Abstract
The vanE operon was characterized from Enterococcus faecalis N00-410 (MIC of vancomycin = 24 μg/ml). The organization of the vanE operon was identical to that of the vanC1 operon from Enterococcus gallinarum, with protein identities ranging from 46 to 63%. An open reading frame located downstream of the vanE operon showed significant homology to a number of integrase genes, all of which are located downstream of the chromosomal GMP synthase gene guaA.
In enterococci, normal peptidoglycan precursors have d-Ala-d-Ala termini that strongly bind vancomycin, whereas in vancomycin-resistant enterococci, alternate biosynthetic pathways lead to precursors with termini that bind vancomycin poorly, thus conferring resistance (4, 18). The vanA (3), vanB (9, 15), and vanD (5, 6, 14) genes code for d-Ala-d-Lac ligases and are responsible for the acquired intermediate- to high-level resistance found mainly in Enterococcus faecalis and Enterococcus faecium. Intrinsic low-level vancomycin resistance is conferred by vanC1, vanC2, and vanC3, which code for d-Ala-d-Ser ligases found on the chromosomes of Enterococcus gallinarum, Enterococcus casseliflavus, and Enterococcus flavescens, respectively (7, 13). The acquired, nontransferable VanE d-Ala-d-Ser ligase found in E. faecalis also confers a low-level resistance phenotype (8). The function of the E. faecalis WCH9 putative vancomycin resistance gene vanG is unknown (11). The identification of the first vanE-containing E. faecalis isolated in Canada has recently been reported (17). In this report, we describe the characterization of the vanE resistance locus and the genes flanking this region.
E. faecalis N00-410 (MIC of vancomycin = 24 μg/ml) (17) and E. faecium ATCC 19434 were grown at 35°C in brain heart infusion broth or cation-adjusted Mueller-Hinton broth. Induction studies were performed as previously described (5). Transfer experiments were attempted by liquid mating with selection on phenol red agar plates (Difco) containing 1% l-arabinose and 5 μg of vancomycin/ml (11). Antimicrobial susceptibilities were determined by using Etest strips (AB Biodisk) or agar dilution according to NCCLS guidelines (12) for high levels of streptomycin and gentamicin. Genomic DNA was extracted from enterococci as previously described (5). An N00-410 DNA library (∼10-kb Sau3A fragments) in ZAP Express (Stratagene) was screened with a vanE PCR product generated with primers VANE1 and VANE2 (8). Labeling and detection of the probe were performed per the manufacturer's instructions (Amersham Pharmacia Biotech). The sequence was obtained by primer walking and by using the EZ::TN<TET-1> insertion kit (Epicentre Technologies). Inverse PCR was carried out with primers vanRE-1 (5′-TCTCGGCTTTTCATGCATC-3′) and vanSE-DN1 (5′-GAATGAAATTAATCATATTCG-3′) and with EcoRV-cut and -religated N00-410 DNA. Primers Eint-DN1 (5′-ATTCAAGGGATATTTTCAATAGC-3′) and guaDN-1 (5′-TTGCACATGTAAACCGTATCG-3′) were used to amplify a 0.9-kb fragment overlapping the inverse PCR product. Homology searches were conducted with BLAST (National Center for Biotechnology Information website [http://www.ncbi.nlm.nih]) (1).
E. faecalis N00-410 (MIC of vancomycin = 24 μg/ml) was isolated from an infected ankle wound of a patient who had no travel history outside of Manitoba for 20 years, had no extensive contact with out-of-province visitors, and had never received vancomycin (17). The strain was susceptible to teicoplanin (0.5 μg/ml), ampicillin (0.75 μg/ml), tetracycline (0.75 μg/ml), chloramphenicol (3 μg/ml), and high levels of streptomycin and gentamicin. This is only the second vanE-containing vancomycin-resistant Enterococcus strain isolated, the first being E. faecalis BM4405, which was isolated from a patient in Chicago who had received vancomycin (8).
