Abstract
DNA gyrase is a prokaryotic type II topoisomerase and a major target of quinolone antibacterials. The majority of mutations conferring resistance to quinolones arise within the quinolone resistance-determining region of GyrA close to the active site (Tyr122) where DNA is bound and cleaved. However, some quinolone resistance mutations are known to exist in GyrB. Present structural data suggest that these residues lie a considerable distance from the quinolone resistance-determining region, and it is not obvious how they affect quinolone action. We have made and purified two such mutant proteins, GyrB(Asp426→Asn) and GyrB(Lys447→Glu), and characterized them in vitro. We found that the two proteins behave similarly to GyrA quinolone-resistant proteins. We showed that the mutations exert their effect by decreasing the amount of quinolone bound to a gyrase-DNA complex. We suggest that the GyrB residues form part of a quinolone-binding pocket that includes DNA and the quinolone resistance-determining region in GyrA and that large conformational changes during the catalytic cycle of the enzyme allow these regions to come into close proximity.
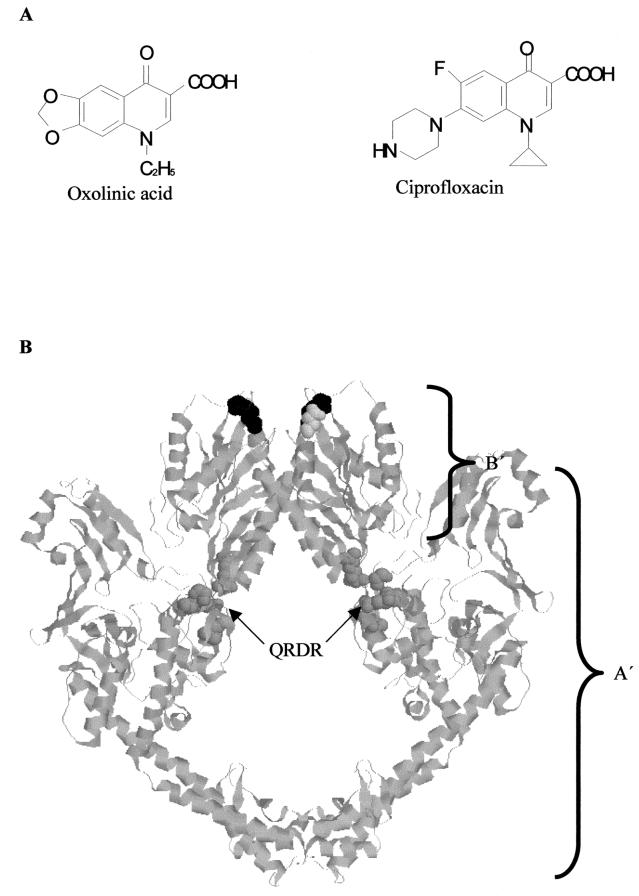
Quinolone drugs are a large and widely used class of synthetic antibacterial compounds (10, 11, 19). First-generation (acidic) quinolones include nalidixic acid and oxolinic acid (OXO).Subsequent generations have been modified to increase spectrum and potency. The most significant modification has been the addition of a fluorine atom at position C-6 in drugs such as ciprofloxacin (CFX), which results in a considerable increase in activity (30). The newer drugs also commonly contain a secondary amine in addition to the carboxylic acid group common to most quinolones, making them amphoteric rather than acidic (Fig. 1A). More recent modifications that increase drug potency include the presence of a methoxy group at C-8 (48).
FIG. 1.
(A) Structures of oxolinic acid and ciprofloxacin. (B) Crystal structure of the 92-kDa fragment of yeast topoisomerase II (5). A′, region corresponding to GyrA; B′, region corresponding to GyrB. Residues corresponding to quinolone resistance residues on GyrA are in space-filling format and colored dark gray: Tyr782 (GyrA Tyr122), Ala766 (GyrA Gln106), Gln743 (GyrA Asp87), Gln739 (GyrA Ser83), and Ala725 (GyrA Arg67). The two residues equivalent to the GyrB quinolone resistance residues Asp426 and Lys447 are Asp451 and Lys477, respectively, in yeast topoisomerase II and are colored light gray and black, respectively. QRDR, quinolone resistance-determining region.
