Abstract
Resistance to azole antifungals continues to be a significant problem in the common fungal pathogen Candida albicans. Many of the molecular mechanisms of resistance have been defined with matched sets of susceptible and resistant clinical isolates from the same strain. Mechanisms that have been identified include alterations in the gene encoding the target enzyme ERG11 or overexpression of efflux pump genes including CDR1, CDR2, and MDR1. In the present study, a collection of unmatched clinical isolates of C. albicans was analyzed for the known molecular mechanisms of resistance by standard methods. The collection was assembled so that approximately half of the isolates were resistant to azole drugs. Extensive cross-resistance was observed for fluconazole, clotrimazole, itraconazole, and ketoconazole. Northern blotting analyses indicated that overexpression of CDR1 and CDR2 correlates with resistance, suggesting that the two genes may be coregulated. MDR1 overexpression was observed infrequently in some resistant isolates. Overexpression of FLU1, an efflux pump gene related to MDR1, did not correlate with resistance, nor did overexpression of ERG11. Limited analysis of the ERG11 gene sequence identified several point mutations in resistant isolates; these mutations have been described previously. Two of the most common point mutations in ERG11 associated with resistance, D116E and E266D, were tested by restriction fragment length polymorphism analysis of the isolates from this collection. The results indicated that the two mutations occur frequently in different isolates of C. albicans and are not reliably associated with resistance. These analyses emphasize the diversity of mechanisms that result in a phenotype of azole resistance. They suggest that the resistance mechanisms identified in matched sets of susceptible and resistant isolates are not sufficient to explain resistance in a collection of unmatched clinical isolates and that additional mechanisms have yet to be discovered.
Candida albicans is a pathogenic yeast that causes oral, vaginal, and systemic infections (for a review, see reference 25). These infections are commonly associated with immune dysfunction, as they are frequently found in AIDS patients and bone marrow transplant patients. Antifungal drugs that are available for the treatment of these infections include the polyene amphotericin B and the azoles, such as fluconazole. In the 1990s, many human immunodeficiency virus (HIV)-infected patients received long-term, low-level azole antifungal therapy, which resulted in azole-resistant isolates of C. albicans (for a review, see reference 41). One study documented azole resistance in up to one-third of the oral C. albicans isolates from HIV-positive patients (12). In recent years, resistance to antifungal drugs has been documented in other patient populations such as bone marrow transplant recipients (9, 19, 20, 22, 24). In a clinical setting, there are many reasons why a fungal infection does not respond to antifungal drugs, including the immune status of the patient, the characteristics of the drug, and the susceptibility of the fungus to the drug (9, 41).
In the last 5 years, several molecular mechanisms by which C. albicans develops resistance to antifungal drugs have been elucidated (for a review, see reference 41). The azole drugs including fluconazole target lanosterol 14α-demethylase, the product of the ERG11 gene. Erg11p is one of the enzymes in the biosynthesis of ergosterol, the major sterol of fungal membranes and an analogue of cholesterol in mammalian systems. Antifungal drug resistance has been associated with point mutations and increased levels of expression of the ERG11 gene. Evidence is accumulating that changes in other enzymes in the ergosterol biosynthetic pathway can also contribute to resistance. Drug efflux from the cells is another component of resistance in C. albicans, as overexpression of two types of efflux pump has been correlated with antifungal resistance. The ABC transporter genes CDR1 and CDR2 encode ATP-dependent efflux pumps that are overexpressed in many azole-resistant isolates. Deletion of these genes results in hypersensitivity to azoles (1, 6, 36, 37). The major facilitator gene MDR1 encodes a pump that uses the proton motive force at the membrane to transport drugs and other compounds across the plasma membrane. Overexpression of this pump is also associated with resistance, and deletion results in hypersensitivity to azole drugs (2, 5, 7). Recently, another major facilitator gene, FLU1, was identified in C. albicans (3). This gene increases the level of azole resistance when it is expressed in Saccharomyces cerevisiae and increases the level of susceptibility when it is deleted from C. albicans, but overexpression of the gene has not yet been correlated with azole resistance in clinical isolates of C. albicans (3).
The known molecular mechanisms of resistance are best illustrated by a series of 17 isolates from an HIV-infected patient (39-42). Resistance developed over time in this series of isolates. The alterations detected in these isolates included mutation and overexpression of the ERG11 gene and overexpression of the efflux pumps encoded by CDR1, CDR2, and MDR1. Changes in the level of susceptibility are associated with each of these molecular alterations. The level of susceptibility for fungal cells is usually measured as the MIC, and the MIC determination method has recently been standardized for reproducibility and interlaboratory consistency (23).
To date, mechanisms of resistance have been determined in clinical isolates in which matched sets of resistant and susceptible isolates of the same strain were analyzed (14, 27, 37, 42). In the most recently published work, the molecular mechanisms of resistance in matched sets of susceptible and resistant isolates of the same strain from an HIV-infected patient population were investigated (27). The study found that 85% of isolates overexpressed efflux pumps, 65% of isolates had mutations in ERG11, and 35% of isolates overexpressed ERG11. Most of the point mutations identified in that study were previously described in a survey of point mutations in ERG11 (18).
The purpose of the present study was to conduct an epidemiologic survey of resistance in an unmatched set of clinical isolates, half susceptible and half resistant, from a variety of clinical settings to determine if the standard molecular techniques and the current understanding of resistance mechanisms are sufficient to explain the resistance phenotypes of arbitrarily selected isolates, like those that might be found in isolate collections in a clinical microbiology laboratory.
MATERIALS AND METHODS
Isolates and growth of cultures.
The C. albicans isolates used in this study represent a collection of 38 arbitrarily selected isolates from 12 hospitals in California. Approximately half of the isolates were susceptible to fluconazole and half were resistant to fluconazole. The isolates were from a variety of body sites. The isolates were initially identified as C. albicans at the hospital of origin and sent to one of the authors (D.A.S.) for MIC determination and further characterization. The isolates were plated on ChromAgar (DRG International, Mountainside, N.J.) to preliminarily characterize them as C. albicans. Two well-characterized isolates, a susceptible isolate (isolate C1 [isolate 2-76]) and a resistant isolate (isolate C17 [isolate 12-99]), from a series of 17 oral isolates from a single patient with AIDS (32) were used as controls.
Cultures were routinely inoculated from single colonies. The isolates were grown at 30°C in YEPD broth (10 g of yeast extract, 20 g of peptone, and 20 g of dextrose per liter) or on YEPD agar plates (10 g of yeast extract, 20 g of peptone, 20 g of dextrose, and 15 g of agar per liter). Isolates were stored at 4°C, subcultured as required, or stored at −80°C in YEPD broth containing 10% glycerol.
Confirmation of isolate species.
Most of the clinical isolates grew as green colonies on ChromAgar plates, suggesting that they are C. albicans. However, C. dubliniensis also grows as dark green colonies on ChromAgar plates and forms hyphae similarly to C. albicans. To determine if any of the isolates were C. dubliniensis, a PCR was performed with two oligonucleotides (oligonucleotides CA-INT-L and CA-INT-R) directed to the transposable intron in the large-subunit (LSU) rRNA gene (rDNA), as described previously (21), and the PCR products were electrophoresed through an agarose gel. The electrophoresis pattern distinguishes three different subgroups of C. albicans, as well as C. dubliniensis.
