or the presence of various concentrations of T-20. Virus infection induces expression of the lucif reporter gene, which was quantified to measure virus entry.
TABLE 1.
Comparison of sensitivities of JC53BL cells and PBMC to HIV-1 infection
| ELISA result for and phenotype of primary HIV-1 isolatea | PBMCb
|
JC53BLc
|
||||
|---|---|---|---|---|---|---|
| Name | p24 (pg/ml) | Phenotype | TCID50 | p24:TCID50 | TCID50 | p24:TCID50 |
| K1 | 9.5 × 105 | NSI | 1.8 × 105 | 5.3 | 2.3 × 105 | 4.1 |
| K2 | 3.2 × 105 | SI | 4.6 × 104 | 6.9 | 1.2 × 105 | 2.7 |
| K3 | 4.2 × 105 | SI | 1.8 × 105 | 2.3 | 4.5 × 105 | 0.9 |
| K4 | 5.5 × 105 | NSI | 4.6 × 104 | 11.9 | 9.5 × 105 | 0.58 |
| K5 | 6.7 × 105 | SI | 4.6 × 104 | 14.6 | 1.8 × 105 | 3.7 |
| K6 | 8.2 × 105 | SI | 4.6 × 104 | 17.8 | 8.5 × 104 | 9.6 |
| K7 | 1.3 × 105 | SI | 1.2 × 104 | 10.8 | 4.5 × 104 | 2.9 |
| K8 | 6.8 × 105 | NSI | 4.6 × 104 | 14.8 | 1.1 × 105 | 6.2 |
| K9 | 4.2 × 105 | NSI | 4.6 × 104 | 9.1 | 1.3 × 105 | 3.2 |
| K10 | 5.3 × 105 | SI | 4.6 × 104 | 11.5 | 1.1 × 105 | 4.8 |
| K11 | 1.2 × 105 | SI | 1.8 × 105 | 0.7 | 4.1 × 105 | 0.29 |
| K12 | 5.4 × 105 | NSI | 4.6 × 104 | 11.7 | 9.0 × 104 | 6.0 |
| K13 | 6.5 × 104 | SI | 1.2 × 104 | 5.4 | 3.8 × 104 | 1.7 |
| K14 | 5.0 × 105 | NSI | 4.6 × 104 | 10.9 | 2.1 × 105 | 2.4 |
| K15 | 2.1 × 105 | NSI | 4.6 × 104 | 4.6 | 1.2 × 105 | 1.8 |
| K16 | 2.2 × 105 | NSI | 4.6 × 104 | 4.8 | 3.6 × 105 | 0.61 |
| K17 | 2.7 × 105 | SI | 1.2 × 104 | 22.5 | 1.0 × 105 | 2.7 |
| K18 | 2.6 × 105 | NSI | 4.6 × 104 | 5.7 | 2.0 × 105 | 1.3 |
| K19 | 5.1 × 105 | NSI | 4.6 × 104 | 11.1 | 2.8 × 105 | 1.8 |
| K20 | 2.1 × 105 | SI | 1.2 × 104 | 17.5 | 4.5 × 104 | 4.7 |
Primary virus isolates were derived by coculture (10 days) of HIV-1-infected patient PBMC with PHA-stimulated normal donor PBMC. Virus concentrations were determined by HIV-1 p24 antigen enzyme-linked immunosorbent assay (ELISA) (Beckman-Coulter), and the syncytium-inducting (SI) or non-syncytium-inducing (NSI) phenotypes were determined on MT2 cells.
Analysis of PBMC-derived HIV-1 infectivity by endpoint dilution culture of PHA-stimulated normal-donor PBMC. Virus titers for 50% tissue culture infectivity doses per milliliter (TCID50/ml) were calculated using the Spearman-Karber formula.
Analysis of PBMC-derived HIV-1 infectivity by counting β-Gal positive JC53BL positive cell colonies under a microscope.
Phenotypic analysis of T-20 env sensitivity.
