Abstract
Susceptibility to macrolides and lincosamides was investigated with 299 consecutive nonduplicate Streptococcus pyogenes clinical isolates collected over a 6-year period (1992 to 1997) from an area of central Italy. During this period, macrolide resistance rates steadily increased (from 9% in 1992 to 53% in 1997; P < 0.001). The increase was caused by isolates with a macrolide-lincosamide-streptogramin B resistance phenotype, carrying mostly erm(B) but also erm(TR) genes, that were not detected in the first 2 years and were detected with increasing prevalence (8, 5, 26, and 37%, respectively) during the following 4 years. During the same period, the prevalence of isolates with a macrolide resistance phenotype, carrying mef(A) determinants, did not vary significantly; on average it was 13%, with modest rate fluctuations in different years and no definite trend. Molecular typing revealed a remarkable clonal diversity among susceptible and resistant isolates and a notable heterogeneity of the genetic environment of the resistance genes. The analysis of clonal diversity in relation with resistance phenotypes and genotypes revealed that increased macrolide resistance rates were due to a complex interplay of different mechanisms, with a relevant contribution played by an “epidemic” spread of genetic elements carrying the erm(B) gene among the circulating streptococcal population.
Streptococcus pyogenes (group A streptococcus) remains one of the leading bacterial pathogens worldwide. Superficial infections caused by group A streptococci, such as pharyngitis and impetigo, are usually mild and self-limiting but ubiquitous and extremely common. On the other hand, the occurrence of severe invasive infections and of nonsuppurative sequelae, although less common, make of S. pyogenes a major public health concern (see reference 3 and references therein).
Macrolide antibiotics are among the preferred drugs for the treatment of group A streptococcal pharyngitis and are largely used in community medicine for empirical chemotherapy of respiratory tract infections, due to their clinical efficacy, good compliance, and low toxicity (34). Resistance to macrolides in S. pyogenes can be caused by two different mechanisms: (i) active drug efflux via a transmembrane pump encoded by horizontally acquired mef genes (7, 35) and (ii) modification of the 23S rRNA target by rRNA adenine methylases encoded by horizontally acquired erm genes (see references 19 and 38 and references therein). The Mef efflux system operates only with 14- and 15-membered ring macrolides (M resistance phenotype) (7), while ribosomal modification by Erm methylases prevents the binding of macrolides, lincosamides, and streptogramins B, leading to resistance to all these compounds (MLS resistance phenotype) (19). The MLS resistance phenotype can be expressed either constitutively (cross-resistance MLS [cMLS] resistance phenotype) or upon induction (inducible MLS [iMLS] resistance phenotype), with variable patterns of activation by different compounds (19, 38, 39).
Since the first report in the 1950s (20), a large number of epidemiological surveys on macrolide resistance in S. pyogenes have been carried out. Results of these studies overall revealed a remarkable variability of resistance rates at different times and in different epidemiological settings (for examples, see references 1, 8, 9, 11, 25, 31, 37, and 41). A clear relationship between resistance rates and the extent of macrolide usage in community medicine was also demonstrated, with relatively rapid variations of the former in response to modifications of prescription policies (6, 11, 30, 31).
Recently, increased macrolide resistance rates in clinical isolates of S. pyogenes have been reported in several countries (1, 6, 8, 25). This phenomenon, which likely reflects the vast popularity of the last generation of macrolides (such as azithromycin, clarithromycin, and roxithromycin) in community medicine, is a cause of considerable concern for antimicrobial chemotherapy.
In this study we investigated, at the level of resistance genes and of clonal diversity of the microbial population, the dynamics underlying the rapid increase of macrolide resistance rates observed in clinical isolates of S. pyogenes from an area of central Italy.
MATERIALS AND METHODS
Group A streptococcal isolates.
The 299 S. pyogenes organisms analyzed in this study were consecutive nonduplicate clinical isolates collected at the Laboratory of Clinical Bacteriology of the Institute of Infectious Diseases, University of Siena, Italy, from 1 January 1992 to 31 December 1997. Most isolates (279; 93%) were from pharyngeal swabs of patients with pharyngitis (in some cases complicated with scarlet fever). The remaining ones were from skin swabs of patients with impetigo (18; 6%) or from invasive infections (one from the lower respiratory tract and one from blood). The streptococcal isolates were identified as belonging to group A according to standard methods (27).
In vitro susceptibility testing.