Susceptibility testing.
MICs were determined by the broth macrodilution method of NCCLS (23) by using laboratory reference resistant and susceptible isolates as internal controls. MIC breakpoints for clotrimazole have not been previously reported but were determined by the same protocols used for the other azole drugs. MICs were redetermined for many of the isolates in the collection; the MICs obtained by all repeat determinations fell within a fourfold range for all isolates tested, consistent with NCCLS standards.
DNA and RNA extractions and Northern and Southern blotting analyses.
Genomic DNAs from the clinical isolates were prepared as described previously (8). Preparation of total RNA, restriction enzyme digestions, gel electrophoresis, Northern blotting, oligonucleotide labeling with polynucleotide kinase, and random priming for radioactive probe preparation were performed by standard published methods (17, 34). Total RNA was prepared from isolates in the mid-logarithmic phase of growth, when the cells had reached an optical density (600 nm) of approximately 1.0 (optical density range, 0.8 to 1.2). All reagents were purchased from Fisher Scientific Co. (Pittsburgh, Pa.) or Sigma Chemical Co. (St. Louis, Mo.), unless otherwise specified.
DNA probes.
Gene fragments and oligonucleotides were used as probes for Northern blotting. For each gene described below, the numbers represent the nucleotide position in the published sequence. If the order of the numbers is reversed, the probe was designed as the reverse complement of the sequence. The GenBank accession number, followed by the reference number for the publication in which the sequence is described, is given in parentheses. Oligonucleotides were prepared to be complementary to the mRNAs for ACT1, FLU1, CDR1, and CDR2. The probe for ACT1, the gene for actin, was a 50mer, positions 2527 to 2478 (GenBank accession no. X16377 [15]); the probe for the FLU1 gene was a 48mer, positions 950 to 902 (GenBank accession no. AF188621 [3]); and the oligonucleotides for the CDR genes were as follows: CDR1, positions 1260 to 1211 (GenBank accession no. X77589 [30]), and CDR2, positions 950 to 902 (GenBank accession no. U63812 [36]). The probes for ERG11 and MDR1 were gene fragments prepared by PCR of genomic DNA. The segments that were amplified were as follows: ERG11, positions 164 to 1589 (GenBank accession no. X13296 [10]), and MDR1, positions 2885 to 3754 (GenBank accession no. X53823 [5]).
After hybridization and washing of the membranes used for Northern blotting, the filters were exposed to a phosphorimaging screen and scanned with a phosphorimager (Storm 860; Molecular Dynamics, Sunnyvale, Calif.). Signal quantification was carried out with the Molecular Dynamics ImageQuant program.
Sequence analysis.
PCRs for sequences spanning the ERG11 gene were performed with genomic DNA. The oligonucleotides used for the reactions and the processing of the PCRs have been described previously (40). The purified PCR fragments were sequenced with an automated DNA sequencer with Taq dye-primer and dye-terminator chemistries (Applied Biosystems, Foster City, Calif.).
Restriction fragment length polymorphism (RFLP) analysis.
To screen the isolates for the D116E mutation within the ERG11gene, two oligonucleotides were used to create a PCR fragment that spans the restriction enzyme site. The oligonucleotides were at positions 133 to 151 and 702 to 683 (GenBank accession no. X13296 [10]). The PCR product is 550 bp in length and is split into fragments of 350 and 200 bp by digestion with HindIII when the D116E mutation is present.
To screen the isolates for the E266D mutation, two different oligonucleotides, from positions 678 to 696 and positions 1255 to 1234 (GenBank accession no. X13296 [10]), were used. The resulting PCR product was 577 bp. It was digested with HpyCH4 IV (New England Biolabs, Beverly, Mass.), an isoschizomer of MaeII. This enzyme digested the PCR product into bands of 198, 64, and 280 bp when the wild-type sequence was present and bands of 198 and 344 bp when the mutant sequence was present.
RESULTS
Collection of isolates.
An initial screen on ChromAgar of the arbitrarily selected collection of isolates used in the present study indicated that two of the isolates were not C. albicans and that four of the isolates were mixtures of C. albicans and C. tropicalis cells. The two non-C. albicans isolates were not analyzed further. The four isolates with mixtures of cells were plated to obtain single colonies, and the C. albicans colonies were isolated and further analyzed. The C. tropicalis cells from the mixed populations were not analyzed further.
Two C. albicans isolates appeared dark green on ChromAgar plates, an indication that the isolates might be C. dubliniensis. Oligonucleotides were used in PCR to obtain a fragment that spans the transposable intron in the LSU rDNA (21) to distinguish the 36 isolates into C. dubliniensis and three subgroups of C. albicans isolates (Table 1). All 36 isolates were C. albicans; none were C. dubliniensis. Twenty-nine of the 36 isolates were subgroup A, while 3 isolates were subgroup B and 4 isolates were subgroup C. There does not appear to be a correlation between the subgroup and the MIC or the mechanism of resistance for this series of isolates, consistent with previous findings (21).
TABLE 1.