In Escherichia coli the major target for the quinolones is DNA gyrase (14, 35). DNA gyrase is a type II topoisomerase that is able to alter the topological state of DNA by cleaving both strands, passing a double strand of DNA through the gap and resealing the ends (6). In the presence of ATP, this results in negative supercoiling, a reaction unique to gyrase. The crystal structures of the 43-kDa N-terminal domain of GyrB (responsible for ATP hydrolysis and capturing a DNA strand) and a 59-kDa fragment of the 64-kDa N-terminal domain of GyrA (containing the residues for DNA binding and cleavage and the quinolone resistance-determining region [QRDR; residues 67 to 106]) have been determined (24, 39). The structure of the 47-kDa C-terminal domain of GyrB is not known but can be inferred from the analogous enzyme from Saccharomyces cerevisiae, topoisomerase II, for which the crystal structures of a 92-kDa region have been solved (5, 12).
Resistance to quinolones commonly arises in DNA gyrase via mutations in the QRDR (44). Such mutations include GyrA(Ser83→Trp), which gives ≈20-fold resistance to a wide range of quinolones (9, 46). This region of GyrA is close to the active site, where the DNA is bound, and close to the tyrosine at position 122, where the phosphotyrosine link between enzyme and DNA is formed. Quinolone drugs bind to gyrase-DNA complexes, but only weakly to either gyrase or DNA (34, 37, 38, 41, 47). It is thought that quinolones bind to a pocket consisting of the QRDR of GyrA and the region of distorted DNA bound to it (17). In such a pocket, the quinolone would interact with elements of both the DNA and the enzyme.
Two clinical isolates resistant to quinolones have mutations in the 47-kDa C-terminal region of GyrB (42, 43). These mutations are GyrB(Asp426→Asn), which has been shown to confer resistance to both acidic and amphoteric quinolones, and GyrB(Lys447→Glu), which has been shown to confer resistance to acidic drugs but slight (≈4-fold) hypersensitivity to amphoteric drugs. Modeling these residues on the analogous parts of the 92-kDa yeast topoisomerase II crystal structure (5, 12) reveals that they lie over 40 Å from the QRDR and the active site and so appear too far from this region to have a direct effect on drug binding (Fig. 1B).
There are several possible explanations for the changes in quinolone sensitivity in these mutants: the mutations may change the conformation of the QRDR indirectly, via conformational changes transmitted through the peptide chain or through effects on the reaction cycle of the enzyme that could alter the frequency of a quinolone-sensitive conformation. A further possibility is that these residues of GyrB are directly involved in the quinolone-binding pocket, something which, based on the topoisomerase II structures, would require a large conformational change. Evidence for such a change can be seen in differences between the positions of the equivalent region in the two topoisomerase II structures (5, 12). The fact that residues in this region, in particular Asp424, are required for DNA cleavage (20), which takes place at the active site of GyrA, is further evidence for major changes in the position of the 47-kDa region of GyrB relative to the active site.
It is possible that the two residues (Asp426 and Lys447) are themselves part of a quinolone-binding pocket (45) and cause the observed resistance or hypersensitivity through electrostatic interactions with the drug molecules. In this scheme, residues 426 and 447 are hypothesized to lie close to each other. In the wild-type enzyme, the negatively charged Asp residue at position 426 is proposed to interact with the positively charged C-7 substituent of amphoteric quinolones while having no effect on the equivalent region of acidic quinolones. The positively charged Lys at 447 is proposed to interact with the negatively charged carboxyl group of Asp426, providing a neutral environment for binding hydrophobic groups. Thus, the wild-type pocket binds both types of quinolone. In the quinolone resistance mutation GyrB(Asp426→Asn), a negative charge is neutralized, and so binding of both types of drug is reduced. In GyrB(Lys447→Glu), the charge on the residue is reversed, repelling the acidic drugs but actually increasing the affinity for the positively charged C-7 substituent of amphoteric drugs.
While providing an explanation for the observed patterns of resistance and hypersensitivity of these two mutants, this model leaves several questions unanswered. The role of DNA, for example, known to be required for quinolone binding to gyrase, is not apparent. It has been suggested that the complete quinolone-binding pocket may involve regions of both GyrB and the QRDR of GyrA (45, 47). In addition, experiments to quantitate binding of quinolones to both GyrB mutants have shown almost identical amounts of enoxacin binding to A2B2447 as to the wild-type protein, while in vitro supercoiling experiments showed that this protein was twofold more sensitive to enoxacin (47) and in vivo experiments suggested it to be fourfold more sensitive (45, 47).