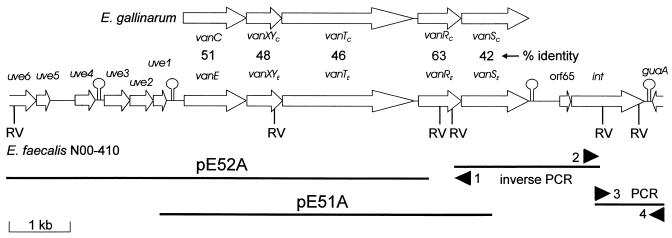
A total of 10,749 bp was characterized by sequence analysis (GenBank accession no. AF430807) of two vanE-positive clones, pE52A (6.9-kb insert) and pE51A (5.4-kb insert), a 2.1-kb inverse PCR fragment, and part of a 0.93-kb PCR product (Fig. 1). Five open reading frames (ORFs) constituting the vanE operon were found: vanE (d-Ala-d-Ser ligase), vanXYE (d,d-dipeptidase/d,d-carboxypeptidase), vanTE (serine racemase), vanRE (response regulator), and vanSE (sensor kinase) (Fig. 1). A comparison with the gene from the vanE type strain BM4405 (8) showed 4.1% nucleotide differences (97% amino acid identity), and thus, the genes may be considered variants. The overall gene order and protein similarity indicate that the vanE operon is the functional equivalent of the vanC1 operon from E. gallinarum (2), a finding which is not unexpected, as the vanE type strain BM4405 was found to synthesize peptidoglycan precursors terminating in d-serine residues (8). As in the vanC1 operon, some of the vanE operon genes overlap one another: vanXYE overlaps vanE, vanTE overlaps vanXYE, and vanRE overlaps vanSE. The percent G+C content of the vanE operon genes is between 30.7 and 35.7%, which is slightly lower than the 37.5% for E. faecalis V583 (http://www.tigr.org). Beginning 31 bp downstream of the vanSE stop codon is a region of dyad symmetry (ΔG = −12.6 kcal/mol) followed by a T-rich region which may play a role in vanE operon transcription termination. Growth studies with N00-410 showed that vancomycin resistance was inducible (data not shown), indicating that expression of the vanE operon may involve not only translational coupling but an active response regulator-sensor kinase system.
FIG. 1.
Genetic organization of the vanE locus from E. faecalis N00-410 showing regions cloned into plasmids and regions isolated by inverse PCR and PCR. The direction of transcription is indicated by arrows, and putative stem-loop structures are indicated. Primers used in inverse PCR and PCR are indicated by black arrowheads: primer 1 is vanRE-1, primer 2 is vanSE-DN1, primer 3 is Eint-DN1, and primer 4 is gua-DN1. The organization of the vanC1 locus from E. gallinarum is shown at the top. Percent identities of the corresponding vanC1 and vanE operon proteins are shown. RV indicates EcoRV sites.
Six ORFs were detected upstream of vanE (uve1 to uve6) (Fig. 1). The uve6 product exhibited 36% identity to the hypothetical Spy1691 protein of Streptococcus pyogenes (GenBank accession no. AE06599). The product of uve1 was 37% identical over its full length to the sigma factor SpoIIIG of Bacillus thuringiensis (accession no. I40582), and the product of uve2 was 26% identical over its full length to the sigma factor SpoIIG of Bacillus subtilis (accession no. M57606). In both cases, however, the enterococcal protein was about half the length of the Bacillus protein. The products of three other ORFs, uve3, uve4, and uve5, had no significant homology to extant proteins. The percent G+C content for the genes in this region was between 27.5 and 32.2%. Two regions of dyad symmetry were detected here: one downstream of uve4 (ΔG = −14.1 kcal/mol) and one downstream of uve1 (ΔG = −17.5 kcal/mol) (Fig. 1).