Identification, isolation, MIC, and resistance mechanism for isolate collection
| Reference isolate no. | Isolate designation | Strain typea | Hospital code | Site of isolation | MIC (μg/ml)c
|
Gene(s) overexpressed | ERG11 sequence analysisd | |||
|---|---|---|---|---|---|---|---|---|---|---|
| FLC | CLT | ITC | KTC | |||||||
| 1 | 95-142 | A | A | Throat | >64 | 8 | >8 | 4 | CDR1, CDR2 | |
| 2 | 95-68 | A | B | Unknown | >64 | 4 | >8 | 16 | CDR1, CDR2 | |
| 3 | 95-158 | A | F | Oropharynx | >64 | 4 | 8 | 4 | Hot spot III | |
| 4 | 98-112 | B | H | Blood | >64 | 2 | >8 | 16 | Complete gene | |
| 5 | 98-145e | A | G | Urine | >64 | 2 | >8 | 16 | Hot spot III | |
| 6 | 95-111e | A | F | Mouth | >64 | 2 | >8 | 4 | CDR1, CDR2 | |
| 7 | 95-16 | A | F | Paracentesis fluid | >64 | 2 | >8 | 4 | Complete gene | |
| 8 | 96-23 | A | A | Throat | >64 | 2 | 1 | 8 | Hot spot III | |
| 9 | 98-136 | C | B | Peritoneal dialysate | >64 | 1 | >8 | 16 | Hot spot III | |
| 10 | 95-188 | A | J | Wrist tissue | 64 | 2 | >8 | 16 | ||
| 11 | 95-165 | A | F | Vaginal | >64 | 4 | >8 | ≤0.5 | Complete gene | |
| 12 | 95-157 | A | I | Tongue | 64 | 4 | 2 | ≤0.5 | ||
| 13 | 95-190 | A | K | Esophageal tissue | 32 | 2 | 4 | 8 | CDR1, CDR2 | |
| 14 | 95-120 | A | A | Blood | 32 | 2 | 1 | 8 | ||
| 15 | 95-133 | A | B | Mouth | 64 | 1 | ≤0.5 | ≤0.5 | CDR1, CDR2 | |
| 16 | 98-24 | C | L | Urine | 8 | 0.5 | 1 | 1 | ||
| 17 | 97-363 | A | L | Abdominal fluid | 2 | 2 | ≤0.5 | 16 | ||
| 18 | 98-144 | A | G | Blood | 2 | 0.5 | >8 | 4 | ||
| 19 | 97-224 | A | F | Blood | 2 | 0.5 | 2 | 1 | ||
| 20 | 98-155 | A | G | Blood | ≤0.5 | 0.5 | 1 | 2 | ||
| 21 | 98-143 | A | A | Urine | ≤0.5 | ≤.25 | >8 | 4 | ||
| 22 | 96-25f | C | A | Abdominal fluid | 32 | 0.5 | 1 | ≤0.5 | ERG11, MDR1 | Hot spot III |
| 23 | 98-141 | C | B | Indwelling catheter | 8 | 2 | ≤0.5 | ≤0.5 | ||
| 24 | 98-7 | A | A | Blood | 4 | ≤.25 | >8 | ≤0.5 | ||
| 25 | 97-119f | A | C | Abdomen | 1 | 1 | ≤0.5 | ≤0.5 | ||
| 26 | 98-216 | A | A | Urine/catheter | 1 | ≤.25 | >8 | ≤0.5 | ||
| 27 | 98-85f | A | A | Peritoneal fluid | ≤0.5 | 0.5 | 1 | ≤0.5 | CDR1, (CDR2) | |
| 28 | 98-125 | A | A | Blood | ≤0.5 | ≤.25 | 2 | ≤0.5 | CDR2 | |
| 29 | 97-87 | B | A | Blood | ≤0.5 | ≤.25 | ≤0.5 | 8 | ||
| 30 | 98-13 | A | E | Paracentesis fluid | ≤0.5 | ≤.25 | ≤0.5 | 1 | ||
| 31 | 95-175 | A | A | Endotracheal aspirate | 32 | ≤.25 | ≤0.5 | ≤0.5 | ||
| 32 | 97-197f | A | B | Blood | 2 | 0.5 | ≤0.5 | ≤0.5 | ||
| 33 | 98-8 | A | B | Oral | ≤0.5 | 0.5 | ≤0.5 | ≤0.5 | ERG11 | |
| 34 | 98-113 | A | D | Peritoneal fluid | ≤0.5 | ≤.25 | ≤0.5 | ≤0.5 | ||
| 35 | 98-126 | A | A | Chest tube | ≤0.5 | ≤.25 | ≤0.5 | ≤0.5 | ||
| 36 | 98-146 | B | A | Pleural tissue | ≤0.5 | ≤.25 | ≤0.5 | ≤0.5 | ||
| C1g | 2-76 | A | Oral | 0.25 | NAh | ≤0.5 | ≤0.5 | |||
| C17g | 12-99 | A | Oral | ≥64 | NA | ≤0.5 | 2 | ERG11, CDR1, CDR2, MDR1 | ||
Strain type, as determined by PCR and restriction enzyme digestion (19).
Isolates were obtained from 12 hospitals throughout California, coded by letter.
FLC, fluconazole; CLT, clotrimazole; ITC, itraconazole; KTC, ketoconazole. Results that indicate that the isolate was resistant according to the breakpoints discussed in the text are underlined.
The ERG11 genes of some of the isolates were sequenced. Either the entire gene or hot spot III, which includes amino acids 405 to 488, was sequenced.
Colonies were dark green on ChromAgar but were confirmed to be C. albicans.
The isolates were originally parts of mixed infections.
C1 and C17, control isolates 1 and 17, respectively, from a well-characterized matched set.
NA, not available.
MICs.
The MICs of four different azoles, fluconazole, clotrimazole, itraconazole, and ketoconazole, were determined (Table 1). For some isolates the MICs of all azoles were relatively high, for other isolates the MICs of one or two azoles were relatively high but the MICs of the other azoles tested were low, and some isolates were susceptible to all azoles tested.
MIC breakpoints that correlate with clinical treatment success have been determined for fluconazole and itraconazole (33). For fluconazole, a resistant isolate is defined as an isolate for which the MIC is ≥64 μg/ml; a susceptible isolate is an isolate for which the MIC is ≤8 μg/ml. Isolates for which MICs are 16 and 32 μg/ml are defined as susceptible, dose dependent (SDD), as they appear to respond to higher doses of drug. For itraconazole, the breakpoints have been defined as follows: susceptible, MIC ≤ 0.125 μg/ml; SDD, MIC = 0.25 to 0.5 μg/ml; and resistant, MIC ≥ 1 μg/ml. The 36 C. albicans isolates included 13 that were resistant, 4 that were SDD, and 19 that were susceptible to fluconazole and 24 that were resistant and 12 that were susceptible or SDD to itraconazole. Clinical breakpoints for ketoconazole have not been proposed, but they are likely to be close to the breakpoints for itraconazole, as the drug concentrations achievable in blood are similar for both drugs when similar doses are administered, and the distributions of resistant and sensitive isolates are similar for both itraconazole and ketoconazole when ≥1 μg/ml is used to define resistance to both drugs. By using an MIC of ≥1 μg/ml to define resistance in this study, the 36 isolates included 20 isolates that were resistant to ketoconazole and 16 isolates that were susceptible or SDD to ketoconazole. The MICs of clotrimazole are not routinely determined, although clotrimazole is commonly used to treat mucosal candidiasis in children with HIV infection. Recently, Pelletier et al. (26) described a collection of isolates from children for which the clotrimazole MIC was ≥1 μg/ml; these isolates represented 10% of the total collection and were cross resistant to other azoles. Amphotericin B was required for successful treatment. By using an MIC of ≥1 μg/ml to define clotrimazole resistance, the 36 isolates included 18 that were resistant and 18 that were susceptible or SDD to clotrimazole. The distribution of isolates in Table 1 shows that 10 isolates (isolates 1 to 10) were resistant to all azoles tested, 4 isolates (isolates 11 to 14) were resistant to three azoles, 7 isolates (isolates 15 to 21) were resistant to two azoles, 9 isolates (isolates 21 to 30) were resistant to one azole, and 6 isolates (isolates 31 to 36) were susceptible to all azoles tested. This distribution of resistance to azoles was broad and appropriate for the testing of the molecular mechanisms of resistance in a collection of clinical isolates.
Molecular mechanisms of resistance.