For T-20 sensitivity testing, Env-specific pseudotyped virus was generated via transfection using calcium phosphate DNA precipitation methods. Briefly, 4 μg of pCDNA3.1-env plasmid (containing the full-length env gene derived from plasma virus) was cotransfected into cultures of 293T cell with 8 μg of the env-deficient pSG3Δenv plasmid DNA. After 18 h the culture medium was replaced. Supernatants were collected 2 days later, clarified by low-speed centrifugation, aliquoted, and frozen at −70°C. The titers of the viral stocks were determined by infecting JC53BL cells with serial fivefold dilutions in the presence of DEAE-dextran (40 μg/ml) for 2 h at 37°C. After 2 days of culture in Dulbecco modified Eagle medium containing 10% fetal bovine serum, the cell monolayers were fixed and stained to detect β-Gal expression as described earlier (27). Viral infectious units were determined by counting the number of β-Gal+ cell colonies, using dilutions that gave between 30 and 300 cell colonies. To analyze the sensitivity of the viral stocks to T-20, 5 × 103 JC53BL-13 cells were plated in each well of a 96-well tissue culture plate. The following day, 1,000 infectious units of pseudotyped virus were added per well in the absence or presence of 25, 5, 1, 0.2, and 0.04 μg of T-20/ml. Quadruplicate wells for each drug concentration were analyzed. After 2 h of incubation at 37°C, 120 μl of Dulbecco modified Eagle medium containing 10% fetal bovine serum was added per well and 37°C incubation was continued for 2 days. The cells were then lysed, and the lucif activity of each well was measured using lucif assay reagent (Promega) and a LUMIstar luminometer (BMG Inc.). Background luminescence was determined from uninfected wells and subtracted from all experimental wells. Assay variability was controlled by accepting only the results which were within acceptable ranges that had been established earlier. Cell viability and toxicity were controlled by monitoring uninfected cultures that were treated with T-20 for changes in basal levels of lucif expression. Relative infectivity (% of control) was calculated by dividing the mean number of lucif units at each T-20 concentration by the mean number from cells containing no drug. Virus susceptibility was determined by plotting relative infectivity on the y axis against T-20 concentration on the x axis.
Site-directed mutagenesis.
Site-directed mutagenesis was done using the Quik-Change kit (Stratagene Inc.). A total of 125 ng of two degenerate complementary primers with mutant sequences [sense, 5′-GGT ACA GGC CAG AC (A/G) (A/T)TT ATT GTC T(G/A)(G/A) TAT G(T/C)G C(A/G)A CAG CAG AAC AAC C-3′); antisense, 5′-GGT TGT TCT GCT GT(T/C) GC(A/G) CTA TA(C/T) (C/T)AG ACA ATA A(T/A)(T/C) GTC TGG CCT GTA CC-3′ (nt 7852 to 7901 in HXB2)] and 20 ng of template pCDNA3.1-env was used for PCR amplification. PCR conditions were as follows: 95°C for 30 s, 58°C for 1 min, and 68°C for 18 min. After 20 cycles, the PCR product was digested with 10 U of DpnI to cleave template DNA at 37°C for 1 h. Mutants were identified by nucleotide sequencing.
Statistical analysis.
To determine the T-20 dose corresponding to the 50% inhibition concentration (IC50) with a 95% confidence interval, a piecewise linear dose-response curve was constructed for each of the virus types analyzed. It was observed that using log IC instead of the inhibition concentration value would enhance the model fit and preserve the normality assumption. After establishing the dose-response curve, an appropriate calibration technique was utilized to estimate the IC50 dose. This approach is appropriate for estimating the IC50 because the distribution of the data in the neighborhood of IC50 reveals a linear dependency. Furthermore, under the linearity assumption the standard error of IC50 can be estimated.
RESULTS
Analysis of plasma vRNA load.
The median viral load at baseline (day 0) for the four individuals treated with 30-mg doses of T-20 was 4.76 log10, and after 14 days of treatment the median change was −0.62 log10 (10). To gain a better understanding of the clinical and virologic responses to T-20, the vRNA load of each patient treated with the 30-mg dose (patients 30-1, 30-2, 30-3, and 30-4) was evaluated separately (Table 2). Prior to treatment (day 0), the vRNA loads of patients 30-1, 30-3, and 30-4 were 511,800, 84,610, and 31,600 copies/ml, respectively. The greatest decline in plasma vRNA among these subjects generally occurred 7 to 10 days after T-20 therapy was initiated. A plateau or increase in vRNA was measured after 14 days of treatment (Table 2). Thus, in contrast to the potent response in the 100-mg group (10), subjects in the 30-mg dose group had suboptimal responses.
TABLE 2.
Plasma vRNA load in patients treated with a 30-mg dose of T-20
| Day | Subjecta
|
|||
|---|---|---|---|---|
| 30-1 | 30-2 | 30-3 | 30-4 | |
| 0 | 5.71 | 4.25 | 4.93 | 4.50 |
| 3 | 5.52 | 3.98 | 4.58 | 4.29 |
| 7 | 5.26 | 4.11 | 4.22 | 3.64 |
| 10 | 5.10 | 4.48 | 4.09 | 3.32 |
| 14 | 5.13 | 4.76 | 4.26 | 3.73 |
Four individuals were administered 30 mg of T-20 twice daily. The numbers represent plasma vRNA loads (log10) over the course of 14 days of treatment, as determined by bDNA (Chiron Inc.) testing.
Genetic analysis of plasma HIV-1 env.
