Abstract
Lipid A is the hydrophobic anchor of lipopolysaccharide (LPS) and forms the major lipid component of the outer monolayer of the outer membrane of gram-negative bacteria. Lipid A is required for bacterial growth and virulence, and inhibition of its biosynthesis is lethal to bacteria. UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase (LpxC) is a metalloenzyme that catalyzes the second step in the biosynthesis of lipid A. Inhibitors of LpxC have previously been shown to have antibiotic activities. We have screened a metalloenzyme inhibitor library for antibacterial activities against an Escherichia coli strain with reduced LpxC activity. From this screen, a series of sulfonamide derivatives of the α-(R)-amino hydroxamic acids, exemplified by BB-78484 and BB-78485, have been identified as having potent inhibitory activities against LpxC in an in vitro assay. Leads from this series showed gram-negative selective activities against members of the Enterobacteriaceae, Serratia marcescens, Morganella morganii, Haemophilus influenzae, Moraxella catarrhalis, and Burkholderia cepacia. BB-78484 was bactericidal against E. coli, achieving 3-log killing in 4 h at a concentration 4 times above the MIC, as would be predicted for an inhibitor of lipid A biosynthesis. E. coli mutants with decreased susceptibility to BB-78484 were selected. Analysis of these mutants revealed that resistance arose as a consequence of mutations in the fabZ or lpxC genes. These data confirm the antibacterial target of BB-78484 and BB-78485 and validate LpxC as a target for gram-negative selective antibacterials.
The clinical efficacy of many existing antibiotics is now being threatened by the emergence of multidrug-resistant pathogens. There is an urgent need for compounds that act on novel molecular targets that circumvent the established resistance mechanisms. Gram-negative bacteria, which are responsible for a large range of infectious diseases, have unique outer membranes that contain lipopolysaccharides which render them impermeable to certain antibiotics and bactericidal compounds (17). Lipid A forms the hydrophobic anchor of lipopolysaccharides and causes many of the toxic side effects associated with gram-negative infections (20). Lipid A from Escherichia coli is a hexaacylated disaccharide of glucosamine sugars, bearing phosphate residues at positions 1 and 4′ (19). Lipid A is required for bacterial growth and virulence, and the inhibition of its biosynthesis is lethal to gram-negative bacteria (7, 13). In addition, certain mutations in the lpxA, lpxC, and lpxD genes have been shown to render bacteria hypersensitive to hydrophobic antibiotics such as erythromycin (19, 24, 25). The enzymes and genes involved in E. coli lipid A synthesis have been extensively studied and characterized (19) and provide new targets in the search for antibacterial agents. The first step of this pathway involves the lpxA gene product, which catalyzes the transfer of an R-3-hydroxymyristoyl moiety from R-3-hydroxymyristoyl acyl carrier protein to the third position of the glucosamine ring of UDP-N-acetylglucosamine (5). This reaction is thermodynamically unfavorable, and consequently the second reaction of the pathway is the committed step of lipid A biosynthesis (1). This step is catalyzed by UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase (LpxC)(27), which removes an acetyl moiety from the 2-N position of glucosamine in the lipid A precursor UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine.
E. coli LpxC contains a bound zinc ion that is required for catalytic activity (10, 11). Chiral phenyloxazoline-based hydroxamates, which are presumed to coordinate the active site metal of LpxC, have been described that have significant antibacterial activities (3, 18). More recently, hydroxamate- and phosphinate-based substrate LpxC inhibitors have been described, though these do not possess significant antibacterial activities (12). The zinc ion-dependent mechanism of LpxC makes it a very attractive target, as there are many potent inhibitors of other zinc metalloenzymes that have been described. These include angiotensin-converting enzyme inhibitors, such as captopril, and inhibitors of the matrix metalloproteinases, such as marimastat (2). Recently, a potent, orally bioavailable antibacterial inhibitor of the metalloenzyme peptide deformylase (PDF) has also been described (4).
We have screened a metalloenzyme inhibitor library for compounds with antibacterial activities. Following this screen, we identified a series of sulfonamide derivatives of the α-(R)-amino hydroxamic acids, exemplified by BB-78484 and BB-78485, which had potent bactericidal gram-negative activities. We have demonstrated that the target of these compounds is LpxC and that decreased susceptibility to these inhibitors can occur through mutations in the fabZ gene, whose gene product is involved in fatty-acid biosynthesis, or in the lpxC target gene. These data further validate LpxC as a target for gram-negative selective antibacterials.
MATERIALS AND METHODS
Bacterial strains, plasmids, enzymes, and chemicals.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown in Mueller-Hinton broth (Oxoid Ltd., Basingstoke, United Kingdom) at 37°C, unless otherwise stated. The medium was supplemented with carbenicillin (100 μg/ml), kanamycin (50 μg/ml), or BB-78484 as needed. Enzymes were purchased from Promega (Southampton, United Kingdom) and used in accordance with the manufacturer's instructions. The potassium salt of UDP-3-O-(R-3-hydroxymyristoyl)GlcNAc was obtained from the Alberta Research Council (Alberta, Canada). Chemical reagents were from Sigma (Poole, United Kingdom), unless otherwise indicated.
TABLE 1.