MICs were determined by a broth microdilution method according to the guidelines of the National Committee for Clinical Laboratory Standards (NCCLS) (22). The breakpoints for susceptibility classification were those recommended by the NCCLS (23). S. pyogenes ATCC 10383 was used for quality control of susceptibility testing. Erythromycin and clindamycin were from Sigma Chemical Co. (St. Louis, Mo.). Josamycin was from ICN Biomedicals (Costa Mesa, Calif.). The triple-disk diffusion test to determine the phenotypes of erythromycin-resistant isolates was performed as described previously (13), except that the disks were placed at the edges of a triangle, spaced from each other by a distance of 20 mm (center to center). Results were interpreted as follows: resistance to only erythromycin indicated an M phenotype; resistance to the three antibiotics indicated a cMLS phenotype; resistance to erythromycin and josamycin associated with a blunting of the clindamycin zone of inhibition or resistance to erythromycin associated with a blunting of the josamycin and clindamycin zones of inhibition indicated an iMLS phenotype.
DNA hybridization experiments.
Colony blot hybridization was performed with cells grown directly on nitrocellulose filters (Schleicher & Schuell, Dassel, Germany) layered onto Columbia blood agar plates supplemented with 20 mM glycine. The streptococcal cell wall was lysed by placing the filter onto filter paper soaked with lysis solution I (0.5 M Tris-HCl [pH 8.0], 50 mM EDTA, 50 μg of hen egg white lysozyme [Sigma, grade I]/ml) for 1 h at 37°C. Otherwise, filter processing and hybridization conditions were as described previously (28). Southern blot hybridization was carried out with nitrocellulose membranes (Schleicher & Schuell) as described previously (28). DNA was extracted from S. pyogenes as described previously (5). The erm(B), erm(TR), and mef(A) probes were amplicons containing partially or entirely the respective genes, generated as described below (see “PCR experiments”). Probes were labeled with 32P by the random priming technique using a commercial kit (Roche Biochemicals, Mannheim, Germany). S. pyogenes 8B27, A200 (32), and 1A77 were included as positive hybridization controls for the erm(B), erm(TR), and mef(A) probes, respectively.
PCR experiments.
PCR for the erm(B) gene was carried out using primers ERMAM-up (5′-CACTTCAGGAGTGATTACATGAA) and ERMAM-dn (5′-CTCATAGAATTATTTCCTCCCGT), targeting amplification of a 765-bp region covering the entire erm(B) open reading frame (ORF), and the following cycling conditions: 94°C for 20 s, 54°C for 60 s, and 72°C for 30 s, repeated for 40 cycles. PCR for the erm(TR) gene was carried out using primers ERMTR-f (5′-CCCGAAAAATACGCAAAATTTCAT) and ERMTR-r (5′-CCCTGTTTACCCATTTATAAACG), targeting amplification of a 590-bp region internal to the erm(TR) ORF (32) and the following cycling conditions: 94°C for 20 s, 48°C for 60 s, and 72°C for 30 s, repeated for 40 cycles. PCR for the mef(A) gene was carried out using primers MEFA-up (5′-GACCAAAAGCCACATTGTGGA) and MEFA-dn (5′-CCTCCTGTCTATAATCGCATG), targeting amplification of a 1,432-bp region covering the entire mef(A) ORF and some flanks, as described previously (24). Restriction analysis was carried out with RsaI and HincII for the erm(B) amplicons (expected sizes of the restriction fragments, 339 + 175 + 98 + 78 + 75 bp and 628 + 137 bp, respectively) and for the erm(TR) amplicons (expected sizes of the restriction fragments, 307 + 203 + 80 bp and 447 + 143 bp, respectively) and with ClaI and BamHI for the mef(A) amplicons (expected sizes of the restriction fragments, 812 + 620 bp and 1,209 + 223 bp, respectively). Restriction enzymes were from Roche Biochemicals.
Analysis of the chromosomal DNA macrorestriction patterns by PFGE.
Chromosomal DNA for macrorestriction analysis was extracted from S. pyogenes isolates as described previously (33), with minor modifications (the PIV buffer contained Tris-HCl [pH 8] and 1 M NaCl; the ESP solution contained EDTA [pH 9] and 0.1% [wt/vol] sarkosyl). Pulsed-field gel electrophoresis (PFGE) separation of the restriction fragments was carried out using a CHEF-DRIII PFGE apparatus (Bio-Rad, Hercules, Calif.) and the following electrophoretic parameters: run time, 23 h; voltage, 6 V/cm; switch time ramp, 10 to 30 s at included angle of 120°. Analysis of the macrorestriction patterns was carried out with the help of the Diversity Database software (version 2.2.0; Bio-Rad), using an electronic database of images of restriction patterns. Clonal relatedness was inferred in consideration of the criteria proposed by Tenover et al. (36).
Statistical analysis.
The significance of the differences of the observed rates of M-type and MLS-type resistant isolates during the study period was evaluated with the χ2 test. Clustering of isolates according to the macrorestriction profiles was carried out according to the Dice coefficient in combination with the UPGMA clustering method. For clustering analysis the band intensity was not weighted.
RESULTS
Antibiotic susceptibility of S. pyogenes isolates.