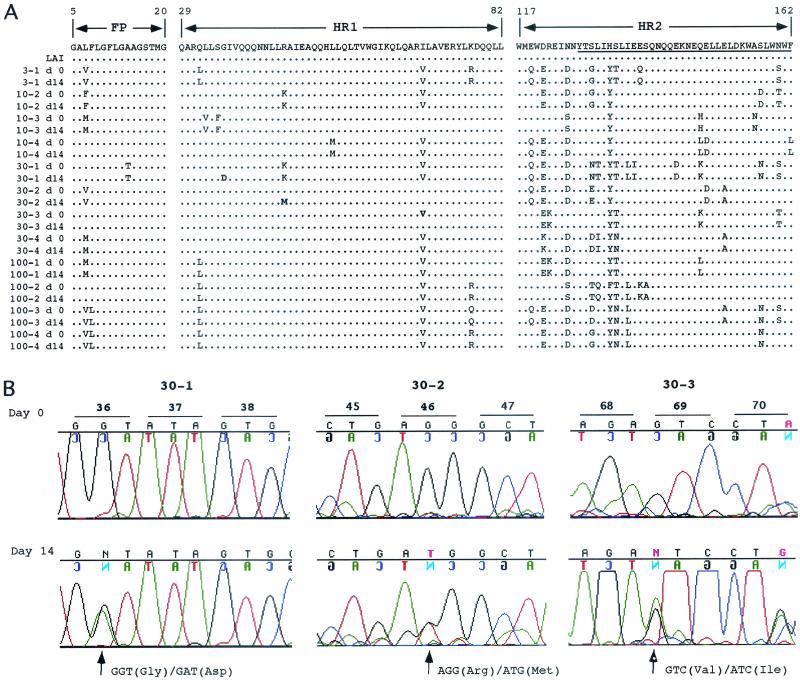
From the plasma virus of subjects enrolled in each of the four T-20 dosage groups, a 692-bp fragment of DNA containing the FP, HR1, and HR2 domains of gp41 was amplified by reverse transcriptase PCR (RT-PCR) directly from plasma and analyzed by population sequencing. Figure 2A compares the amino acid sequences of the FP, HR1, and HR2 regions prior to (day 0) and after (day 14) treatment with T-20. No differences in the viral population sequence were detected pre- and posttherapy for any of the patients in the 3-, 10-, or 100-mg arms of the study, although each patient's virus, as expected, was unique. However, in the 30-mg arm, day 14 plasma virus from three of four patients contained a mutation within the HR1 domain. The DNA sequence chromatograms illustrated that approximately one-half of the plasma virus from subject 30-1 contained an aspartic acid residue in place of glycine at position 36 (G36D). Similarly, approximately one-half of the plasma virus from patient 30-2 contained a methionine in place of arginine at position 46 and virus from subject 30-3 contained an isoleucine in place of valine at position 69 (Fig. 2B). While the G36D mutation has been shown to confer resistance to T-20 in vitro (5, 6), the mutations at positions 46 and 69 were not known to play a role in resistance to T-20. In particular, the isoleucine substitution at position 69 represents a conservative change and is present in the HIV-1 strain LAI consensus sequence (Fig. 2A).
FIG. 2.
Population sequence analysis of plasma HIV-1 env. (A) Comparison of the FP, HR1, and HR2 amino acid sequences pre- and posttreatment with T-20. The plasma virus from one subject (3-1) in the 3-mg dose group, two subjects (10-2 and 10-3) in the 10-mg dose group, and four subjects each in the 30- and 100-mg groups were analyzed by population sequencing from before day 0 (d 0) and after 14 days of treatment (d14) with T-20. The sequences were analyzed using Sequencher software and aligned against the LAI consensus sequence. The FP, HR1, and HR2 regions are depicted. The region within HR2 to which T-20 binds is underlined. (B) Quantitative detection of mutant plasma virus by automated population DNA sequencing. DNA sequence chromatograms of specified regions within HR1 are shown for patients 30-1 (residues 36 to 38), 30-2 (residues 45 to 47), and 30-3 (residues 68 to 69). Chromatograms are shown for each subject from before day 0 and after 14 days (Day 14) of T-20 treatment. The sequences shown were obtained from, and therefore are presented as, the minus (noncoding) DNA strand. The minus-strand sequence corresponds to the plus strand (coding), which is indicated on each chromatogram. Codon changes are indicated as plus-strand substitutions.