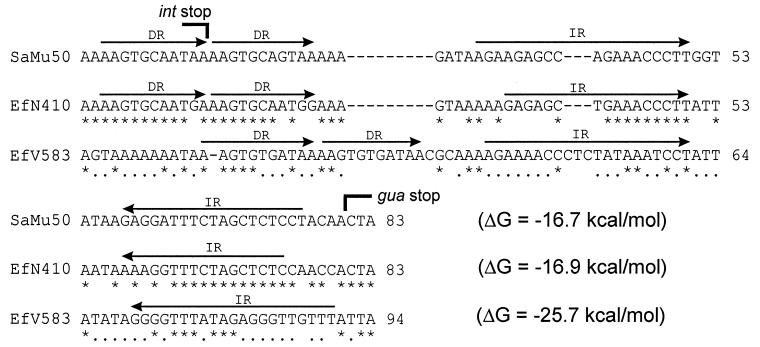
Three ORFs were found downstream of the vanE operon: orf65, int410, and the 3′ end of guaA, encoding GMP synthase (Fig. 1). The Int410 protein had 69% identity to the integrase of Tn5801 (Int5801, Orf SAV0392) from Staphylococcus aureus Mu50 (10) and the Int459 integrase of the tet(M) element from Clostridium perfringens CW459 (16). The Orf65 protein was 80% identical to the product of the SAV0393 gene, orf66 (66 amino acids), found just upstream of the int5801 gene. Furthermore, we identified a gene identical to orf66 in exactly the same location upstream of int439, although it was not annotated in the GenBank entry for the C. perfringens tet(M) element (accession no. AF329848). A comparison of the complete C. perfringens tet(M) element with Tn5801 revealed that they share >97% sequence identity (data not shown) and that in both organisms, the elements are inserted downstream of guaA. Using int410 to query the E. faecalis V583 genome (http://www.tigr.org), we identified an integrase gene (int583) downstream of guaA whose product had 53% identity to the Int410 protein. Furthermore, overlapping int583 by 8 bp was orf70 (70 amino acids), whose product exhibited 52 and 47% identity to Orf65 and Orf66, respectively. The location, size, and basic pI values of the proteins of orf65, orf66, and orf70 indicate that they may be excisionase proteins. Thus, it appears that the transposon-like elements described above belong to a group with integrative functions recognizing sequences downstream of guaA. Analysis of the int-guaA intergenic regions allowed for the identification of some common features (Fig. 2). In S. aureus and C. perfringens, the sequences are identical and the same length as the N00-410 region (67 bp), with which they share 67% identity. The E. faecalis V583 region (78 bp) exhibits 58% identity to the N00-410 region and 49% identity to the S. aureus and C. perfringens regions. Inverted repeats in each region may be involved in transcription termination of the convergently orientated int and guaA genes. Direct repeats of 11 bp in S. aureus, C. perfringens, and N00-410 involve a duplication of the end of the int genes and in E. faecalis V583 are found just downstream of the int gene. A direct repeat consensus sequence, AAGTGYRRTRR (Y = pyrimidine, R = purine), can be postulated and may be involved in the integration of these elements, as may the inverted repeats. Since guaA is a housekeeping gene, we assume it to be outside of the transposon-like elements.
FIG. 2.
Alignments of the int-guaA intergenic regions from S. aureus Mu50 (SaMu50), E. faecalis N00-410 (EfN410), and E. faecalis V583 (EfV583). The sequence from the equivalent region in C. perfringens CW459 is identical to that of S. aureus Mu50. Asterisks indicate an identical nucleotide in all three sequences, and periods indicate an identical nucleotide in two out of three sequences. The ΔG values of the inverted repeats are indicated after each sequence. DR, direct repeat; IR, inverted repeat.
The transfer of vancomycin resistance to E. faecium ATCC 19434 was unsuccessful. Transfer to an E. faecalis strain was not carried out due to the lack of a suitable recipient strain. Fines et al. (8) were unable to demonstrate the transfer of vancomycin resistance from E. faecalis BM4405 to E. faecalis JH2-2. Whether Tn5801 is transferable is unknown, while in C. perfringens CM459, the tet(M) gene appears to be nontransferable (16).