Two hundred ninety-nine nonduplicate clinical isolates of S. pyogenes, consecutively collected during a 6-year period (from January 1992 to December 1997) at the Laboratory of Clinical Bacteriology of the Institute of Infectious Diseases of the University of Siena (central Italy), were investigated in this study.
In vitro susceptibilities to erythromycin (a 14-membered ring macrolide), josamycin (a 16-membered ring macrolide), and clindamycin (a lincosamide) were evaluated by microdilution assay for all the isolates. Of these, 214 (72%) were found to be susceptible to all three antibiotics, 21 (7%) were resistant to all of them, 21 (7%) were resistant to erythromycin and josamycin but not to clindamycin, and 43 (14%) were resistant to erythromycin only (Table 1).
TABLE 1.
Susceptibility patterns and resistance determinants of the 299 S. pyogenes isolates analyzed in this study
| Susceptibility categorya (no. of isolates) | MIC range (μg/ml) of: |
Resistance phenotypeb (no. of isolates) | Resistance determinantsc (no. of isolates) | ||
|---|---|---|---|---|---|
| Erythromycin | Josamycin | Clindamycin | |||
| Susceptible (214) | ≤0.06-0.25 | ≤0.06-0.5 | ≤0.06-0.5 | NAd | Nonee |
| Ermr Josr Clir (21) | >64 | >64 | >64 | cMLS (21) | erm(B) (19); erm(TR) (2) |
| Ermr Josr Clis (21) | >64 | >64 | 0.12-0.5 | iMLS (21) | erm(B) (19); erm(TR) (2) |
| Ermr Joss Clis (43) | 2->64 | ≤0.06-0.5 | ≤0.06-0.5 | M (40) | mef(A) (40) |
| iMLS (3) | erm(TR) (3) | ||||
According to the results of the broth microdilution assay. Superscripts: S, susceptible; R, resistant.
According to the results of the triple-disk diffusion assay (see text for more details).
The presence of resistance determinants was investigated by colony blot hybridization with specific probes and confirmed by PCR followed by restriction analysis of the amplification products (see text for more details).
NA, not applicable.
The presence of resistance determinants was screened by colony blot hybridization in 70 (33%) randomly selected susceptible isolates.
The 85 erythromycin-resistant isolates were then analyzed by a triple-disk diffusion test to determine the nature of their resistance phenotype. Results of this test assigned (i) a cMLS phenotype to the 21 isolates resistant to the three antibiotics, (ii) an iMLS phenotype to the 21 isolates resistant to erythromycin and josamycin and to 3 isolates resistant to erythromycin only, and (iii) an M phenotype to the remaining 40 isolates resistant to erythromycin only (Table 1).
The MICs of macrolides and clindamycin for the susceptible isolates were always ≤0.5 μg/ml. The M-type resistant isolates showed a low-level resistance to erythromycin (MICs, 2 to 16 μg/ml) and were susceptible to josamycin and clindamycin, with MICs similar to those of the susceptible isolates. The cMLS-type resistant isolates always exhibited a high-level resistance to both macrolides and clindamycin (>64 μg/ml). The iMLS-type resistant isolates showed a high-level resistance to the inducing compounds (erythromycin and, usually, also josamycin [>64 μg/ml]), while the MICs of the noninducing antibiotics (clindamycin and, occasionally, josamycin) were similar to those for the susceptible isolates (Table 1).
Resistance determinants.
The occurrence of genomic sequences related to the erm(B), erm(TR), and mef(A) genes, which are the macrolide resistance determinants so far identified in group A streptococci, was investigated in all the resistant isolates by means of colony blot hybridization and confirmed by PCR experiments.
In colony blot hybridization the erm(B) probe recognized 38 MLS-type resistant isolates (19 cMLS and 19 iMLS, all of which had a resistance phenotype inducible by erythromycin and josamycin), while it did not recognize any of the M-type resistant isolates. The erm(TR) probe recognized seven MLS-type resistant isolates (two cMLS and five iMLS, of which two were inducible by erythromycin and josamycin and three were inducible by erythromycin only), while it did not recognize any of the M-type resistant isolates. Hybridization of the same MLS-type resistant isolate with both erm probes was never observed. The mef(A) probe recognized all the isolates with an M phenotype, while it did not recognize any of the MLS-type resistant isolates. The three probes were also tested with 70 (33%) randomly selected susceptible isolates and always yielded negative results (Table 1).
PCR amplification of the resistance determinants, carried out with specific sets of primers as described in Materials and Methods, yielded amplification products of the expected sizes [765 bp for erm(B), 590 bp for erm(TR), and 1,432 bp for mef(A)] in the various resistant isolates. Results were always consistent with those of the colony blot assay. Restriction analysis of the amplicons revealed, in all cases, a pattern compatible with that of the respective sequences (data not shown), suggesting that the erm(B), erm(TR), and mef(A) determinants carried by the resistant isolates were either identical or closely related, at the sequence level, to those previously described (14, 32, 35).