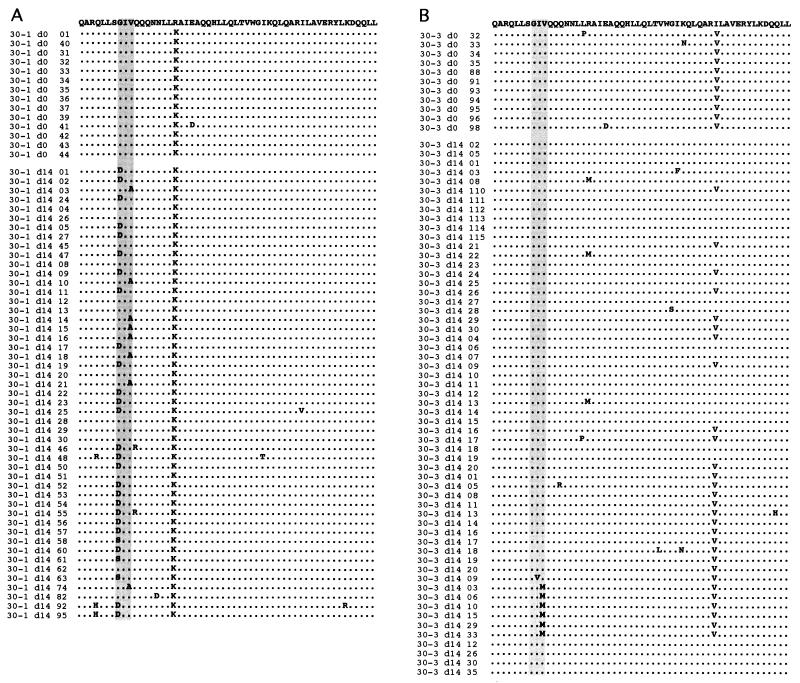
To obtain a detailed molecular genetic analysis of each patient's viral quasispecies in the 30-mg study arm, a HindIII/BamHI DNA fragment containing the entire env gene was amplified from plasma virus using RT PCR and cloned in the pCDNA3.1 plasmid. Multiple clones from each patient were derived and sequenced. A comparative analysis of the day 0 and 14 sequences revealed that the plasma virus of patients 30-1 and 30-3 contained amino acid substitutions within the GIV sequence (Fig. 3). None of 47 env clones from patient 30-2 and none of 16 env clones from patient 30-4 contained mutations in this motif (data not shown). Analysis of 49 clones derived from the day 14 plasma of patient 30-1 indicated that 36 clones contained a single mutation at either position 36 or 38 (Fig. 3A). Twenty-five of the clones (51%) contained the G36D mutation, three (6%) contained the G36S mutation, and eight (16%) contained the V38A mutation. Interestingly, none of the sequences contained a double mutation at residues 36 and 38. The only other mutation detected in more than one of the env clones was located at either position 32 or 39, and these always occurred together with the G36D mutation. Of the 58 env clones derived from the day 14 plasma virus of patient 30-3 that were analyzed, 7 (12%) contained mutations within the GIV sequence. These included one I37V and six V38M mutants (Fig. 3B) which were not apparent from population sequencing. Thirteen of 33 clones (39%) derived from the day 14 plasma of patient 30-2 contained an arginine-to-methionine substitution at position 46 (R46M). Two of 12 (17%) of the day 0 clones also contained the R46M mutation (data not shown). No remarkable amino acid substitutions were detected among the clones derived from patient 30-4.
FIG. 3.
Sequence analysis of env clones derived from plasma virus. Prior to (d0) and 14 days after (d14) treatment with T-20, the full-length env gene was amplified from viral RNA by RT-PCR and cloned into pCDNA3.1. The sequences of multiple clones derived from subjects 30-1 (A) and 30-3 (B) were analyzed using Sequencher software and aligned against the LAI consensus sequence. The sequence of the entire HR1 domain is illustrated. The vertical shaded rectangles highlight the GIV motif.
Phenotypic analysis of plasma virus susceptibility to T-20.
Of four patients treated with the 30-mg dose of T-20, our genetic analysis identified mutant plasma virus for two whose virus load was incompletely suppressed, which is highly suggestive of in vivo selection. To analyze the biologic affect of specific mutations on HIV-1 replication, we analyzed the susceptibility of various env clones to inhibition by T-20 by using a broadly sensitive HIV entry assay capable of quantifying infection within a single cycle of replication. This assay is based on the JC53BL indicator cell line, which is highly sensitive to infection with both R5 and X4 primary viruses. DNA fragments containing the entire env gene were subcloned from the pCR-XL-TOPO plasmid into the pCDNA3.1 plasmid under control of the cytomegalovirus promoter. The pCDNA3.1-env expression plasmids were cotransfected into 293T cells with the HIV-1 env− pSG3Δenv proviral clone. The titers of the transfection-derived pseudotyped virions were determined and then analyzed for sensitivity to T-20 by using the JC53BL indicator cell line. An overview of our approach for analyzing phenotypic resistance is illustrated in Fig. 1C.