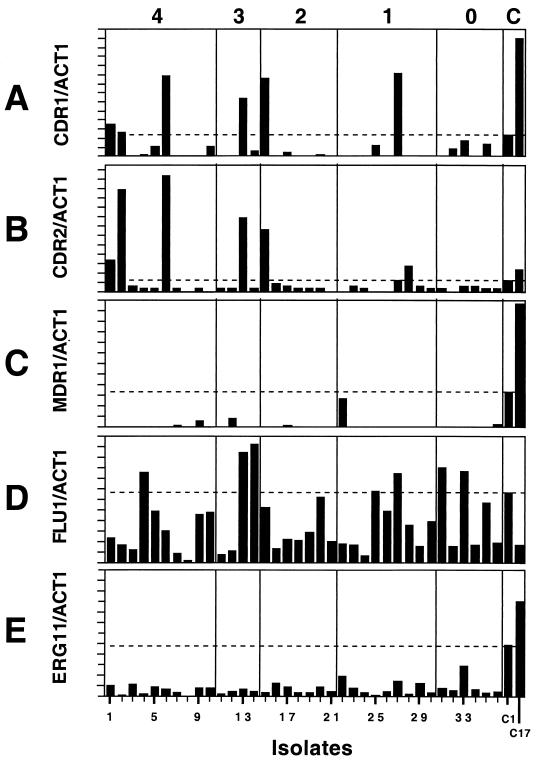
Previous work on the analysis of resistance mechanisms has routinely used matched sets of susceptible and resistant clinical isolates of the same strain (14, 27, 37, 42). This is necessary, as C. albicans is mostly clonal in nature (31) and isolates might differ considerably in their levels of expression of different genes. Matched sets of isolates were not available for the collection of isolates described above, nor will matched sets be available in most clinical settings. The purpose of the experiments described below is to use standard molecular techniques to screen for known resistance mechanisms without the aid of matching the isolates with pretreatment isolates and to determine if these methods are sufficient to determine mechanisms of resistance in unmatched isolates. Controls for these experiments included previously characterized matched susceptible and resistant isolates from the series of 17 isolates discussed above and hybridization with ACT1 as a control to normalize the amount of RNA from each isolate. Since the specific activities of the probes vary, the relative intensities of the gene-specific hybridization signals compared to that for ACT1 are presented, but the absolute values or a numerical value for the relative intensities are not significant, as they will vary with the specific activities of the probes. The relative levels of expression of the hybridization signals by the isolates are presented in Fig. 1 for each probe. The isolates are numbered on the basis of their levels of resistance (Table 1), so the first 10 isolates on the left side of Fig. 1 are resistant to all four azoles, and the last 6 isolates on the right side of the figure are susceptible to all four azoles.
FIG. 1.
Levels of gene expression relative to ACT1 gene expression, as determined by Northern blotting analysis. Total RNA from the 36 isolates and 2 controls was hybridized with gene-specific probes on Northern blots. The mRNA signals for the two probes were quantitated with a Storm phosphorimager, and the ratio of the two intensities was computed and graphed. The x axis represents the 36 isolates, arranged in numerical order. The last two isolates on the right (isolates 37 and 38) are the control isolates (isolates C1 and C17, respectively) from the well-characterized matched series. The dotted lines indicate the level of expression of each of the genes by susceptible control isolate C1. As indicated at the top, isolates 1 to 10 are resistant to four azoles, isolates 11 to 14 are resistant to three azoles, isolates 15 to 21 are resistant to two azoles, isolates 22 to 30 are resistant to one azole, and isolates 31 to 36 are susceptible to all azoles tested. C, control.
CDR efflux pump overexpression.
The CDR genes are commonly overexpressed in resistant isolates (14, 27, 37). In general, the CDR genes are not normally expressed at high levels in susceptible isolates (16). Total RNA was prepared from the 36 isolates while they were in the mid-logarithmic phase of growth, which has been standard procedure in previous studies. The RNA was electrophoresed through an agarose gel, blotted onto nitrocellulose, and hybridized with oligonucleotides specific for CDR1 and CDR2.
Six isolates from the collection expressed CDR1 at higher levels than susceptible control isolate C1 did (Fig. 1A). The isolates overexpressing CDR1 included isolates 1, 2, 6, 13, 15, and 27. These isolates came from four different hospitals and were isolated from the mouth, throat, esophagus, and peritoneal fluid (the site of isolation of isolate 2 was unknown) (Table 1). Of the six isolates that overexpressed CDR1, three isolates were resistant to all four azoles, one isolate was resistant to three azoles and SDD to the other azole, one isolate was resistant to two azoles, and one isolate was resistant to one azole (itraconazole).
Six isolates expressed CDR2 at higher levels than susceptible control isolate C1 did. The isolates overexpressing CDR2 included isolates 1, 2, 6, 13, 15, and 28 (Fig. 1B). Five of the six isolates overexpressed both CDR1 and CDR2, and these five isolates were resistant to most, if not all, azoles tested (Table 1). The level of expression of CDR2 in isolate 27, which overexpresses CDR1, is approximately the same as that in isolate C1, suggesting that CDR1 and CDR2 are jointly overexpressed in six different isolates from this collection. When all isolates that overexpressed CDR1 and CDR2 are considered, there is no preference for resistance to one azole over that to any of the other azoles tested.
MDR1 efflux pump overexpression.
The MDR1 gene for the major facilitator efflux pump has been shown to be expressed in several resistant clinical isolates (14, 27, 37, 39). The MDR1 gene is not normally expressed in susceptible isolates under normal conditions, as determined by Northern blotting analysis (16). Northern blotting analysis was performed as described above with an MDR1-specific probe (Fig. 1C). Only one isolate, isolate 22, showed significant levels of expression of the MDR1 gene by Northern blotting analysis, although its level of expression was still lower than the level of MDR1 expression in isolate C1. Isolate 22, originally taken from abdominal fluid, was mildly resistant to itraconazole, but it was susceptible to the other azoles tested (Table 1). Clearly, MDR1 overexpression during the mid-logarithmic phase of growth is not closely correlated with resistance in this collection. Two other isolates, isolates 9 and 12, had low levels of expression of MDR1 (Fig. 1C). Isolate 9 was resistant to all four azoles tested; isolate 12 was resistant to all the azoles tested but ketoconazole (Table 1).
FLU1 efflux pump overexpression.
When the recently described FLU1 gene is deleted from Saccharomyces or C. albicans, the resulting cells are hypersusceptible to azole drugs (3). However, overexpression of the gene has not been linked to resistance in clinical isolates. Northern blotting analysis was performed, and FLU1 gene expression was quantitated as described above using an FLU1-specific probe (Fig. 1D). In this Northern blotting analysis, isolate C1 expressed more FLU1 than isolate C17 did, consistent with previous data from our laboratory (data not shown). Seven isolates (isolates 4, 13, 14, 25, 27, 31, and 33) expressed more FLU1 than the other isolates did. There does not appear to be a correlation between expression of high levels of FLU1 and resistance in these isolates.
ERG11 target enzyme overexpression.