Properties and/or descriptions of the bacterial strains and plasmids used in this study
| Strain or plasmid | Property and/or description | Source |
|---|---|---|
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Life Technologies |
| D21 | proA23 lac-28 tsx-81 trp-30 his-51 rpsL173(strR) ampCp-1 | CGSCa |
| D22 | lpxC101 proA23 lac-28 tsx-81 trp-30 his-51 rpsL173(strR) tufA1 ampCp-1 | CGSCa |
| BL21 (DE3) | F−ompT hsdS(rB− mB−) gal dcm λ(DE3) | CN Biosciences |
| XL10-Gold | TetrΔ (mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F′ proAB lacIqZΔM15 Tn10 (Tetr) Amy Camr] | Stratagene |
| Plasmids | ||
| pET24a | T7 expression vector, Kanr | CN Biosciences |
| pPCR-Script-Amp | Cloning vector, Ampr | Stratagene |
| pFC1 | fabZ cloned in pPCR-Script-Amp | This study |
| pFC2 | lpxC cloned in pPCR-Script-Amp | This study |
CGSC, E. coli Genetic Stock Center.
Recombinant DNA techniques.
Plasmid DNAs were prepared using a Wizard Plus Minipreps DNA purification kit (Promega). All other DNA manipulations and cell transformations were performed using previously described methods (22).
PCR and sequencing.
PCRs were performed in a Touchdown thermocycler (Hybaid, Ashford, United Kingdom) using Pfu polymerase. DNA sequencing was performed using dye terminator cycle sequencing and an ABI377 DNA Sequencer (PE Applied Biosystems, Warrington, United Kingdom).
Construction of plasmid pFC1.
The E. coli fabZ gene was amplified by PCR, using the appropriate forward and reverse primers. The forward primer (5′-TA GAATTCCAAGCGTCTGAAATCGCTTGAGCGCAAGGT-3′) started upstream of the sequence of the predicted promoter and the reverse primer (5′-TA GAATTCTCAGGCCTCCCGGCTACGAGCACACATCAT-3′) introduced an EcoRI site downstream from the translational stop codon. (The italics indicate the restriction enzyme recognition sequence.) The PCR product was ligated into pPCR-Script-Amp (Stratagene, Amsterdam, The Netherlands) to give plasmid pFC1. The ligation mixture was used to transform Epicurian Coli XL10-Gold-competent E. coli cells (Stratagene), and the sequence of the PCR insert was verified by DNA sequencing.
Construction of expression plasmid for LpxC.
The gene encoding E. coli LpxC was amplified by PCR, using the appropriate forward and reverse oligonucleotide primers. The forward primer (5′-AT CATATGATCAAACAAAGGACACTTAAACGTA-3′) introduced an NdeI site overlapping with the start codon and the reverse primer (5′-AT GAATTCTTATGCCAGTACAGCTGAAGGCGCTTTGAA-3′) introduced an EcoRI site downstream from the stop codon. The PCR product was digested with the corresponding restriction enzymes, isolated from an agarose gel, and ligated into pET24a (CN Biosciences, Nottingham, United Kingdom). The ligation mixture was used to transform E. coli DH5α-competent cells, and the colonies were selected on agar plates containing kanamycin (50 μg/ml). After verifying the DNA sequence, plasmid DNA was transformed into E. coli BL21 (DE3) cells. pET24a contains the T7 promoter and is designed to allow protein expression in the presence of T7 RNA polymerase. This polymerase is provided by the BL21 (DE3) host strain, which contains a chromosomal copy of the T7 RNA polymerase gene under control of the lacUV5 promoter. Expression is induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG), which provides a tightly regulated bacterial expression system (23).
Purification of E. coli LpxC.
A 3-liter culture of BL21 (DE3) cells containing the pET-24a/LpxC plasmid was grown to an optical density at 600 nm of 0.6. IPTG was added to a final concentration of 0.4 mM, and the cells were incubated for a further 3 h at 25°C. The cells were harvested by centrifugation at 5,000 × g for 10 min and the cell pellet was resuspended and washed once with ice-cold 10 mM potassium phosphate (pH 8.0) (buffer A) and stored at −70°C. The cell pellet was resuspended in 100 to 120 ml of buffer A and sonicated on ice for five 60-s bursts by using a medium probe and a 22-μm amplitude setting. The cell debris was pelleted by centrifugation at 15,000 × g for 25 min. The soluble fraction was loaded onto a preequilibrated (with buffer A) Q-Sepharose anion exchange column (2.5 by 6 cm) at 8 ml/min. The column was washed with the same buffer until the A280 returned to baseline and was eluted using a step gradient of 0 to 1 M NaCl in buffer A. The protein eluted at around 0.2 M NaCl was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The fractions containing the protein were pooled (30 ml) and concentrated to approximately 6 ml by using a 5-kDa-cutoff Centricon ultrafiltration device (Millipore, Watford, United Kingdom). The protein sample was loaded onto a Superdex 75 column (1.6 by 70 cm) and eluted at 1 ml/min with buffer A at 4°C. Multiple runs were carried out on this column, as a maximum of only 2 ml could be loaded per run. The fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the cleanest were pooled and concentrated. The protein concentration was determined at A280, using a calculated extinction coefficient of 24,500 M−1 cm−1 (10).
LpxC assay.