Variation of the resistance rates, phenotypes, and determinants during the study period.
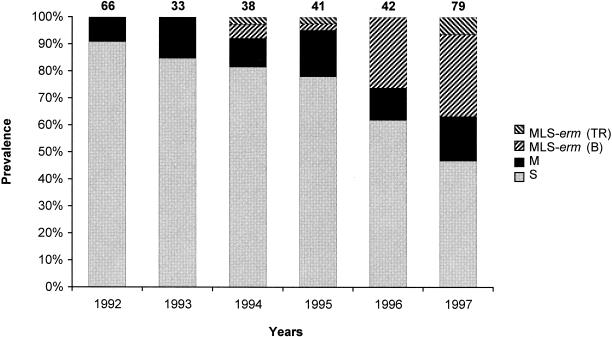
The prevalence of erythromycin-resistant isolates increased steadily, from 9% in 1992 to 53% in 1997, during the study period (Fig. 1).
FIG. 1.
Prevalence of macrolide resistance phenotypes and genotypes observed during the study period in the 299 S. pyogenes isolates. S, susceptible phenotype (░⃞); M, M resistance phenotype (▪); MLS-erm(B), MLS resistance phenotype with an erm(B) gene (▨); MLS-erm(TR), MLS resistance phenotype with an erm(TR) gene (▧). The number above each column represents the total number of isolates in the indicated year. The prevalence differences observed during the study period were not significant for the M-type resistant isolates and were statistically significant (P < 0.001) for the MLS-type resistant isolates.
The resistance phenotypes showed a different chronological distribution. Isolates with an M phenotype were encountered throughout the study period with modest rate fluctuations and no definite trend (Fig. 1). On the other hand, the isolates with an MLS phenotype have been encountered only since 1994 and their prevalence underwent a remarkable increase from 1996 onward (Fig. 1). The progressive increase of resistance rates observed during the study period, therefore, was essentially contributed by a net increase of MLS-type resistant isolates.
Concerning the distribution of the resistance determinants, both the erm(B) and the erm(TR) genes have been detected since 1994, but the erm(B)+ isolates were by far the most prevalent and constituted the principal cause for the increased resistance rates observed during the last 2 years (Fig. 1).
Clonal diversity of the group A streptococcal isolates.
Clonal relationships were investigated, by comparison of the PFGE profiles of genomic DNAs digested with SmaI, with all the resistant isolates and with a sample of 49 susceptible isolates collected at different times (14 of those collected in 1997 and 7 per year of those collected in the other years, selected at random). A readable restriction profile was obtained with all the 45 MLS-type resistant isolates, with 26 of the 40 M-type resistant isolates, and with 48 of the 49 susceptible isolates. The numbers of detectable bands ranged from 5 to 12, and their sizes ranged from 45 to 380 kb (data not shown). In the 15 isolates whose chromosomal DNA was resistant to restriction by SmaI, a macrorestriction profile could be obtained with SfiI (data not shown), suggesting that the presence of a specific DNA modification system rather than the quality of the DNA preparation was likely the cause of resistance to digestion by SmaI.
The SmaI PFGE profiles of the various isolates were remarkably heterogeneous. Comparison of the restriction profiles was used to generate a similitude tree. Considering as indicative of clonal relatedness a similitude coefficient higher than 73% (which usually corresponded to a difference of not more than four bands in the restriction profiles), the 120 genotyped isolates could be clustered into 46 different clonal lineages, with a variable number of minor variants within most of them. An analysis of the distribution of the various clones according to the year of isolation and to the resistance phenotype and genotype (Table 2) revealed the following aspects: (i) a notable clonal heterogeneity was found among either the susceptible isolates (27 clones among the 48 typeable isolates), or the M-type resistant isolates (10 clones among the 26 typeable isolates), or the MLS-type resistant isolates (14 clones among the 45 typeable isolates), although the degree of heterogeneity was apparently lower among the resistant isolates (the ratios of clones to isolates were 0.31 and 0.38 for the M- and MLS-type isolates, respectively, versus 0.60 for the susceptible isolates); (ii) some clones (either susceptible or resistant) were isolated over prolonged periods; (iii) the M- and MLS-type isolates were found to be clonally unrelated to each other except in one case (clone P); (iv) among the MLS-type isolates, the erm(B) and erm(TR) genes were sometimes found in clonally related isolates (e.g., clones D, XXI, and XXIII); (v) susceptible isolates clonally related to resistant ones carrying either an erm(B), or an erm(TR), or an mef(A) determinant were found (e.g., clones M, W, I, II, XX, XXII, and XXIII); (vi) the net increase of erm(B)+ isolates observed during the last two years was apparently contributed both by a phenomenon of clonal expansion and by the appearance of new clones.