The gene for the azole target enzyme, ERG11, is expressed during growth of both susceptible and resistant isolates (16). Northern blotting analysis was performed and ERG11 gene expression was quantitated with total RNA from cells in the mid-logarithmic phase of growth using an ERG11-specific probe (Fig. 1E). There was considerable variation in the levels of ERG11 expression within this collection of isolates, but the level of expression did not appear to be related to the azole resistance of the isolates. All of the isolates expressed ERG11 at levels below the level at which isolate C1 expressed it. The highest relative level of ERG11 expression was found by isolate 33, which is an isolate that is susceptible to all azoles tested, suggesting that absolute levels of ERG11 expression are not a priori linked to azole resistance. One isolate (isolate 22) is SDD to fluconazole and mildly resistant to itraconazole, and it had a relatively high level of ERG11 expression. It is noteworthy that this isolate is the one isolate in the collection with significant levels of expression of MDR1. The distribution of ERG11 expression suggests that high levels of ERG11 mRNA are not necessarily associated with increased azole resistance. There is no obvious link between the site of isolation in the human body and the level of ERG11 expression.
ERG11 sequence analysis.
Point mutations within the ERG11 gene have been identified in several matched isolates (18, 27, 35, 40). In a recent compilation of mutations, Marichal et al. (18) noted that there were three diffuse hot spots within the amino acid sequence of the ERG11 gene, including amino acid regions 105 to 165, 266 to 287, and 405 to 488. Sequence analysis of the entire ERG11 genes of three resistant isolates (isolates 4, 7, and 11) was performed. These isolates did not overexpress any of the genes tested, including ERG11, CDR1, CDR2, and MDR1. The genomic DNAs were amplified by PCR, and the sequences of the PCR products were determined as described previously (40) and compared to the published sequence (10). As expected from unrelated clinical isolates, frequent silent mutations that do not change the protein sequence were identified (data not shown). In addition, since C. albicans is a diploid organism, allelic differences were observed upon sequencing of DNA from single isolates (data not shown). The sequence alterations that resulted in changes in the protein sequence are listed in Table 2. Amino acid substitutions are usually denoted by the amino acid position, preceded by the single-letter amino acid designation for the wild type and followed by the single-letter amino acid designation for the mutant (e.g., D116E). Five mutations, located in the three hot spots, were observed in the three complete sequences; these mutations included D116E, K128T, K143R, E266D, and V437I (Table 2). Careful inspection of the sequence data distinguished between point mutations in both alleles (the isolate is homozygous for the mutation) and mutations in only one allele (the isolate is heterozygous for the mutation). Isolate 11 contained three homozygous mutations (D116E, K128T, and K143R), all within the first hot spot. Isolate 7 contained two heterozygous mutations (D116E/D and K128T/K). Both of these mutations were also found in isolate 11 and were present in the first hot spot. Isolate 4 was heterozygous for D116E/D in the first hot spot, homozygous for E266D in the second hot spot, and heterozygous for V437I/V in the third hot spot.
TABLE 2.
Protein sequence alterations associated with resistancea
| Mutation | Result for isolates whose complete gene was sequenced
|
Result for isolates for which the hot spot III region was sequenced
|
||||||
|---|---|---|---|---|---|---|---|---|
| 4 | 7 | 11 | 3 | 5 | 8 | 9 | 22 | |
| D116E | Heterozyg. | Heterozyg. | Homozyg. | |||||
| K128T | WT | Heterozyg. | Homozyg. | |||||
| K143R | WT | WT | Homozyg. | |||||
| E266D | Homozyg. | WT | WT | |||||
| V437I | Heterozyg. | WT | WT | WT | Homozyg. | WT | WT | Homozyg. |
| G448R | WT | WT | WT | WT | WT | Homozyg. | WT | WT |
Point mutations that resulted in amino acid changes identified by sequencing of ERG11 from the indicated isolates. WT, the amino acid at that position was wild type (no change occurred in either allele); Heterozyg., the amino acid at that position was mutated as indicated in one allele; Homozyg., the amino acid at that position was mutated as indicated in both alleles.
As the mutations identified in these three isolates had previously been identified in other collections of clinical isolates, limited sequence analysis of the third hot spot region was also performed for five other resistant isolates. The V437I mutation was homozygous in isolates 5 and 22 (Table 2). An additional homozygous mutation, G448R, was identified in isolate 8. Of these six mutations, the D116E, K128T, K143R, E266D, and V437I mutations have been described in isolates from other studies (18). The sixth mutation, G448R, has not been described previously, but a similar mutation, G448E, has been identified previously (13). D116E, K128T, and E266D were three of the most common mutations identified in a recent survey (18). Sequence analysis of the eight isolates from the present collection which had limited pump expression failed to identify additional mutations within the ERG11 gene. Therefore, sequencing of additional isolates with unknown mechanisms of resistance was not performed.
ERG11 RFLP analysis.
The D116E mutation in the first hot spot of ERG11 was identified in each of the three isolates whose complete sequences were determined, and it was the most common mutation identified in a recent survey of mutations (18). The D116E mutation can be identified by HindIII restriction enzyme digestion of PCR products that span the mutation, since HindIII will recognize the sequence of the mutant allele but not the sequence of the wild-type allele. All 36 isolates were screened for the mutation (Table 3). Four isolates were homozygous for the mutation at amino acid position 116, and 19 isolates were heterozygous for the mutation at amino acid position 116. The four isolates that were homozygous were resistant to at least two azoles, although the numbers of isolates are insufficient to be significant.
TABLE 3.
RFLP analysis of the sequences at positions 116 and 266 in ERG11a
| Amino acid present | Isolate(s) resistant to the following no. of azoles:
|
||||
|---|---|---|---|---|---|
| Four | Three | Two | One | None | |
| 116D/D | 2, 5, 8, 9 | 15, 18, 20, 21 | 22, 23, 30 | 31, 32 | |
| 116D/E | 1, 3, 4, 7, 10 | 13, 14 | 17, 19 | 24, 25, 26, 27, 28, 29 | 33, 34, 35, 36 |
| 116E/E | 6 | 11, 12 | 16 | ||
| 266E/E | 1, 2, 3, 5, 6, 7, 8, 9, 10 | 11, 13, 14 | 15, 17, 18, 19, 21 | 22, 24, 25, 26, 27, 28, 30 | 31, 33, 34 |
| 266E/D | 4 | 16, 20 | 29 | 36 | |
| 266D/D | 12 | 23 | 32, 35 | ||
RFLP analysis was performed with the 36 isolates to assess the sequences at two locations, corresponding to amino acids 116 and 266, as described in Materials and Methods. The possible amino acids at those positions can be distinguished by RFLP analysis.
The E266D mutation in the second hot spot of ERG11 is also a frequent mutation in the ERG11 gene (18). This mutation can be screened for by restriction enzyme digestion of PCR fragments that span this region. MaeII and its isoschizomers digest the wild-type sequence but do not digest the mutant sequence. PCR products that include the sequence were digested and analyzed by agarose gel electrophoresis. The results (Table 3) indicate that there is no correlation between the E266D mutation and azole resistance.