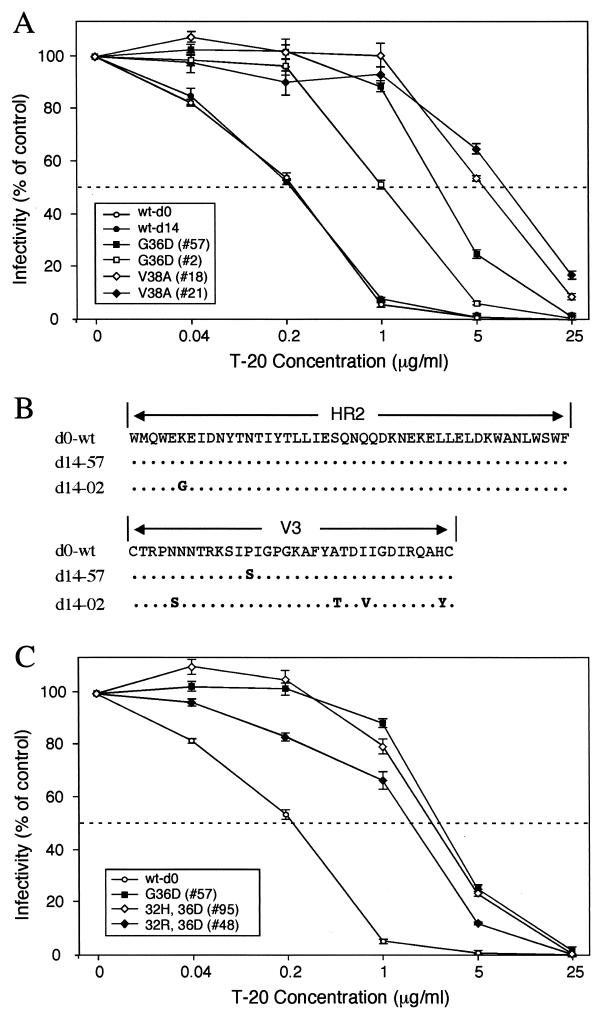
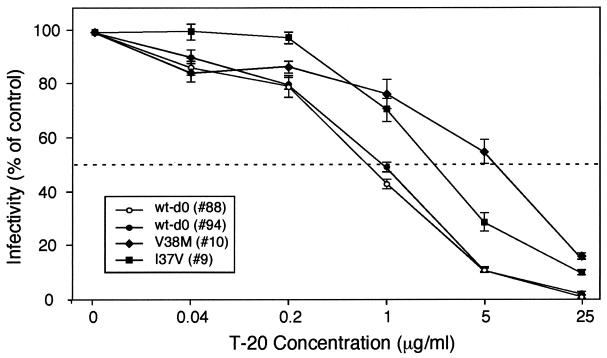
Viral envelopes from patient 30-1 containing the G36D or V38A mutation were analyzed. Two different clones of each mutant were included in the analysis. Similarly, two wild-type (wt) env clones were analyzed, one derived from day 0 and the other from day 14 plasma virus. The mean IC50 for the two V38A Env mutants was 11.2 μg/ml compared with 0.25 μg/ml for the wt Envs (P < 0.001), an increase of 45-fold (Fig. 4A). The G36D mutants also exhibited reduced levels of sensitivity to T-20. The IC50 for G36D mutant no. 57 was 3.4 μg/ml, a statistically significant difference of 13.6-fold (P < 0.001). In repeated experiments the IC50 of G36D mutant no. 2 was 1.13, an increase of 4.5-fold compared to that of the wt. It may be noteworthy that env no. 2 but not env no. 57 contained an arginine-to-glycine (R122G) substitution within the HR2 domain (Fig. 4B). Since determinants of coreceptor specificity contained within the gp120 V3 loop can modulate sensitivity to T-20 (6), the V3 amino acid sequences of Envs 2 and 57 were compared. While some amino acid differences were noted in V3 (Fig. 4B), based on the study by Derdeyn et al. (6), these differences appear not to affect susceptibility to T-20. Three of the 30-1 env clones contained a double mutation involving residues 32 and 36: two contained Q32H plus G36D (Q32H/G36D) and one contained Q32R/G36D. Our phenotypic analysis of these double mutations indicated a loss in sensitivity similar to that of the single G36D mutation (Fig. 4C). This result suggested that neither the Q32R nor the Q32H mutation causes a further loss in viral susceptibility to T-20. A more comprehensive analysis that includes longer treatment trials and greater numbers of subjects will be necessary to delineate how mutations outside of the GIV sequence contribute to the emergence of T-20-resistant virus.
FIG. 4.