TABLE 2.
Distribution of resistant and susceptible isolates subjected to PFGE genotyping according to clonal relatedness, resistance genotype and/or phenotype, and year of isolation
| Clone | Resistance characteristic(s)a of isolate(s) collected in: |
|||||
|---|---|---|---|---|---|---|
| 1992 | 1993 | 1994 | 1995 | 1996 | 1997 | |
| A | • | |||||
| B | • | |||||
| C | • | |||||
| D | ▸✚ | |||||
| E | • | |||||
| F | • | |||||
| G | ▪ | |||||
| H | ••• | • | ||||
| J | ▸ | |||||
| L | ▪ | |||||
| M | ▪ | •• | ||||
| N | ▪ | |||||
| O | ▪ | |||||
| P | ✚ | ▪▪ | ||||
| Q | • | |||||
| R | • | |||||
| S | ✚ | |||||
| T | • | |||||
| U | • | • | ||||
| W | •• | •• | • | ▸•• | •• | |
| Y | • | |||||
| Z | ▸ | |||||
| I | • | • | ✚ | • | ||
| II | • | ▪ | ||||
| III | ••• | • | • | |||
| IV | ▪▪ | ▪▪▪ | ▪▪▪▪ | |||
| V | • | |||||
| VI | ▪ | ▪ | ||||
| VII | • | |||||
| VIII | • | |||||
| IX | • | |||||
| X | •• | |||||
| XI | • | |||||
| XII | ▪ | ▪ | ▪ | |||
| XIII | ▪ | |||||
| XIV | ✚✚ | |||||
| XV | ✚✚ | |||||
| XVI | ✚✚✚ | |||||
| XVII | • | |||||
| XVIII | • | |||||
| XIX | ✚ | |||||
| XX | • | ✚✚✚✚ | ||||
| XXI | ✚ | ✚✚✚✚✚ | ▸✚✚✚✚✚ | |||
| XXII | •▪▪▪ | ▪▪ | ||||
| XXIII | • | ✚ | ▸▸✚✚✚✚✚✚✚✚✚✚ | |||
| RRb | □□□ | □ | ○□□□□ | □□□□□□ | ||
•, susceptible isolate; ○, restriction-resistant susceptible isolate; ▪, M-type resistant isolate; □, restriction-resistant M-type isolate; ✚, MLS-type resistant isolate with an erm(B) gene; ▸, MLS-type resistant isolate with an erm(TR) gene.
RR, group of nontypeable restriction-resistant isolates.
Among the 14 M-type isolates whose chromosomal DNA was resistant to SmaI, the SfiI macrorestriction profiles revealed the presence of several different clones (data not shown).
Analysis of the genetic environment of the resistance determinants.
In streptococci, the mef(A) and erm(B) genes are known to be carried on transposable elements that are usually chromosome borne and have been variously characterized (4, 12, 17, 18, 21, 29), while no information is currently available on the genetic support of erm(TR). The genetic environments of the mef(A) and erm(B) genes carried by the resistant isolates were analyzed by Southern blot experiments in which the genomic DNAs, digested with EcoRI [for mef(A)+ isolates] or with HindIII [for erm(B)+ isolates], were hybridized to the respective probes. The above enzymes were selected since they do not cut into the respective resistance genes but cut within the mobile elements carrying the respective genes that have thus far been characterized among streptococci (12, 14, 18, 21, 29, 35).
In the mef(A)+ isolates the probe always recognized a single EcoRI fragment, although it varied in size (2.0, 3.7, or 10.2 kb) (data not shown). The 2.0-kb hybridization pattern was found in the 14 isolates whose genomic DNA was resistant to digestion with SmaI, as well as in 5 typeable isolates belonging to three clones (P, IV, and VI). The 3.7-kb hybridization pattern was found in eight isolates belonging to the same three clones (although, usually, in different clonal variants) and in eight isolates belonging to six additional clones (G, L, O, II, XII, and XIII). These two patterns were the most prevalent and were found during the whole study period. The 10.2-kb hybridization pattern was less common, being found in isolates of a separate clone (XXII) detected since 1996. The 2.0-kb hybridization pattern could be consistent with the structure of Tn1207.1, an mef(A)-containing genetic element recently characterized from Streptococcus pneumoniae (29).