We have characterized the vanE locus from an E. faecalis strain isolated in Canada. The gene order and protein similarity indicate a close relationship to the vanC1 operon from E. gallinarum. Downstream is an integrase gene, which may have been involved in the initial acquisition of the vanE operon. The N00-410 vanE-containing element, the C. perfringens tet(M) element, Tn5801 from S. aureus Mu50, and a transposon-like element from E. faecalis V583 belong to a family of elements that can integrate downstream of guaA genes.
Acknowledgments
We acknowledge the excellent technical assistance of Tim Du, Romeo Hizon, Isabelle Paquin, and the DNA Core Facility, National Microbiology Laboratory. Preliminary sequence data for E. faecalis V583 was obtained from the website of The Institute for Genomic Research (http://www.tigr.org).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias, C. A., P. Courvalin, and P. E. Reynolds. 2000. vanC cluster of vancomycin-resistant Enterococcus gallinarum BM4174. Antimicrob. Agents Chemother. 44:1660-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur, M., C. Molinas, F. Depardieu, and P. Courvalin. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur, M., P. Reynolds, and P. Courvalin. 1996. Glycopeptide resistance in enterococci. Trends Microbiol. 4:401-407. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, D. A., J. Conly, H. Dedier, G. Peters, L. Robertson, E. Slater, and M. R. Mulvey. 2000. Molecular characterization of the vanD gene cluster and a novel insertion element in a vancomycin-resistant Enterococcus isolated in Canada. J. Clin. Microbiol. 38:2392-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadewall, B., and P. Courvalin. 1999. Characterization of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339. J. Bacteriol. 181:3644-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutka-Malen, S., C. Molinas, M. Arthur, and P. Courvalin. 1992. Sequence of the vanC gene of Enterococcus gallinarum BM4147 encoding a d-alanine:d-alanine ligase-related protein necessary for vancomycin resistance. Gene 112:53-58. [DOI] [PubMed] [Google Scholar]
- 8.Fines, M., B. Perichon, P. Reynolds, D. Sahm, and P. Courvalin. 1999. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. Antimicrob. Agents Chemother. 43:2161-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handwerger, S., and J. Skoble. 1995. Identification of chromosomal mobile element conferring high-level vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 39:2446-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 11.McKessar, S. J., A. M. Berry, J. M. Bell, J. D. Turnidge, and J. C. Paton. 2000. Genetic characterization of vanG, a novel vancomycin resistance locus of Enterococcus faecalis. Antimicrob. Agents Chemother. 44:3224-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.Navarro, F., and P. Courvalin. 1994. Analysis of genes encoding d-alanine-d-alanine ligase-related enzymes in Enterococcus casseliflavus and Enterococcus flavescens. Antimicrob. Agents Chemother. 38:1788-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perichon, B., P. Reynolds, and P. Courvalin. 1997. VanD-type glycopeptide resistant Enterococcus faecium BM4339. Antimicrob. Agents Chemother. 41:2016-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quintiliani, R., and P. Courvalin. 1996. Characterization of Tn1547, a composite transposon flanked by IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene 172:1-8. [DOI] [PubMed] [Google Scholar]
- 16.Roberts, A. P., P. A. Johanesen, D. Lyras, P. Mullany, and J. I. Rood. 2001. Comparison of Tn5397 from Clostridium difficile, Tn916 from Enterococcus faecalis and the CW459tet(M) element from Clostridium perfringens shows that they have similar conjugation regions but different insertion and excision modules. Microbiology 147:1243-1251. [DOI] [PubMed] [Google Scholar]
- 17.Van Caeseele, P., S. Giercke, J. Wylie, D. Boyd, M. Mulvey, S. Amin, and M. Ofner-Agostini. 2001. Identification of the first vancomycin-resistant Enterococcus faecalis harbouring vanE in Canada. Can. Commun. Dis. Rep. 27:101-104. [PubMed] [Google Scholar]
- 18.Woodford, N. 1998. Glycopeptide-resistant enterococci: a decade of experience. J. Med. Microbiol. 47:849-862. [DOI] [PubMed] [Google Scholar]