LpxC assays were performed in black 96-well Optiplates (Packard BioScience, Pangbourne, United Kingdom) with reaction mixtures (0.1 ml) containing 40 mM sodium morpholinoethanesulfonic acid (MES) buffer (pH 6.0), 0.02% Brij 35, 80 μM dithiothreitol, 25 μM UDP-3-O-(R-3-hydroxymyristoyl)GlcNAc, 50 ng (approximately 1.5 nM) of E. coli LpxC (>95% purity)/ml, and 2% (vol/vol) dimethyl sulfoxide (DMSO) vehicle with or without inhibitor. Reactions were started by the addition of enzyme and allowed to proceed for 30 min at 37°C, prior to stopping with 40 μl of 0.625 M sodium hydroxide. After 10 min at 37°C to hydrolyze the 3-O-acyl ester (10), 40 μl of 0.625 M acetic acid was added and the UDP-glucosamine was detected with 0.12 ml of o-phthaldialdehyde (OPA)-2-mercaptoethanol (250 nmol) in 0.1 M borax (pH 9.5) (21). Fluorescence (340 nm excitation/460 nm emission) was measured using a series 400 Fluostar fluorometer (BMG Labtechnologies, Aylesbury, United Kingdom). In some experiments, the acetic acid neutralization step was omitted and the detection reagents were added in 0.16 ml of 0.2 M boric acid at its natural pH. LpxC inhibitors were prepared in DMSO and diluted to 10% (vol/vol) DMSO for assay.
Antibiotic susceptibility determination.
Organisms were tested by broth or agar microdilution, using Mueller-Hinton broth or agar, as described by the National Committee for Clinical Laboratory Standards (16). The MIC of the compound was defined as the lowest concentration that prevented visible growth of the bacteria after incubation at 37°C for 20 h. Antimicrobial combinations were assessed using the checkerboard method (6).
Time-kill analysis.
The test strains were grown overnight at 37°C in Mueller-Hinton broth and then diluted with fresh broth to yield a starting inoculum of approximately 106 CFU/ml. BB-78484 and control antibiotics were added at a final concentration four- or eightfold above their MICs, and the cultures were incubated with agitation at 37°C. A parallel culture containing no antibiotic served as a control. Colony counts were determined at intervals by serial dilution onto Tryptone Soya agar plates. Antibiotic carryover was eliminated by using a dilution factor of at least 100.
Isolation of BB-78484-resistant mutants and determination of mutation frequencies.
Approximately 109 CFU of exponentially growing bacteria were plated onto Mueller-Hinton agar containing concentrations of BB-78484 from 1 to 64 μg/ml. After 48 h of incubation, the colonies were counted and the mutation frequencies were determined relative to the total number of viable organisms plated.
RESULTS
Metalloenzyme inhibitor library screen.
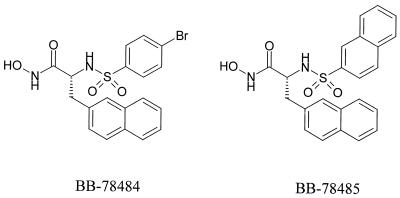
A proprietary low-molecular weight library of compounds featuring metal chelating groups was screened for antibacterial activities against E. coli D22 (lpxC101). This strain was predicted to be differentially hypersensitive to LpxC inhibitors, as it carries a mutation in the lpxC gene which leads to impaired activity of LpxC (27). The strain also shows increased sensitivity to a range of antibiotics due to the reduced biosynthesis of lipid A (Table 2). A number of compounds with MICs of less than 1 μg/ml against E. coli D22 were identified from this screen. The most potent compounds (MIC < 0.1 μg/ml) derived from a series of sulfonamide derivatives of the α-(R)-amino hydroxamic acids, of which BB-78484 and BB-78485 are examples (Fig. 1 and Table 2). Against a wider panel of pathogens, BB-78484 and BB-78485 showed predominantly gram-negative activities (Table 3), with activities against members of the Enterobacteriaceae, Serratia marcescens, Morganella morganii, Moraxella catarrhalis, Haemophilus influenzae, and Burkholderia cepacia. Neither compound showed activity against Pseudomonas aeruginosa; however, against a “leaky” P. aeruginosa strain, some activity was seen with BB-78485. BB-78484 and BB-78485 had little or no activity against the gram-positive bacteria tested.
TABLE 2.
MICs of antibiotics against D21 and D22 (lpxC101) and the results of synergy studies
| Antibiotic | MIC (μg/ml)
|
Synergy with BB-78484a | |
|---|---|---|---|
| D21 | D22 (lpxC101) | ||
| BB-78484 | 4 | 0.016 | |
| BB-78485 | 2 | 0.016 | |
| Gentamicin | 2 | 1 | additive |
| Rifampin | 4 | 0.031 | additive |
| Tetracycline | 2 | 0.5 | additive |
| Chloramphenicol | 8 | 2 | no effect |
| Carbenicillin | 32 | 4 | no effect |
| Ceftriaxone | 0.25 | 0.063 | no effect |
| Ciprofloxacin | 0.031 | 0.016 | no effect |
Antimicrobial combinations were assessed using the checkerboard method (6).
FIG. 1.
Structures of the LpxC inhibitors BB-78484 and BB-78485.