In the erm(B)+ isolates the Southern blot profiles were more heterogeneous. In most isolates the probe recognized a single HindIII fragment whose sizes were quite variable (2.8, 6.7, 9.7, 17, or 28 kb), while in a limited number of isolates it recognized two fragments (2.8 + 9.7 or 2.8 + 5.8 kb) (data not shown). No clear relationship was evident between clonality and the hybridization patterns, with identical patterns being consistently found in clonally unrelated isolates and different patterns in clonally related ones (although in this case the different patterns were usually found in different clonal variants). The 6.7- and 2.8-kb single-banded patterns were the first to be detected (in 1994 and in 1995, respectively) and persisted during the following years. The number of circulating patterns increased in the last 2 years, with four different patterns over a total of 11 erm(B)+ isolates in 1996 and seven different patterns over a total of 24 erm(B)+ isolates in 1997. Some hybridization patterns (5.8, 6.7, and 17 kb) could be consistent with the structures of some erm(B)-containing transposons previously identified in streptococci (Tn1545, Tn3872, and Tn3701, respectively) (4, 17, 21).
DISCUSSION
Resistance determinants and clonal relatedness among macrolide-resistant isolates of S. pyogenes have been recently investigated in several studies (2, 13, 15, 16, 37, 40), but a combined analysis of these aspects in consecutive clinical isolates from a defined epidemiological setting while macrolide resistance rates were rapidly increasing was not reported previously.
Analysis of the resistance phenotypes and determinants revealed that, in our area, the increased macrolide resistance rates observed in S. pyogenes were essentially contributed by MLS-type isolates, while the rates of M-type isolates did not vary significantly during the study period. Such findings were different from those observed elsewhere which showed that the increased resistance rates were mostly or almost uniquely contributed by M-type resistant isolates (16, 25, 26, 40), and this discrepancy underscores the variability that can occur in different epidemiological settings. The reasons for this variability, which could be relevant to the formulation of guidelines for empirical chemotherapy, remain as yet poorly understood. Different conditions of selective pressure (due to the use of different antimicrobials in clinical practice) and/or a variable linkage of the macrolide resistance determinants with other resistance or virulence genes (10) could be among the causes. Similar reasons could also account for the fact that, notwithstanding that both types of erm determinants have been detected since 1994, erm(TR) remained considerably less common than erm(B) during the following years and could account for the simultaneous presence of mef and erm genes in the same isolate, an occurrence which was never detected in this study but was reported elsewhere (2, 9, 13, 16).
The resistance phenotypes could always account for the presence of known resistance determinants: mef(A) was detected in all the M-type isolates, while erm(B) or the more recently described erm(TR) was found in the MLS-type isolates, with the possibility of constitutive or inducible expression for either type of methylase gene. The presence of cryptic erm or mef determinants was never observed in susceptible isolates from the same area, suggesting that a molecular approach based on detection of the resistance genes could have been highly predictive for testing macrolide resistance in group A streptococci. Analysis of the genetic environment of the mef(A) and erm(B) genes revealed a notable heterogeneity, especially for erm(B), suggesting a remarkable diversity of the cognate genetic elements present in the group A streptococcal population. Similar findings have also been reported for erm(B)-containing elements from pneumococci (21), although in that case the strains had originally been selected to represent different clones.
PFGE genotyping showed a remarkable clonal diversity in the group A streptococci circulating during the study period, in spite of a relatively low-stringency criterion adopted for the definition of clonal relatedness. This finding is consistent with those reported in other studies on isolates from either defined (9, 25, 26) or diverse (16) epidemiological settings and confirms the notion that the group A streptococcal populations circulating in human communities tend to be highly polyclonal. On the other hand, the finding of clonally related isolates at different times indicates that members of some lineages can circulate for years among the population of a certain area.
When microbial drug resistance is caused by acquired resistance determinants, as in this case, two mechanisms can variably contribute to increased resistance rates: (i) a clonal spread of strains that acquired the resistance determinants; (ii) an epidemic spread of the resistance determinants among the circulating microbial population by horizontal transfer. Analysis of clonal diversity in relation with resistance phenotypes and genotypes provided some insight on the mechanisms underlying the increased macrolide resistance rates observed among group A streptococci in our setting. Resistant strains were, at least in part, generated following the acquisition of resistance genes by circulating susceptible strains, as suggested by the consistent finding (for any type of resistance determinant) of resistant isolates that were clonally related to susceptible ones circulating during the same period. In fact, since only a fraction of the susceptible isolates were analyzed, a similar phenomenon could have been relatively common, although it is also possible that some resistant strains could have been de novo introduced into the circulating population from external sources. A clonal spread of resistant strains was also apparent, especially with some of the erm(B)+ strains, suggesting that a similar mechanism could have played a relevant role in the increase of resistance rates observed during the last 2 years. However, (i) the diversity of the erm(B) genetic environments, (ii) the heterogeneity at the subclonal level observed within the “expanded” clones, and (iii) the consistent finding of identical genetic environments in clonally unrelated strains suggest that the observed clonal spread most likely resulted from the acquisition of different elements carrying the resistance gene by members of a circulating susceptible clone, rather than from the spreading of a single strain that had originally acquired the resistance determinant. It could also be possible that the different erm(B) genetic environments observed within the same clone were the result of genetic rearrangements which occurred after the acquisition of the resistance determinant, but the finding of discrete patterns of genetic environments in different clones does not favor this mechanism as a major cause for the remarkable intraclonal diversity of the genetic elements bearing the erm(B) gene. In fact, the remarkable diversity of the genetic elements carrying erm(B) genes among the resistant isolates points to a scenario in which different mobile elements have been recruited in the circulating population of susceptible clones from external sources rather than to one in which a single mobile element arrived in the population of group A streptococci and then spread among the population. In conclusion, the rapid increase of macrolide resistance rates observed in our setting was likely due to a complex interplay of different mechanisms, among which an epidemic spread of genetic elements carrying the erm(B) gene among the circulating streptococcal population was apparently a relevant one. An investigation into the nature of these elements is currently under way.