The details of the interaction between quinolone drugs, DNA, and gyrase remain unclear. Such information will be useful for designing new and improved drugs to combat the ongoing threat posed by bacterial pathogens. The aim of this work was to further investigate the in vitro resistance and hypersensitivity of the two GyrB mutants and to establish a more complete model of how these residues are involved in interaction with quinolone drugs.
MATERIALS AND METHODS
Proteins and DNA.
Wild-type gyrase was made as separate subunits (GyrA and GyrB) as described previously (22); GyrA was a gift from A. J. Howells (University of Leicester). Point mutations encoding GyrB(Asp426→Asn) and GyrB(Lys447→Glu) were cloned into pAG111, the plasmid encoding GyrB (16), and transformed into E. coli JM109 cells (Stratagene). Mutations were verified by DNA sequencing (Protein and Nucleic Acid Chemistry Laboratory, University of Leicester). Protein was purified to homogeneity using the same method as for wild-type GyrB.
Plasmid pBR322 in relaxed and supercoiled forms was a gift from A. J. Howells. A 140-bp fragment containing the major preferred gyrase cleavage site from pBR322 (13) was synthesized by PCR using primers 5′-TCG GGG AAT TCG CAT GGC G, and 5′-TGG ACA GCA TGG CCT GCA A. All oligonucleotides were made by Protein and Nucleic Acid Chemistry Laboratory (University of Leicester).
Supercoiling and relaxation assays.
The supercoiling activity of GyrB in the presence of GyrA was determined using a method similar to that described elsewhere (32). GyrA and GyrB were added to 30-μl reaction mixtures containing 35 mM Tris-HCl (pH 7.5), 24 mM KCl, 4 mM MgCl2, 1.8 mM spermidine, 5 mM dithiothreitol (DTT), 6.5% (wt/vol) glycerol, 0.36 mg of bovine serum albumin per ml, 9 μg of tRNA per ml, 1.4 mM ATP, and 10 μg of pBR322 DNA per ml. Reaction mixtures were incubated at 37°C for 30 min, and reactions were stopped by the addition of 30 μl of chloroform-isoamyl alcohol (24:1) and 15 μl of 40% sucrose-0.1 M Tris-HCl (pH 8)-0.1 M EDTA-1% bromophenol blue. Samples (15 μl) of the aqueous phase were loaded onto 1% agarose gels and typically run at 80 V for 2 h. Relaxation assays were identical to supercoiling assays except that ATP and spermidine were omitted and supercoiled pBR322 was used instead of the relaxed form.
Quinolone-induced DNA cleavage assays.
Conditions for quinolone-induced DNA cleavage assays were as for supercoiling reactions except that ATP and spermidine were omitted and ciprofloxacin or oxolinic acid was included at a range of concentrations, depending on the nature of the assay. Cleaved DNA was revealed after digestion of gyrase (100 nM wild type, 200 nM mutant) by the addition of 0.2% sodium dodecyl sulfate (SDS) and 0.1 mg of proteinase K per ml and incubation for 30 min at 37°C. Different amounts of mutant and wild-type gyrases were used to give equivalent, measurable amounts of submaximal cleavage. Reactions were stopped as for supercoiling reactions and analyzed by agarose gel electrophoresis. The amount of DNA in each was determined by densitometry, and the amount of DNA in the cleaved band was expressed as a percentage of the total.
ATPase assays.
ATPase reactions were followed using an enzyme-linked assay system that links the hydrolysis of ATP to the production of NAD+ in a stoichiometric fashion (1). Reactions were carried out in a volume of 150 μl at 25°C in a solution of 50 mM Tris-HCl (pH 7.5), 100 mM KCl, 2 mM ATP, 10% (wt/vol) glycerol, 1 mM EDTA, 5 mM DTT, 5 mM MgCl2, 400 μM phosphoenolpyruvate, 250 μM NADH, and 1.5 μl of pyruvate kinase/lactate dehydrogenase enzyme mix (50% [wt/vol] glycerol, 100 mM KCl, 10 mM HEPES [pH 7.0], 0.1 mM EDTA; Sigma). Reactions were initiated by the addition of ATP, and the decrease in A340 was measured over time. ATPase activities of GyrB were measured in the absence and presence of excess GyrA and linear pBR322.