DISCUSSION
The goal of the series of experiments described here was to use standard techniques to identify molecular mechanisms of resistance in a collection of arbitrarily selected susceptible and resistant isolates. The results of the study do not define any specific correlations. Six strains overexpressed CDR1 and CDR2. This is much lower than the frequencies observed in oral isolates from patients with AIDS. Five of the six strains overexpressing CDR1 and CDR2 were resistant to one or more of the four azoles tested (Table 1), although the results are not statistically significant for any drug. The one strain that overexpressed MDR1 was resistant to only one azole, while low-level expression of MDR1 occurred in two strains resistant to three or four azoles (Fig. 1). Similarly, overexpression of FLU1, overexpression of ERG11, or frequent mutations within the ERG11 genes were not correlated with resistance in this collection. Overall, this suggests that standard molecular analyses of unmatched isolates is not sufficient to identify resistance mechanisms.
An important initial analysis of the collection of isolates used in the present study was confirmation that the isolates were C. albicans. Misidentification is not an uncommon problem, and experience indicates that isolates should always be rechecked for species identification and for the possibility that mixtures of isolates are present (data not shown). Rechecking of the hospital isolates indicated that two of the isolates were not C. albicans, and ChromAgar plate cultures of four of the isolates contained mixtures of C. albicans and C. tropicalis. Failure to detect these non-C. albicans isolates would have had important effects on the molecular analyses, as the oligonucleotides used as probes and primers for PCR would not react with the nucleic acids from the other species. Mixed infections would also have had consequences for MIC determinations and molecular characterizations. The possibility that mixtures of C. albicans strains were present was not addressed, although single colonies were used for the molecular analyses.
Recent identification and characterization of C. dubliniensis indicate that the species is easily confused with C. albicans. Limited techniques are available to quickly differentiate between C. albicans and C. dubliniensis. PCR with oligonucleotides that span a transposable element in the LSU rDNA of these species is a simple and specific test (21). Use of this test with the isolate collection indicated that all of the isolates were C. albicans. Furthermore, most of the isolates were identified as members of subgroup A of C. albicans, as defined previously (21).
The isolates in this collection were obtained from 38 different patients in 12 different hospitals throughout California over a 4-year period, making it highly unlikely that these isolates were related strains of C. albicans. This was confirmed by the distribution of the RFLP types presented in Table 3. The isolates in this collection were taken from a variety of body sites, including oral and vaginal mucosae, blood, urine, and abdominal fluid. While the HIV infection status of these patients is not known, it is likely that a large number of these patients infected with either resistant or susceptible organisms are not HIV-positive or AIDS patients.
The relationship between the MIC and clinical success or failure is an important issue. Breakpoints between susceptibility and resistance for isolates from patients with oropharyngeal candidiasis have been determined for itraconazole and fluconazole (33). However, these breakpoints may not apply to isolates from patients with systemic infections. Breakpoints for ketoconazole and clotrimazole have not been proposed. The MICs of four azoles, fluconazole, clotrimazole, itraconazole, and ketoconazole, were determined for all of the isolates (Table 1).
The clotrimazole MIC was greater than 1 μg/ml for all 13 fluconazole-resistant isolates; this MIC has been suggested to be associated with clotrimazole resistance (26). In addition, of four isolates that were SDD to fluconazole, the clotrimazole MIC was >1 μg/ml for two of them. Sixteen isolates were considered susceptible to both fluconazole and clotrimazole, and the fluconazole and clotrimazole susceptibilities were discordant for only 3 of 36 isolates (fluconazole susceptible, clotrimazole resistant), demonstrating a strong correlation between fluconazole and clotrimazole resistance. This has important consequences for AIDS patients being treated with clotrimazole, especially HIV-infected children.
The MICs of itraconazole and ketoconazole were consistent with those of fluconazole and clotrimazole. Only 2 of the 17 isolates resistant or SDD to fluconazole would have been considered itraconazole susceptible and only 5 of the 17 isolates would have been considered ketoconazole susceptible by using the definitions of resistance proposed in this study. Ten isolates were resistant to all azoles tested, and most isolates were resistant to at least one of the azoles tested.
The collection of resistant isolates used in the present study was screened for the currently characterized molecular mechanisms of azole resistance. The expression of CDR1 and CDR2 in this collection (Fig. 1A and B) shows that several resistant isolates overexpress both genes. However, it was also found that at least one isolate resistant to only one azole also expressed large amounts of CDR1 and CDR2 (isolate 27 for CDR1, isolates 27 and 28 for CDR2). The fact that CDR1 and CDR2 are overexpressed simultaneously in six isolates suggests that the genes are coregulated. It raises the possibility that other efflux pumps are also coregulated, but the contributions of these other pumps to resistance cannot be predicted. The oligonucleotide probes specific for CDR1 and CDR2 do not cross-hybridize to each other, to any other CDR genes, or to other genomic DNA (data not shown). Therefore, cross-hybridization cannot explain the coregulation.
The gene for the MDR1 efflux pump is not expressed at high levels in cells in the mid-logarithmic phase of growth (16). The overexpression of MDR1 in the present collection of isolates was limited to three isolates, all of which were resistant to one or more azole drugs. It has previously been shown that the MDR1 message is variable in its pattern of expression even within a single isolate, possibly due to message instability (16). This may contribute to the expression or lack of expression of MDR1 observed in these isolates.
The patterns of expression of the resistance genes in the present panel of isolates have been characterized only during the mid-logarithmic phase of growth, characterized as an optical density of approximately 1.0 in a single medium in the absence of azole drugs (Fig. 1), which is the standard methodology for the study of resistance mechanisms. The timing of expression of most of these genes has been studied during cellular growth (from the early logarithmic to the late stationary phase) in the control series of isolates (16). However, study of the timing of expression of each of these isolates in the presence and absence of azole drugs would be an extensive analysis that was beyond the scope of the present study.
The gene for the FLU1 pump has not previously been associated with resistance (3), but its expression in this collection was evaluated because of the phenotypes of Saccharomyces and Candida cells from which the gene was deleted. The levels of expression do not clearly correlate with the levels of resistance of the isolates, adding additional support to the hypothesis that the gene is not involved in resistance in clinical isolates.
The pattern of ERG11 expression is also variable and appears to have no correlation with resistance. The one resistant isolate that overexpressed MDR1 also overexpressed ERG11, which might provide a hint that there is a link between the two. It is noteworthy that the levels of expression of ERG11 in isolate C1 are elevated compared to those in all of the other isolates. This suggests that ERG11 expression in the control susceptible isolate may already have been altered slightly in response to azole drugs but had not resulted in resistance.
The levels of mRNA for the genes associated with resistance were determined during the mid-logarithmic phase of growth in the absence of azole drugs, as has been the case previously (41). Certainly, the azole drugs will have additional effects on the levels of expression of these genes, which will be dependent on the concentration of the drug and the MIC for the cell. These factors add considerable complexity to the analysis and are beyond the scope of the present study.
Sequence analysis of the isolates was not extensive, but sufficient sequences were obtained to indicate that many of the point mutations identified in the present collection have already been identified in other isolates that have been sequenced previously (18). Therefore, it was likely that additional sequencing of the present series of isolates would not be informative. The mutations identified in the three complete sequences are located within the three hot spots identified by Marichal et al. (18).