TABLE 3.
Activities of LpxC inhibitors against a range of pathogens
| Organism | MIC (μg/ml)
|
||
|---|---|---|---|
| BB-78484 | BB-78485 | Imipenem | |
| Escherichia coli ATCC 25922 | 2 | 1 | 0.25 |
| Escherichia coli ATCC 12014 | 2 | 1 | 0.125 |
| Klebsiella pneumoniae ATCC 13883 | 4 | 2 | 1 |
| Enterobacter cloacae ATCC 13047 | 32 | 4 | 0.25 |
| Pseudomonas aeruginosa ATCC 27853 | >32 | >32 | 4 |
| Pseudomonas aeruginosa C53 | >32 | 4 | 0.125 |
| Morganella morganii ATCC 36030 | 8 | 4 | 4 |
| Haemophilus influenzae Syn 1390 | >32 | 4 | 2 |
| Moraxella catarrhalis Syn 2412 | >32 | 4 | <0.125 |
| Burkholderia cepacia Syn 2072 | 16 | 1 | 4 |
| Serratia marcescens ATCC 8100 | 2 | 1 | 1 |
| Staphylococcus aureus ATCC 29213 | >32 | 32 | <0.125 |
| Staphylococcus epidermidis ATCC 27626 | >32 | >32 | <0.125 |
| Micrococcus luteus ATCC 9341 | >32 | >32 | <0.125 |
In vitro inhibition of LpxC.
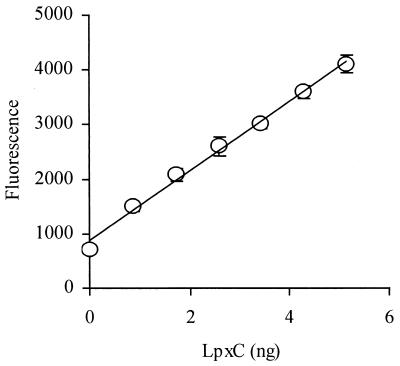
In order to confirm the molecular target of this class of inhibitor, we developed a novel homogeneous fluorometric E. coli LpxC assay. Cleavage of the natural substrate, UDP-3-O-(R-3-hydroxymyristoyl)GlcNAc, was measured using OPA to detect the sugar amine product (see Materials and Methods). As shown in Fig. 2, there was a linear LpxC-dependent increase in fluorescence over a range of 0 to 5 ng (0 to 1.5 nM) for this enzyme. In the absence of substrate, there was no such increase in fluorescence, indicating that the concentration of protein in the assay was too low to cause a significant background with OPA (data not shown). Based on a calibration using glucosamine 1-phosphate, the specific activity of E. coli LpxC was estimated to be 1.2 μmol/min/mg of protein. This value is somewhat lower than might be expected based on a kcat of 3.3 s−1, probably because E. coli LpxC requires 0.5 mg of bovine serum albumin/ml to achieve maximal activity (10).
FIG. 2.
LpxC activity (arbitrary units) as a function of enzyme concentration.
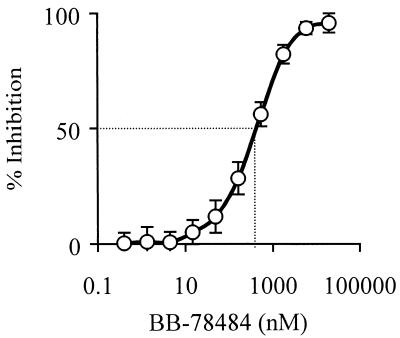
Using this assay, LpxC was inhibited by BB-78484 with a 50% inhibitory concentration (IC50) value of 400 ± 90 nM (0.18 μg/ml) (Fig. 3). The S-2-naphthyl compound, BB-78485, gave an IC50 value of 160 ± 70 nM (0.08 μg/ml), consistent with its improved activity on whole cells (Table 3). For comparison, the LpxC inhibitor L-161,240 was also tested. This compound exhibits an IC50 value of 26 nM when assayed with 3 μM UDP-3-O-(R-3-hydroxymyristoyl)GlcNAc (18). Characteristically for a competitive inhibitor, this value increased to 440 ± 10 nM in our assay due to the higher substrate concentration (25 μM).
FIG. 3.
In vitro inhibition of E. coli LpxC by BB-78484. The IC50s for BB-78484, BB-78485, and L161,240 were 400 ± 90, 160 ± 70, and 440 ± 10, respectively.
The bacterial metalloenzyme PDF has been shown to be inhibited by hydroxamate-based compounds (4). Neither BB-78484 nor BB-78485 showed any activity in a PDF enzyme assay at concentrations up to 1 μM.
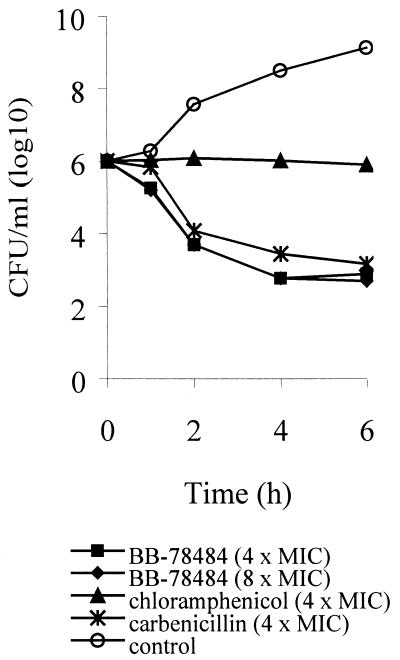
Antibacterial activity is bactericidal.