Acknowledgments
We are grateful to Janne Kataja for kindly providing us with the S. pyogenes erm(TR)+ strain A200.
This work was supported in part by quota servizi P.A.R, from Università di Siena.
S.C. and M.L. contributed equally to this work.
REFERENCES
- 1.Baquero, F., J. A. Garcia-Rodríguez, J.G. de Lomas, L. Aguilar, and The Spanish Surveillance Group for Respiratory Pathogens. 1999. Antimicrobial resistance of 914 beta-hemolytic streptococci isolated from pharyngeal swabs in Spain: results of a 1-year (1996-1997) multicenter surveillance study. Antimicrob. Agents Chemother. 43:178-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bingen, E., F. Fitoussi, C. Doit, R. Cohen, A. Tanna, R. George, C. Loukil, N. Brahimi, I. Le Thomas, and D. Deforche. 2000. Resistance to macrolides in Streptococcus pyogenes in France in pediatric patients. Antimicrob. Agents Chemother. 44:1453-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisno, A. L., and D. S. Stevens. 2000. Streptococcus pyogenes, p. 2101-2116. In G. L. Mandell, J. E. Bennet, and R. Dolin (ed.), Principles and practice of infectious diseases, 5th ed. Churchill Livingstone, Inc., New York, N.Y.
- 4.Caillaud, F., C. Carlier, and P. Courvalin. 1987. Physical analysis of the conjugative shuttle transposon Tn1545. Plasmid 17:58-60. [DOI] [PubMed] [Google Scholar]
- 5.Chassy, B. M. 1976. A gentle method for the lysis of oral streptococci. Biochem. Biophys. Res. Commun. 68:603-608. [DOI] [PubMed] [Google Scholar]
- 6.Cizman, M., M. Pokoron, K. Seme, A. Orazem, and M. Paragi. 2001. The relationship between trends in macrolide use and resistance to macrolides of common respiratory pathogens. J. Antimicrob. Chemother. 47:475-477. [DOI] [PubMed] [Google Scholar]
- 7.Clancy, J., J. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retsema. 1996. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 8.Cornaglia, G., M. Ligozzi, A. Mazzariol, L. Masala, G. Lo Cascio, G. Orefici, The Italian Surveillance Group for Antimicrobial Resistance, and R. Fontana. 1998. Resistance of Streptococcus pyogenes to erythromycin and related antibiotics in Italy. Clin. Infect. Dis. 27:S87-S92. [DOI] [PubMed] [Google Scholar]
- 9.De Azavedo, J. C. S., R. H. Yeung, D. J. Bast, C. L. Duncan, S. B. Borgia, and D. E. Low. 1999. Prevalence and mechanisms of macrolide resistance in clinical isolates of group A streptococci from Ontario, Canada. Antimicrob. Agents Chemother. 43:2144-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Facinelli, B., C. Spinaci, G. Magi, E. Giovanetti, and P. E. Varaldo. 2001. Association between erythromycin resistance and ability to enter human respiratory cells in group A streptococci. Lancet 358:30-33. [DOI] [PubMed] [Google Scholar]
- 11.Fujita, K., K. Murono, M. Yoshinkawa, and T. Murai. 1994. Decline of erythromycin resistance of Group A streptococci in Japan. Pediatr. Infect. Dis. J. 13:1075-1078. [DOI] [PubMed] [Google Scholar]
- 12.Gay, K., and D. S. Stephens. 2001. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 184:56-65. [DOI] [PubMed] [Google Scholar]
- 13.Giovanetti, E., M. P. Montanari, M. Mingoia, and P. E. Varaldo. 1999. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob. Agents Chemother. 43:1935-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horinouchi, S., W. H. Byeon, and B. Weisblum. 1983. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J. Bacteriol. 154:1252-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kataja, J., P. Huovinen, M. Skurnik, The Finnish Study Group for Antimicrobial Resistance, and H. Seppälä. 1999. Erythromycin resistance genes in group A streptococci in Finland. Antimicrob. Agents Chemother. 43:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kataja, J., P. Huovinen, The Macrolide Resistance Study group, and H. Seppälä. 2000. Erythromycin resistance genes in group A streptococci of different geographical origins. J. Antimicrob. Chemother. 46:789-792. [DOI] [PubMed] [Google Scholar]