The RFLP analysis for D116E and E266D performed in the present study is important, as it is the first time that these point mutations have been screened for in a large collection of isolates, both susceptible and resistant. The fortuitous presence of a restriction enzyme recognition site at both locations allows easy screening of these isolates. Table 3 shows that both mutations are present in both susceptible and resistant isolates, at least in a heterozygous state. The absence of a susceptible isolate homozygous for 116E may be due to random sampling. These data indicate that two of the most frequent mutations identified in the survey are the result of allelic polymorphisms and are not likely to be associated with resistance.
To date, analyses of resistance have focused on oral isolates from HIV-infected patients, from whom matched susceptible and resistant isolates are most easily obtained (14, 27, 37, 42). The present analysis used the same techniques with a collection of unmatched isolates that included many isolates that were isolated from blood, urine, and other body fluids and locations. As isolates from HIV patients are usually oral, the isolates in this collection are unlikely to be associated with HIV infection. In addition, this survey documents the levels of resistance to three other azoles. A collection of isolates selected to include approximately equal numbers of isolates resistant and susceptible to fluconazole were shown to comprise a spectrum of susceptibility to the four azoles.
The most recent analysis of matched sets of oral isolates from HIV-infected patients described pump overexpression by 85% of fluconazole-resistant isolates and ERG11 overexpression by 35% of fluconazole-resistant isolates (27). Estimates from the present survey showed that pump overexpression was found in 31% of fluconazole-resistant isolates (35% if isolates SDD to fluconazole are included), but it was also found in 11% of fluconazole-susceptible isolates. Similar percentages were found for the other azoles. The levels of expression of ERG11 represent a gradual continuum, so it is difficult to define overexpression of this gene without a matched control.
The methods used in this analysis included Northern blotting analysis, DNA sequencing, and RFLP analysis, all of which can be done relatively quickly with a collection of isolates. Additional techniques could be applied to these isolates, although such techniques require extensive effort for each isolate. For instance, biochemical analysis of ergosterol synthesis could be performed to determine the sensitivity of the ergosterol synthesis pathway to the azole drugs, as reported previously (40). Microsomal analysis of Erg11p could also be performed to determine the susceptibility of an isolate (11). Drug accumulation can also be monitored, as described previously (4), by using fluorescent dyes. However, previous work suggests that the CDR pumps are primarily responsible for the efflux of rhodamine 6G, so the data from studies with fluorescent dyes may be limited. Finally, chromosomal copy numbers can be altered in resistant isolates compared to the number in matched susceptible isolates (28). However, Candida chromosome patterns are frequently used to differentiate strains (29), so differences in chromosome size or copy number between strains in a collection may not be specifically associated with resistance. Finally, ERG11 genes from resistant isolates can be expressed in Saccharomyces, in which they can be analyzed in greater detail (27, 35). Such an analysis is best controlled by use of a susceptible version of the enzyme, which was not available from the present collection of isolates, and the techniques used to express the genes eliminate the allelic variation that was shown to occur in the collection of isolates.
The known mechanisms of azole resistance were previously defined in matched sets of susceptible and resistant isolates, each from a single strain (for a review, see reference 41). Alterations in the resistant isolates were identified because they differ from the susceptible isolates of the same strain. However, these matched sets are rarely available in a clinical setting. It was of interest to use the methods that have been applied to matched sets of isolates in an analysis of a random collection of isolates for which matched susceptible and resistant isolates were not available. In this collection, mRNA levels could not be compared between susceptible and resistant isolates; they could only be normalized to the levels of mRNA for actin. Therefore, we are only comparing absolute levels in this collection, not increases within matched sets. The lack of correlation between resistance and levels of mRNA for CDR1, CDR2, MDR1, and ERG11 in this collection is a lack of correlation of absolute levels and not a result of differences in levels of mRNA expression between susceptible and resistant isolates. Similarly, point mutations in the isolates could be associated with resistance, or they could be the result of allelic variations between different strains. C. albicans is known to reproduce predominantly by asexual fission (31) and to contain allelic differences, including balanced lethals, within each strain (38). In addition, several other factors contribute to resistance. Multiple molecular mechanisms have been associated with resistance, strains can differ in their levels of resistance to four different azole drugs, and each C. albicans strain reproduces clonally, resulting in highly unrelated strains. Therefore, it is difficult to make statistical arguments about correlations between molecular mechanisms and resistance phenotypes. Because of the differences associated with the analysis of mechanisms of resistance in matched sets of isolates and random isolates and because of the complexity of resistance in C. albicans strains, mechanisms of resistance identified in matched sets of isolates are not easily identified in random isolates.
Molecular tests based on our understanding of resistance mechanisms are unlikely to be sufficient to define clinical resistance in C. albicans. It may be important to consider alternate molecular tests that would monitor the general health or status of the fungal cell in the presence of drug in a shorter time than the present MIC determinations allow. Such tests might be able to accurately identify a resistant fungal isolate quickly, without the need for detailed knowledge about the exact molecular mechanisms of resistance.
Acknowledgments
This research was supported by NIH NIDR grant R01 DE-11367. T.C.W. was supported by a New Investigator Award from the M. J. Murdock Charitable Trust and was the recipient of a New Investigator Award in Molecular Pathogenic Mycology from the Burroughs Wellcome Fund. D.A.S. was supported for this work by a grant from the Alza Corp.