Sensitivity of GIV mutant Env to inhibition by T-20. One thousand infectious units of HIV-1 pseudotyped with mutant or wt Envs derived from patient 30-1 was used to infect cultures of JC53BL cells containing 0, 0.04, 0.2, 1, 5, and 25 μg of T-20 per ml. Two days later, virus entry was measured by analyzing the indicator cells for lucif activity. Virus infectivity was calculated by dividing the mean lucif activity value at each of the drug concentrations by the mean value of the control (0 mg). Resistance to T-20 is depicted by plotting relative infectivity on the y axis against T-20 concentration on the x axis. (A) HIV-1/SG3Δenv was pseudotyped with the G36D mutant Envs 57 and 2, the V38A mutant Envs 18 and 21, and wt Envs (wt-d0 and wt-d14). (B) Sequence analysis of the HR2 and V3 Env domains for the wt (d0-wt), no. 2 (d14-02), and no. 57 (d14-57) env clones. (C) HIV-1/SG3Δenv stocks were pseudotyped with G36D mutant Env 57, the 32H/36D double mutant Envs (32H, 36D), the 32R/36D double mutant Env (32R, 36D), and the wt Env (wt-d0). The results depicted in panels A and B were highly reproducible and are representative of three independent experiments.
Although it was not apparent from population sequencing of plasma virus (Fig. 2), Env mutations, including V38M and I37V, were detected by clonal analysis in the day 14 plasma virus of patient 30-3 (Fig. 3B). The IC50 of T-20 for the V38 M Env mutant was 7.6 μg/ml compared with a mean IC50 of 0.99 μg/ml for the wt Envs, a statistically significant increase of approximately 7.7-fold (P < 0.001) (Fig. 5). The IC50 of the I37V mutant was 3.0, which is threefold greater than the mean of those of the wt Envs. None of the other mutations that were detected in the HR1 domain, including R46M and V69I, were found to affect sensitivity to T-20 (data not shown).
FIG. 5.
Sensitivity of GIV mutant Env to inhibition by T-20. HIV-1 pseudotyped with mutant or wt env derived from patient 30-3 was used to infect cultures of JC53BL cells exactly as described for Fig. 4. Pseudotyped HIV-1/SG3Δenv stocks were prepared with the V38M and I37V mutant and wt (wt-d0) Envs. The results are representative of three independent experiments.
Site-directed mutagenesis was used to confirm that single mutations in the GIV sequence were sufficient to confer resistance to T-20. A wt, day 0 env clone derived from patient 30-1 was mutated using the Quik-Change site-directed mutagenesis kit (Stratagene Inc.) and degenerative primers. Virus pseudotyped with the wt Env had an IC50 of 0.24, while that pseudotyped with the G36D site-directed mutant Env had an IC50 of 1.4, a 5.8-fold increase (Table 3). The occurrence of Q32R in combination with G36D (G36D/Q32R) did not increase the IC50 beyond that of G36D alone. Interestingly, the V38A/Q32R mutant Env exhibited an IC50 of 1.5 μg/ml, supporting the earlier data for a role of the V38A mutation in resistance to T-20. These results indicated that single mutations within the GIV sequence were sufficient to confer HIV-1 resistance to T-20 in vivo.
TABLE 3.
Site-directed mutagenesisa
| Mutation | IC50 | Fold change |
|---|---|---|
| wt | 0.24 | |
| G36D | 1.4 | 5.8 |
| G36D/Q32R | 1.3 | 5.4 |
| V38A/Q32R | 1.5 | 6.3 |
Mutations were introduced into a wt, day 0 env clone (pcDNA3.1-env) derived from patient 30-1 by using the Quik-Change kit (Stratagene Inc.) and degenerate oligonucleotide primers. Mutations were identified by nucleotide sequence analysis.
Detection of T-20-resistant HIV-1 cultured from plasma.
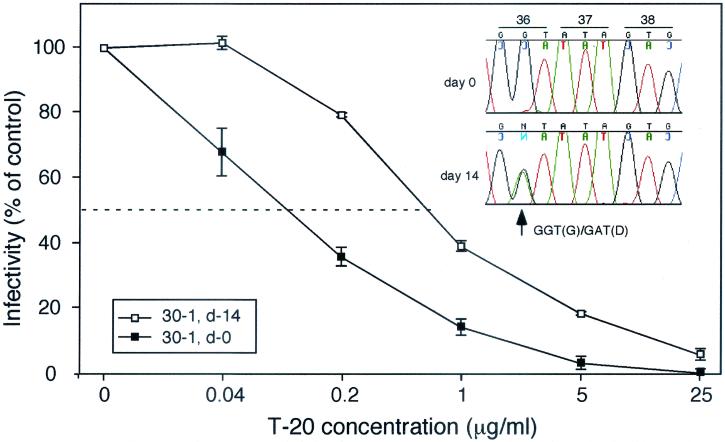
Both our genetic and phenotypic analyses indicated rapid in vivo selection of HIV-1 with reduced sensitivity to T-20. Since the plasma HIV-1 load of patient 30-1 remained relatively high during therapy and approximately 50% of the plasma virus contained a mutation in the GIV sequence, it seemed likely that resistant virus could be isolated directly from the plasma. Plasma taken from patient 30-1 prior to T-20 administration and after 14 days of treatment was cocultured with PHA-stimulated PBMC. After 18 days of culture, the supernatants were collected and the infectious virus titers were determined using JC53BL cells. The titers of both virus stocks were greater than 105 per ml. The baseline IC50 (day 0 virus) was 0.13 μg/ml. The day 14 virus had an IC50 of 0.79 μg/ml, a statistically significant increase of 6.1-fold (Fig. 6). Population sequence analysis confirmed that approximately 50% of the virus derived by coculture contained the G36D env mutation (see insert). This result confirmed the presence of T-20-resistant, replication-competent virus. It also suggested that the in vivo susceptibility of HIV-1 to T-20 can be analyzed from isolates of primary virus.