LpxC inhibitors inhibited the growth of E. coli ATCC 25922 in a bactericidal manner. Using BB-78484 at both 4 and 8 times above the MIC, viable bacterial counts were decreased by more than 3 log units in 4 h (Fig. 4). This killing was comparable to the action of the known bactericidal agent carbenicillin and contrasted with that of chloramphenicol, a known bacteriostatic agent.
FIG. 4.
Time-kill kinetics against E. coli ATCC 25922. A comparison of the effects of the addition of BB-78484 (4 and 8 × MIC) to those of chloramphenicol (4 × MIC) and carbenicillin (4 × MIC) on CFU/ml is shown.
Synergy studies.
Certain mutations in the lpxC gene have been shown to lead to increased susceptibility to other antibiotics, due to increased permeability in the gram-negative outer membrane (19) (Table 2). It has been previously suggested that inhibitors of lipid A biosynthesis may act synergistically with other antibiotics, facilitating their entry into the pathogen (18). This possibility was examined in a series of checkerboard experiments using E. coli D21 and BB-78484. Despite the increased susceptibility of E. coli D22 (lpxC101) to a range of antibiotics, no significant synergistic effects were detected with these antibiotics, although additive effects were seen with gentamicin, rifampin, and tetracycline (Table 2).
Development of resistance to BB-78484.
The spontaneous mutation frequencies of BB-78484 against E. coli ATCC 25922 and a laboratory-derived K-12 strain, DH5α, were determined. Bacteria were plated on antibiotic-containing plates at 8 times the agar MIC for each organism and incubated for 48 h at 37°C. BB-78484-resistant mutants arose at frequencies of 4 × 10−8 for E. coli DH5α and 2 × 10−9 for E. coli ATCC 25922. Analysis of the resistant mutants revealed they had 4- to 64-fold decreases in susceptibility to BB-78484 (Table 4) but remained sensitive to a range of other antibiotics (data not shown).
TABLE 4.
Genotypic characterizations of BB-78484-resistant mutants
| E. coli strain(s) | Altered gene | Triplet change | Amino acid change | BB-78484 MIC (μg/ml) |
|---|---|---|---|---|
| DH5α | 1 | |||
| 1, 3, 5, 8, 9 | fabZ | GCA → GTA | A69 → V | 32 |
| 2, 6 | fabZ | GCG → GTG | A78 → V | 32 |
| 11 | fabZ | AAG → CAG | K102 → Q | 64 |
| 7, 10, 12 | fabZ | ATG → GTG | M144 → V | 32 |
| 4 | lpxC | ATC → ACC | I38 → T | 16 |
| ATCC 25922 | 2 | |||
| 11 | fabZ | CCT → CTT | P56 → L | 32 |
| 8 | fabZ | CCG → ACG | P59 → T | 32 |
| 1 | fabZ | ATT → AGT | I60 → S | 128 |
| 5, 6, 7 | fabZ | GCA → GTA | A69 → V | 32 |
| 2, 3, 4, 12 | fabZ | CAG → CAT | Q72 → H | 64-128 |
| 9 | ? | ? | 8 |
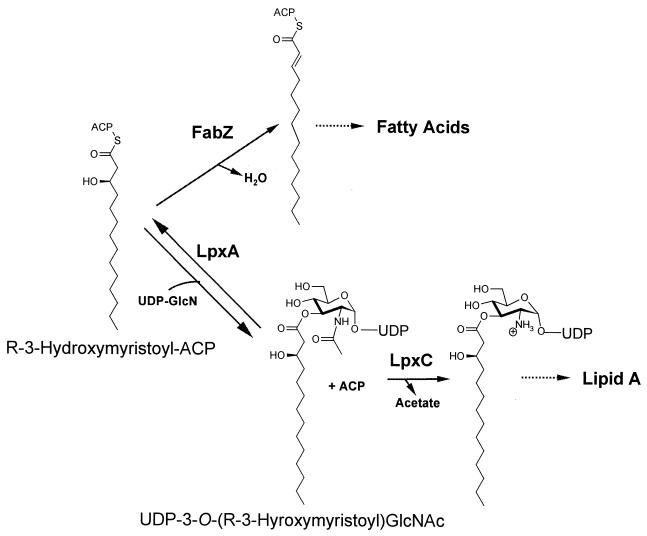
The mechanism of resistance was investigated by sequencing candidate genes involved in lipid A biosynthesis. The sequences of the fabZ, lpxA, lpxB, lpxC, and lpxD genes of the DH5α parent strain and of 12 DH5α BB-78484-resistant mutants were determined. The wild-type DH5α gene sequences were identical to those of the E. coli genome database entries. The majority of resistant strains harbored a mutation within the fabZ gene, while one strain contained a mutation within the lpxC gene. All other sequenced genes were identical to those of the wild type strain (Table 4). DNA sequence analysis of the fabZ and lpxC genes of the E. coli ATCC 25922 mutants revealed a similar pattern, with 10 of the 11 strains analyzed harboring a mutation in the fabZ gene. The remaining mutant was wild type for both lpxC and fabZ. The fabZ gene encodes R-3-hydroxymyristoyl acyl carrier protein dehydrase, which acts on a precursor common between lipid A and fatty acid biosynthesis (Fig. 5). FabZ mutants have previously been described as suppressors of certain lpxC mutations (15) and may function by increasing the concentration of the LpxC substrate available for lipid A biosynthesis. Eight independent mutations in fabZ were identified, with each mutant possessing a single base pair change resulting in a single amino acid substitution (Table 4). Certain mutations were found more frequently than others; FabZ A69V occurred in eight independently obtained BB-78484-resistant mutants. The MICs of BB-78484 against all the fabZ mutants were similar, leading to an average 32-fold decrease in susceptibility. The lpxC mutant I38T had a 16-fold decrease in susceptibility to BB-78484.