- 17.Le Bouguénec, C., G. de Cespédès, and T. Horaud. 1988. Molecular analysis of a composite chromosomal conjugative element (Tn3701) of Streptococcus pyogenes. J. Bacteriol. 170:3930-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Bouguénec, C., G. de Cespédès, and T. Horaud. 1990. Presence of chromosomal elements resembling the composite structure Tn3701 in streptococci. J. Bacteriol. 172:727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leclercq, R., and P. Courvalin. 1991. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowbury, E. J. L. 1958. Symposium on epidemiological risks of antibiotics hospital infections. Proc. R. Soc. Med. 51:807-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDougal, L. K., F. C. Tenover, L. N. Lee, J. K. Rasheed, J. E. Patterson, J. H. Jorgensen, and D. J. LeBlanc. 1998. Detection of Tn917-like sequences within a Tn916-like conjugative transposon (Tn3872) in erythromycin-resistant isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2312-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 23.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial disk susceptibility testing; 11th informational supplement. Approved standard M100-S11. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.Oster, P., A. Zanchi, S. Cresti, M. Lattanzi, F. Montagnani, C. Cellesi, and G. M. Rossolini. 1999. Pattern of macrolide resistance determinants among community-acquired Streptococcus pneumoniae isolates over a 5-year period of decreased macrolide susceptibility rates. Antimicrob. Agents Chemother. 43:2510-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palavecino, E. L., I. Riedel, X. Berrios, S. Bajaksouzian, D. Johnson, E. Kaplan, and M. R. Jacobs. 2001. Prevalence and mechanism of macrolide resistance in Streptococcus pyogenes in Santiago, Chile. Antimicrob. Agents Chemother. 45:339-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Trallero, E., J. M. Marimon, M. Montes, B. Orden, and M. de Pablos. 1999. Clonal differences among erythromycin-resistant Streptococcus pyogenes in Spain. Emerg. Infect. Dis. 5:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruoff, K. L., R. A. Whiley, and D. Beighton. 1999. Streptococcus, p. 283-296. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Santagati, M., F. Iannelli, M. R. Oggioni, S. Stefani, and G. Pozzi. 2000. Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2585-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seppälä, H., T. Klaukka, R. Lehtonen, E. Nenonen, and P. Huovinen. 1995. Outpatient use of erythromycin: link to increased erythromycin resistance in group A streptococci. Clin. Infect. Dis. 21:1378-1385. [DOI] [PubMed] [Google Scholar]
- 31.Seppälä, H., T. Klaukka, J. Vuopio-Varkila, A. Muotiala, H. Helenius, K. Lager, P. Huovinen, and The Finnish Study Group for Antimicrobial Resistance. 1997. The effect of changes in the consumption of macrolide antibiotics on erythromycin resistance in group A streptococci in Finland. N. Engl. J. Med. 337:441-446. [DOI] [PubMed] [Google Scholar]
- 32.Seppälä, H., M. Skurnik, H. Soini, M. C. Roberts, and P. Huovinen. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Single, L. A., and D. R. Martin. 1992. Clonal differences within M-types of the group A Streptococcus revealed by pulsed field gel electrophoresis. FEMS Microbiol. Lett. 91:85-90. [DOI] [PubMed] [Google Scholar]
- 34.Steigbigel, N. H. 1995. Macrolides and clindamycin, p. 334-346. In G. L. Mandell, J. E. Bennet, and R. Dolin (ed.), Principles and practice of infectious diseases, 4th ed. Churchill Livingstone, Inc., New York, N.Y.
- 35.Sutcliffe, J., A. Tait-Kamradt, and L. Wondrack. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzelepi, E., G. Kouppari, A. Mavroidi, A. Zaphiropoulou, and L. S. Tzouvelekis. 1999. Erythromycin resistance amongst group A β-haemolytic streptococci isolated in a pediatric hospital in Athens, Greece. J. Antimicrob. Chemother. 43:745-746. [DOI] [PubMed] [Google Scholar]
- 38.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisblum, B. 1995. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 39:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan, J. J., H. M. Wu, A. H. Huang, H. M. Fu, C. T. Lee, and J. J. Wu. 2000. Prevalence of polyclonal mefA-containing isolates among erythromycin-resistant group A streptococci in Southern Taiwan. J. Clin. Microbiol. 38:2475-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.York, M. K., L. Gibbs, F. Perdreau-Remington, and G. F. Brooks. 1999. Characterization of antimicrobial resistance in Streptococcus pyogenes isolates from the San Francisco Bay area of Northern California. J. Clin. Microbiol. 37:1727-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]