FIG. 6.
Analysis of T-20 resistance from cultured plasma virus. One thousand infectious units of HIV-1 derived by the coculture of plasma from patient 30-1, prior to (d-0) and after 14 days (d-14) of T-20 treatment, was used to infect JC53BL indicator cells in either the absence or presence of different concentrations (0, 0.04, 0.2, 1, 5, and 25 μg/ml) of T-20. Two days later, the indicator cells were lysed and analyzed for lucif activity. These results are representative of two independent experiments. The DNA sequence chromatogram specific for the GIV sequence is shown (upper right) for the day 14 plasma virus that was isolated after 18 days of cocultivation with normal-donor PBMC.
DISCUSSION
The phase I T-20 clinical trial consisted of four dose groups: 3, 10, 30 and 100 mg administered twice daily. Those enrolled in the 3- and 10-mg dose groups had no significant changes in viral load, suggesting the lack of a pharmacological effect. On the other hand, the plasma vRNA loads of all subjects in the 100-mg group declined to less than 500 copies/ml (median change, −1.96 log10 by ultrasensitive assays) (10). Subjects in the 30-mg arm experienced partial declines in viral load, but the median change on day 14 was only −0.62 log10. One individual in the 30-mg arm did not have sustained suppression below baseline. The vRNA load of each of the 30-mg subjects remained well above (at least 10-fold) the limits of detection during T-20 treatment. It was notable that the greatest decline in viral load in three of the four subjects occurred on day 10, with a modest increase on day 14 (Table 1). Sequence analysis provided evidence for the selection of resistant virus. Of multiple env clones that were derived from subjects in all dose groups at baseline, none contained mutations in the GIV motif. However, after 14 days of treatment a significant proportion of env clones derived from two of the four subjects that received the 30-mg dose of T-20 contained mutations in the GIV sequence. These same two subjects had the highest viral load at baseline. Our analyses demonstrated that a population of resistant virus had emerged in 2 of 16 HIV-1 infected adults enrolled in the phase I T-20 clinical trial during 14 days of treatment.
It has recently been shown that coreceptor specificity contained within the gp120 V3 loop can modulate sensitivity to T-20 (6). Viruses dependent on CCR5 coreceptor usage are less sensitive to inhibition by T-20 compared with those that utilize CXCR4 (6). To understand whether coreceptor specificity may have affected the emergence of resistant virus in subjects 30-1 and 30-3, we analyzed coreceptor usage by using GHOST indicator cells and found that the viral Envs from all four subjects in the 30-mg dose group, including 30-1 and 30-3, used CCR5 exclusively (data not shown). Therefore, in this study it is unclear what, if any, contribution virus coreceptor usage had on the emergence of T-20-resistant virus.
Despite a relatively conserved target in the gp41 sequence, our findings demonstrate that in certain clinical settings, HIV-1 membrane fusion inhibitors may select for resistance-conferring mutations. This determination is similar to those for the presently available classes of antiretroviral therapies: a single mutation in the RT sequence that conveys resistance to the nucleoside analog lamivudine (3TC) is detectable within weeks in some individuals (20), monotherapy with nonnucleoside RT inhibitors may select for one or two critical resistance-conferring mutations in less than 6 weeks (17), and single-agent therapy with protease inhibitors may select for a series of stepwise mutations over months that convey cross-resistance to multiple agents within this class (4). The present phase I trial of T-20 was short (14 days), yet clear-cut virologic resistance developed; it seems likely that with longer periods of treatment, the proportion of patients with virologic resistance would rise (10a). The GIV sequence appears to be of central importance, but our analysis also suggested that mutations in adjacent residues (such as residues 39 and 42) also contribute to viral resistance.
Finally, in this report we present data showing that the sensitivity of the JC53BL-13 cell line to infection by R5 and X4 viruses is equal to, or in some cases greater than, that of normal PHA-stimulated human lymphocytes. This observation, together with the viral envelope glycoprotein pseudotype experiments showing entry inhibition by and escape from T-20, suggests that the entry assay described herein may be useful for detecting and characterizing, at the biological and molecular levels, naturally occurring neutralizing antibodies to HIV-1.