FIG. 5.
The involvement of a common precursor in the synthesis of lipid A and saturated fatty acids. ACP, acyl carrier protein; UDP-GlcN, UDP-glucosamine; UDP-3-O-(R-3-Hydroxymyristoyl)GlcNAc, UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine.
To confirm that the mutations in fabZ were responsible for the decreased susceptibility to BB-78484, the E. coli DH5α mutant strains FabZ A69V and FabZ A78V were transformed with the wild-type fabZ gene cloned into the plasmid pPCR-Script-Amp. The presence of the plasmid copy of the wild-type fabZ gene restored the susceptibility to BB-78484 of the strains, indicating the recessive nature of these mutations. Conversely, introduction of a plasmid copy of the lpxC gene had no effect on the susceptibility of the lpxC or fabZ mutant strains (data not shown).
DISCUSSION
Metalloenzymes represent a significant proportion of bacterial genomes, but to date no marketed antibiotics target metalloenzymes. We have developed a library of inhibitors that feature metal binding groups and have used it to screen for antibacterial activity. From this screen, a series of sulfonamide derivatives of the α-(R)-amino hydroxamic acids, exemplified by BB-78484 and BB-78485, were identified as having significant antibacterial activities. In particular, an E. coli strain carrying the lpxC101 mutation, which leads to a significant reduction in LpxC activity (27), was hypersensitive to these inhibitors, showing a 250-fold increase in sensitivity (Table 2). LpxC has recently been identified as a metalloenzyme (10), and phenyloxazoline hydroxamate LpxC inhibitors had been shown to have antibacterial activities (18). This result suggested that the metalloenzyme LpxC was the target of this novel class of compound. However, strains carrying the lpxC101 mutation also show increased susceptibility to a range of other antibiotics due to the perturbation of the outer membrane (Table 2).
To confirm that BB-78484 and BB-78485 were LpxC inhibitors, we developed an in vitro LpxC assay. Until recently (26), LpxC has been assayed using a procedure based upon UDP-3-O-(R-3-hydroxymyristoyl)GlcNAc labeled at either the α-phosphate or the acetyl group with 32P (13) or 3H (9), respectively. Such procedures require a complex protocol to prepare the labeled substrate and require the separation of product and substrate prior to measurement. For this report, we used synthetic (nonradioactive) UDP-3-O-(R-3-hydroxymyristoyl)GlcNAc and developed a novel homogeneous fluorometric assay using OPA to measure the formation of the sugar amine product. Using this assay, BB-78484 and BB-78485 showed IC50 values of 400 ± 90 nM and 160 ± 70 nM, respectively, which compared favorably with that of 440 ± 10 nM for the previously described phenyloxazoline-based hydroxamic acid inhibitor L-161,240 (18).
The hydroxamate-based LpxC inhibitors presumably act by analogy with other zinc metalloenzymes by binding the catalytic Zn2+ ion and displacing the Zn2+-bound water molecule (8). Derivatives of BB-78484 in which the hydroxamate is replaced by a carboxyl group show no significant antibiotic or LpxC-inhibitory activities (data not shown). BB-78484 and BB-78485 showed activity against a range of gram-negative pathogens tested and showed promising activity against members of the Enterobacteriaceae, Serratia marcescens, and Burkholderia cepacia. Although no antibacterial activity was seen against wild-type Pseudomonas aeruginosa, some activity was seen against a “leaky” P. aeruginosa strain, suggesting that access to this target rather than lack of LpxC inhibitory activity may limit the potency of these compounds. Previous studies with the hydroxamate phenyloxazoline inhibitor L-161,240 have shown that compared with the E. coli enzyme, it is significantly less potent against the P. aeruginosa enzyme (18) and has no activity against S. marcescens and P. aeruginosa whole cells. The spectrum of activities seen with the sulfonamide hydroxamate series suggests the possibility of obtaining LpxC inhibitors with broad-spectrum gram-negative activities.