REFERENCES
- 1.Balan, I., A. M. Alarco, and M. Raymond. 1997. The Candida albicans CDR3 gene codes for an opaque-phase ABC transporter. J. Bacteriol. 179:7210-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Yaacov, R., S. Knoller, G. A. Caldwell, J. M. Becker, and Y. Koltin. 1994. Candida albicans gene encoding resistance to benomyl and methotrexate is a multidrug resistance gene. Antimicrob. Agents Chemother. 38:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calabrese, D., J. Bille, and D. Sanglard. 2000. A novel multidrug efflux transporter gene of the major facilitator superfamily from Candida albicans (FLU1) conferring resistance to fluconazole. Microbiology (United Kingdom) 11:2743-2754. [DOI] [PubMed] [Google Scholar]
- 4.Clark, F. S., T. Parkinson, C. A. Hitchcock, and N. A. R. Gow. 1996. Correlation between rhodamine 123 accumulation and azole sensitivity in Candida species: possible role for drug efflux in drug resistance. Antimicrob. Agents Chemother. 40:419-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fling, M. E., J. Kopf, A. Tamarkin, J. A. Gorman, H. A. Smith, and Y. Koltin. 1991. Analysis of a Candida albicans gene that encodes a novel mechanism for resistance to benomyl and methotrexate. Mol. Gen. Genet. 227:318-329. [DOI] [PubMed] [Google Scholar]
- 6.Franz, R., S. Michel, and J. Morschhauser. 1998. A fourth gene from the Candida albicans CDR family of ABC transporters. Gene 220:1-2. [DOI] [PubMed] [Google Scholar]
- 7.Goldway, M., D. Teff, R. Schmidt, A. B. Oppenheim, and Y. Koltin. 1995. Multidrug resistance in Candida albicans: disruption of the BENr gene. Antimicrob. Agents Chemother. 39:422-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 9.Holmberg, K., and D. A. Stevens. 1999. Resistance to antifungal drugs: current status and clinical implications. Curr. Opin. Anti-Infective Investig. Drugs 1:306-317. [Google Scholar]
- 10.Lai, M. H., and D. R. Kirsch. 1989. Nucleotide sequence of cytochrome P450 L1A1 (lanosterol 14 alpha-demethylase) from Candida albicans. Nucleic Acids Res. 17:804.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamb, D. C., D. E. Kelly, T. C. White, and S. L. Kelly. 2000. The R467K amino acid substitution in Candida albicans sterol 14α-demethylase causes drug resistance through reduced affinity. Antimicrob. Agents Chemother. 44:63-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Law, D., C. B. Moore, H. M. Wardle, L. A. Ganguli, M. G. Keaney, and D. W. Denning. 1994. High prevalence of antifungal resistance in Candida spp. from patients with AIDS. J. Antimicrob. Chemother. 34:659-668. [DOI] [PubMed] [Google Scholar]
- 13.Loffler, J., S. L. Kelly, H. Hebart, U. Schumacher, C. Lass Florl, and H. Einsele. 1997. Molecular analysis of cyp51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol. Lett. 151:263-268. [DOI] [PubMed] [Google Scholar]
- 14.Lopez Ribot, J. L., R. K. McAtee, L. N. Lee, W. R. Kirkpatrick, T. C. White, D. Sanglard, and T. F. Patterson. 1998. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 42:2932-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Losberger, C., and J. F. Ernst. 1989. Sequence of the Candida albicans gene encoding actin. Nucleic Acids Res. 17:9488.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyons, C. N., and T. C. White. 2000. Transcriptional analyses of antifungal drug resistance in Candida albicans. Antimicrob. Agents Chemother. 44:2296-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Marichal, P., L. Koymans, S. Willemsens, D. Bellens, P. Verhasselt, W. Luyten, M. Borgers, F. C. S. Ramaekers, F. C. Odds, and H. Vanden Bossche. 1999. Contribution of mutations in the cytochrome P450 14 alpha-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology (United Kingdom) 10:2701-2713. [DOI] [PubMed] [Google Scholar]
- 19.Marr, K. A., C. N. Lyons, T. R. Rustad, R. A. Bowden, and T. C. White. 1998. Rapid, transient fluconazole resistance in Candida albicans is associated with increased mRNA levels of CDR. Antimicrob. Agents Chemother. 42:2584-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marr, K. A., T. C. White, J. A. H. van Burik, and R. A. Bowden. 1997. Development of fluconazole resistance in Candida albicans causing disseminated infection in a patient undergoing marrow transplantation. Clin. Infect. Dis. 25:908-910. [DOI] [PubMed] [Google Scholar]
- 21.McCullough, M. J., K. V. Clemons, and D. A. Stevens. 1999. Molecular and phenotypic characterization of genotypic Candida albicans subgroups and comparison with Candida dubliniensis and Candida stellatoidea. J. Clin. Microbiol. 37:417-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mori, T., M. Matsumura, Y. Kanamaru, S. Miyano, T. Hishikawa, S. Irie, K. Oshimi, T. Saikawa, and T. Oguri. 1997. Myelofibrosis complicated by infection due to Candida albicans: emergence of resistance to antifungal agents during therapy. Clin. Infect. Dis. 25:1470-1471. [DOI] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard. NCCLS document M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.Nolte, F. S., T. Parkinson, D. J. Falconer, S. Dix, J. Williams, C. Gilmore, R. Geller, and J. R. Wingard. 1997. Isolation and characterization of fluconazole- and amphotericin B-resistant Candida albicans from blood of two patients with leukemia. Antimicrob. Agents Chemother. 41:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odds, F. C. 1988. Candida and Candidosis: a review and bibliography. Bailliere Tindall, London, United Kingdom.
- 26.Pelletier, R., J. Peter, C. Antin, C. Gonzalez, L. Wood, and T. J. Walsh. 2000. Emergence of resistance of Candida albicans to clotrimazole in human immunodeficiency virus-infected children: in vitro and clinical correlations. J. Clin. Microbiol. 38:1563-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perea, S., J. L. Lopez Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perepnikhatka, V., F. J. Fischer, M. Niimi, R. A. Baker, R. D. Cannon, Y. K. Wang, F. Sherman, and E. Rustchenko. 1999. Specific chromosome alterations in fluconazole-resistant mutants of Candida albicans. J. Bacteriol. 181:4041-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfaller, M. A., C. J. Rhine, S. W. Redding, J. Smith, G. Farinacci, A. W. Fothergill, and M. G. Rinaldi. 1994. Variations in fluconazole susceptibility and electrophoretic karyotype among oral isolates of Candida albicans from patients with AIDS and oral candidiasis. J. Clin. Microbiol. 32:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prasad, R., W. P. De, A. Goffeau, and E. Balzi. 1995. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet. 27:320-329. [DOI] [PubMed] [Google Scholar]
- 31.Pujol, C., J. Reynes, F. Renaud, M. Raymond, M. Tibayrenc, F. J. Ayala, F. Janbon, M. Malli'e, and J. M. Bastide. 1993. The yeast Candida albicans has a clonal mode of reproduction in a population of infected human immunodeficiency virus-positive patients. Proc. Natl. Acad. Sci. USA 90:9456-9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Redding, S., J. Smith, G. Farinacci, M. Rinaldi, A. Fothergill, C. J. Rhine, and M. Pfaller. 1994. Resistance of Candida albicans to fluconazole during treatment of oropharyngeal candidiasis in a patient with AIDS: documentation by in vitro susceptibility testing and DNA subtype analysis. Clin. Infect. Dis. 18:240-242. [DOI] [PubMed] [Google Scholar]
- 33.Rex, J. H., M. A. Pfaller, J. N. Galgiani, M. S. Bartlett, A. Espinel-Ingroff, M. A. Ghannoum, M. Lancaster, F. C. Odds, M. G. Rinaldi, T. J. Walsh, and A. L. Barry. 1997. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro/in vivo correlation data for fluconazole, itraconazole, and Candida infections. Clin. Infect. Dis. 24:235-247. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143:405-416. [DOI] [PubMed] [Google Scholar]
- 37.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whelan, W. L., and D. R. Soll. 1982. Mitotic recombination in Candida albicans: recessive lethal alleles linked to a gene required for methionine biosynthesis. Mol. Gen. Genet. 187:477-485. [DOI] [PubMed] [Google Scholar]
- 39.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White, T. C. 1997. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α-demethylase in Candida albicans. Antimicrob. Agents Chemother. 41:1488-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White, T. C., M. A. Pfaller, R. G. Rinaldi, J. Smith, and S. W. Redding. 1997. Stable azole drug resistance associated with a substrain of Candida albicans from an HIV-infected patient. Oral Dis. 3:S102-S109. [DOI] [PubMed] [Google Scholar]