Acknowledgments
This research was supported by National Institute of Health grants CA73470, R41 AI46112, and AI35467; and facilities of the central AIDS virus, biostatistical, genetic sequencing, and flow cytometry cores of the Birmingham Center for AIDS Research (P30-AI-27767).
We thank Trimeris, Inc., Durham, N.C., for providing T-20 and for helpful comments regarding the manuscript. We also thank David Kabat for JC53 cells and Tranzyme Inc. for use of the JC53BL-13 reporter cell line.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 3.Chen, C.-H., T. J. Matthews, C. B. McDanal, D. P. Bolognesi, and M. L. Greenberg. 1995. A molecular clasp in the human immunodeficiency virus (HIV) type 1 TM protein determines the anti-HIV activity of gp41 derivatives: implications for viral fusion. J. Virol. 69:3771-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condra, J. H., et al. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 5.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, Z. Zhang, W. A. O'Brien, L. Ratner, G. M. Shaw, and E. Hunter. 2001. Sensitivity of human immunodeficiency virus type 1 to fusion inhibitors targeted to the gp41 first heptad repeat involves distinct regions of gp41 and is consistently modulated by gp120 interactions with the coreceptor. J. Virol. 75:8605-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hart, T. K., R. Kirsh, H. Helleos, R. W. Street, D. M. Lambert, S. R. Petteway, J. Leary, and P. Bugelski. 1991. CD4 HIV-1 interactions: binding of soluble CD4 (sT4) to HIV-1 and HIV-1 infected cells induces shedding of envelope gp120. Proc. Natl. Acad. Sci. USA 88:2189-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunter, E. 1997. Viral entry and receptors, p. 71-120. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. [PubMed]
- 9.Jiang, S., K. Lin, N. Strick, and A. R. Neurath. 1993. HIV-1 inhibition by a peptide. Nature 365:113.. [DOI] [PubMed] [Google Scholar]
- 10.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 10a.Kilby, J. M., J. P. Lalezari, J. J. Eron, M. Carlson, C. Cohen, R. C. Arduino, J. C. Goodgame, J. E. Gallant, P. Volberding, R. L. Murphy, F. Valentine, M. S. Saag, E. L. Nelson, P. R. Sista, and A. Dusek. The safety, plasma pharmacokinetics, and antiviral activity of subcutaneous enfuvirtide (T-20), a peptide inhibitor of gp41-mediated virus fusion, in HIV-infected adults. AIDS Res. Hum. Retrovir., in press. [DOI] [PubMed]
- 11.Li, Y., J. C. Kappes, J. A. Conway, R. W. Price, G. M. Shaw, and B. H. Hahn. 1991. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J. Virol. 65:3973-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 13.Moore, J. P., R. A. M. Keating, R. A. Weiss, and Q. J. Sattentau. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139.. [DOI] [PubMed] [Google Scholar]
- 14.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. A. Verma, and D. Trono. 1996. In vivo gene delivery and stable gene transduction of nondividing cells by a lentivirus vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 15.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infection by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72:986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saag, M. S., E. A. Emini, O. L. Laskin, J. Douglas, W. I. Lapidus, W. A. Schleif, R. J. Whitley, C. Hildebrand, V. W. Byrnes, J. C. Kappes, et al. 1993. A short-term clinical evaluation of L-697,661, a non-nucleoside inhibitor of HIV-1 reverse transcriptase. New Eng. J. Med. 329:1065-1072. [DOI] [PubMed] [Google Scholar]
- 18.Sattentau, Q. J., and J. P. Moore. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sattentau, Q. J., and J. P. Moore. 1993. The role of CD4 in HIV binding and entry. Philos. Trans. R. Soc. Lond. B Biol. Sci. 342:59-66. [DOI] [PubMed] [Google Scholar]
- 20.Schuurman, R., M. Nijhuis, R. van Leeuwen, P. Schipper, D. de Jong, P. Collis, S. A. Danner, J. Mulder, C. Loveday, C. Christopherson, et al. 1995. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC). J. Infect. Dis. 171:1411-1419. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan, N., Y. Sun, J. Li, L. Hoffmann, and J. Sodroski. 1995. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J. Virol. 69:4413-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 invection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 23.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the extodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 24.Wild, C., T. Greenwell, and T. Matthews. 1993. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res. Hum. Retrovir. 9:1051-1053. [DOI] [PubMed] [Google Scholar]
- 25.Wild, C. T., T. Oas, C. B. McDanal, D. Bolognesi, and T. J. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA 89:10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of HIV-1 gp41 are potent inhibitors of virus infection Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hunter, and J. C. Kappes. 1997. Functional RT and IN incorporated into HIV-1 particles independently of the Gag-Pol precursor protein. EMBO J. 16:5113-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]