Strains with increased resistance to BB-78484 were obtained against a laboratory-derived E. coli K-12 strain and a wild-type strain with frequencies of 4 × 10−8 and 2 × 10−9, respectively. To determine the nature of these mutants, we selected a set of candidate genes that could potentially be implicated in decreased susceptibility to BB-78484, namely those involved in lipid A biosynthesis and neighboring pathways. Sequence analysis of mutants derived from E. coli DH5α and ATCC 25922 revealed that the majority of sequence changes occurred in the fabZ gene, with one mutant containing a mutation within the presumed target gene lpxC (Table 4). The fabZ gene encodes a dehydrase involved in the metabolic pathway responsible for fatty acid biosynthesis. The FabZ substrate, R-3-hydroxymyristoyl acyl carrier protein, is situated at an important biosynthetic branch point, as it is also involved in the synthesis of lipid A (Fig. 5). Mutations in fabZ which lead to a reduction in dehydrase activity have previously been isolated as suppressors of lpxC mutations (15). It has been proposed that such mutations initially cause an increase in the pool of R-3-hydroxymyristoyl-ACP, the substrate of LpxA and the first step in lipid A biosynthesis. This increase in turn would lead to an increase in the concentration of the LpxC substrate, UDP-3-O-(R-3-hydroxymyristoyl)GlcNAc and thus would suppress the inhibition of LpxC by mass action (Fig. 5). These mutations are predicted to be recessive in nature, as restoration of full dehydrase activity would lead to a normal turnover of the R-3-hydroxymyristoyl acyl carrier protein. The fabZ mutations found in this study (Table 4) differ from those of the previously reported lpxC suppressor mutants, R100H, F101Y, P104S, and R121H (14, 15). It is assumed that the mutations we report also lead to a decrease in dehydrase activity, although this has not been demonstrated. However, consistent with the predicted recessive nature of such mutations, introduction of the wild-type fabZ gene back into the strains harboring the fabZ A69V and A78V mutations restored the susceptibility of these strains to BB-78484.
In addition to the fabZ mutations, one strain with a mutation in the lpxC gene was also isolated. This mutation, LpxC I38T, does not occur within a conserved region of LpxC, and the role of this residue awaits further structural information on the LpxC protein. However, the mutant strain maintains its sensitivity to a range of diverse antibiotics (data not shown), suggesting that it does not significantly reduce LpxC activity, thus producing the same effect on the outer-membrane integrity of the cell as found with some alleles of lpxC (e.g., lpxC101). Instead, it is probable that the I38T mutation affects the ability of LpxC to bind BB-78484. It is assumed that such a mutation will be dominant, and as expected, introduction of the wild-type lpxC gene into this strain did not significantly affect its susceptibility to BB-78484.
As predicted for an inhibitor of lipid A biosynthesis, BB-78484 had a bactericidal mode of action, achieving 3-log killing in 4 h against a wild-type E. coli strain. Similar activities are observed in conditional mutants of lpxA at the restrictive temperature (7). The time-kill kinetics was similar to that seen for carbenicillin and contrasted with that seen for chloramphenicol, a bacteriostatic agent. It has been suggested that inhibitors of LpxC activity may act synergistically with other classes of antibiotics because of their ability to reduce lipid A biosynthesis and hence outer-membrane integrity (18). Despite the increased sensitivity of a range of antibiotic classes against E. coli D22 (lpxC101), no synergy was seen in standard checkerboard in vitro assays with BB-78484 against the parental strain E. coli D21. This suggests that only a narrow window for enhanced uptake of a second antibiotic exists with these inhibitors and may reflect a limited dose response range on lipid A biosynthesis in whole cells.
In conclusion, using a metalloenzyme inhibitor library we have discovered a distinct class of LpxC inhibitors that have potent bactericidal gram-negative selective activities. These data further confirm the potential of LpxC as a target for gram-negative selective antibacterials. Development of knowledge regarding the structure activity relationships of LpxC inhibitors and the structure of the enzyme will facilitate the design of potent and broad-spectrum inhibitors as useful antibiotics.
Acknowledgments
We thank Sameeh Salama at NAEJA Pharmaceutical Inc. for some of the antimicrobial susceptibility testing.
REFERENCES
- 1.Anderson, M. S., H. G. Bull, S. M. Galloway, T. M. Kelly, S. Mohan, K. Radika, and C. R. H. Raetz. 1993. UDP-N-acetylglucosamine acyltransferase of Escherichia coli. The first step of endotoxin biosynthesis is thermodynamically unfavorable. J. Biol. Chem. 268:19858-19865. [PubMed] [Google Scholar]
- 2.Beckett, R. P., A. H. Davidson, A. H. Drummond, P. Huxley, and M. Whittaker. 1996. Recent advances in matrix metalloproteinase inhibitor research. Drug Discov. Today 1:16-26. [PubMed] [Google Scholar]
- 3.Chen, M. H., M. G. Steiner, S. E. de Laszlo, A. A. Patchett, M. S. Anderson, S. A. Hyland, H. R. Onishi, L. L. Silver, and C. R. H. Raetz. 1999. Carbohydroxamido-oxazolidines: antibacterial agents that target lipid A biosynthesis. Bioorg. Med. Chem. Lett. 9:313-318. [DOI] [PubMed] [Google Scholar]
- 4.Clements, J. M., R. P. Beckett, A. Brown, G. Catlin, M. Lobell, S. Palan, W. Thomas, M. Whittaker, S. Wood, S. Salama, P. J. Baker, H. F. Rodgers, V Barynin, D. W. Rice, and M. G. Hunter. 2001. Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor. Antimicrob. Agents Chemother. 45:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman, J., and C. R. H. Raetz. 1988. First committed step of lipid A biosynthesis in Escherichia coli: sequence of the lpxA gene. J. Bacteriol. 170:1268-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eliopoulous, G. M., and R. C. Moellering. 1996. Antimicrobial combinations, p. 813-834. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. The Williams & Wilkins Co., Baltimore, Md.
- 7.Galloway, S. M., and C. R. H. Raetz. 1990. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J. Biol. Chem. 265:6394-6402. [PubMed] [Google Scholar]
- 8.Holmes, M. A., and B. W. Matthews. 1981. Binding of hydroxamic acid inhibitors to crystalline thermolysin suggests a pentacoordinate zinc intermediate in catalysis. Biochemistry 20:6912-6920. [DOI] [PubMed] [Google Scholar]
- 9.Hyland, S. A., S. S. Eveland, and M. S. Anderson. 1997. Cloning, expression and purification of UDP-3-o-Acyl-GlcNAc deacetylase from Pseudomonas aeruginosa: a metalloamidase of the lipid A biosynthesis pathway. J. Bacteriol. 179:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackman, J. E., C. R. H. Raetz, and C. A. Fierke. 1999. UDP-3-O-(R-3-Hydroxymyrystoyl)-N-acetylglucosamine deacetylase of Escherichia coli is a zinc metalloenzyme. Biochemistry 38:1902-1911. [DOI] [PubMed] [Google Scholar]
- 11.Jackman, J. E., C. R. H. Raetz, and C. A. Fierke. 2001. Site-directed mutagenesis of the bacterial metalloamidase UDP-(3-O-acyl)-N-acetylglucosamine deacetylase (LpxC). Identification of the zinc binding site. Biochemistry 40:514-523. [DOI] [PubMed] [Google Scholar]
- 12.Jackman, J. E., C. A Fierke, L. N. Tumey, M. Pirrung, T. Uchiyama, S. H. Tahir, O. Hindsgaul, and C. R. H. Raetz. 2000. Antibacterial agents that target lipid A biosynthesis in gram-negative bacteria. Inhibition of diverse UDP-3-O-(r-3-hydroxymyristoyl)-n-acetylglucosamine deacetylases by substrate analogs containing zinc binding motifs. J. Biol. Chem. 275:11001-11009. [DOI] [PubMed] [Google Scholar]
- 13.Kelly, T. M., S. A. Stachula, C. R. H. Raetz, and M. S. Anderson. 1993. The firA gene of Escherichia coli encodes UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase. The third step of endotoxin biosynthesis. J. Biol. Chem. 268:19866-19874. [PubMed] [Google Scholar]
- 14.Kloser, A., M. Laird, M. Deng, and R. Misra. 1998. Modulations in lipid A and phospholipid biosynthesis pathways influence outer membrane protein assembly in Escherichia coli K-12. Mol. Microbiol. 27:1003-1008. [DOI] [PubMed] [Google Scholar]
- 15.Mohan, S., T. M. Kelly, S. S. Eveland, C. R. H. Raetz, and M. S. Anderson. 1994. An Escherichia coli gene (fabZ) encoding R-3-hydroxymyristoyl acyl carrier protein dehydrase. Relation to fabA and suppression of mutations in lipid A biosynthesis. J. Biol. Chem. 269:32896-32903. [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. NCCLS document no. M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 17.Nikaido, H. 1996. Outer membrane, p. 29-47. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, Jr., B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 18.Onishi, H. R., B. A. Pelak, L. S. Gerckens, L. L. Silver, F. M. Kahan, M. H. Chen, A. A. Patchett, S. M. Galloway, S. A. Hyland, M. S. Anderson, and C. R. H. Raetz. 1996. Antibacterial agents that inhibit lipid A biosynthesis. Science 274:980-982. [DOI] [PubMed] [Google Scholar]
- 19.Raetz, C. R. H. 1996. Lipopolysaccharide biosynthesis, p. 1035-1063. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, Jr., B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 20.Raetz, C. R. H. 1993. Bacterial endotoxins: extraordinary lipids that activate eucaryotic transduction. J. Bacteriol. 175:5745-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth, M. 1971. Fluorescence reaction for amino acids. Anal. Chem. 4:880-882. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 24.Vaara, M. 1993. Antibiotic-supersusceptible mutants of Escherichia coli and Salmonella typhimurium. Antimicrob. Agents Chemother. 37:2255-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vuorio, R., and M. Vaara. 1992. The lipid A biosynthesis mutation lpxA2 of Escherichia coli results in drastic antibiotic supersusceptibility. Antimicrob. Agents Chemother. 36:826-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, W., M. Maniar, R. Jain, J. Jacobs, J. Trias, and Z. Yuan. 2001. A fluorescence-based homogeneous assay for measuring activity of UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase. Anal. Biochem. 290:338-346. [DOI] [PubMed] [Google Scholar]
- 27.Young, K., L. L. Silver, D. Bramhill, P. Cameron, S. S. Eveland, C. R. H. Raetz, S. A. Hyland, and M. S. Anderson. 1995. The envA permeability/cell division gene of Escherichia coli encodes the second enzyme of lipid A biosynthesis, UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase. J. Biol. Chem. 270:30384-30391. [DOI] [PubMed] [Google Scholar]