DNA binding assays.
DNA binding experiments were carried out with 1 nM 32P-radiolabeled 140-bp DNA containing the preferred binding site for gyrase from pBR322 (13) and various concentrations of gyrase in a total of 10 μl of a solution of 50 mM Tris-HCl (pH 7.5), 55 mM KCl, 5 mM DTT, 4 mM MgCl2, and 5% (wt/vol) glycerol. Samples were loaded onto a 5% polyacrylamide gel (5% [vol/vol] of 30% [37.5:1] Protogel acrylamide and 1% [wt/vol] ammonium persulfate in 90 mM Tris-borate-4 mM MgCl2), and run at 150 V for 45 min. Gels were dried and exposed to a phosphorimager plate, typically for 16 h.
Quinolone binding assays.
The binding of quinolone drugs to DNA gyrase was assessed using rapid gel filtration (41). Samples contained 50 mM Tris-HCl (pH 7.5), 55 mM KCl, 4 mM MgCl2, 5 mM DTT, 5% (wt/vol) glycerol, 34 or 40 nM 140-bp DNA fragment, 400 nM gyrase, and various amounts of [3H]CFX (Amersham). Samples were incubated at 25°C for 3 h, and 80 μl was withdrawn and applied to a Sephadex G50 column (Pharmacia). Columns were centrifuged at 1,700 rpm in a Mistral bench-top centrifuge (MSE) for 10 min at 4°C. The volumes of the eluants were measured, and the amount of tritiated drug retained was determined by scintillation counting. The value for 100% drug bound was taken from the plateau level of the resulting binding curves. To measure OXO binding, enzyme was incubated with saturating amounts of [3H]CFX, which was competed with unlabeled OXO.
Proteolysis.
Proteolysis of DNA gyrase was carried out using trypsin, as described previously (18). Samples contained 0.3 mg of GyrA and GyrB per ml, 0.4 mg of pBR322 per ml, 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 4 mM MgCl2, 4 mM DTT, and glycerol (6.5%, wt/vol). Samples were incubated for 2 h at 25°C with CFX or OXO at a range of concentrations. Trypsin (10 μg/ml) was then added to the samples, and they were incubated for 1 h at 37°C. Reactions were stopped by the addition of an equal volume of a solution of 62 mM Tris-HCl (pH 7.5), 50 mM KCl, 4 mM MgCl2, 4 mM DTT, 6.5% (wt/vol) glycerol, 2% (wt/vol) SDS, 5% β-mercaptoethanol, and 0.001% (wt/vol) bromophenol blue, and boiling for 5 min. Proteolysis products were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
RESULTS
GyrB(Asp426→Asn) and GyrB(Lys447→Glu) show reduced supercoiling and relaxation activities and altered ATPase activities.
The supercoiling activity of GyrB can be calculated by titrating it into a supercoiling reaction at a fixed concentration of GyrA (Table 1). Such an experiment reveals that A2B2426 is ≈18-fold less active than the wild-type enzyme and A2B2447 is ≈6-fold less active. Keeping the enzyme concentration constant and analyzing the topological state of the DNA as the supercoiling reaction proceeds gives a similar result (data not shown). The activities of both mutants (≈0.4 × 104 to 1 × 104 U/mg), are similar to the activities found previously (2 × 104 to 4 × 104 U/mg [47]). A similar assessment of relaxation activities (in which ATP is omitted from the reaction mix and supercoiled DNA is used as the substrate) showed that both mutants have a ≈40-fold reduction in activity (Table 1). These results suggest that the mutations, in addition to their effects on quinolone sensitivity, also inhibit the catalytic activity of the enzyme.
TABLE 1.
Supercoiling and relaxation activities of wild-type and mutant gyrases and the IC50s of OXO and CFX
| Gyrase | Activity (U/mg)
|
IC50 (μM)
|
||||
|---|---|---|---|---|---|---|
| Supercoiling | Relaxation | Supercoiling
|
Relaxation
|
|||
| CFX | OXO | CFX | OXO | |||
| A2Bwt2 | 7.7 × 104 (± 1 × 104) | 1.0 × 104 (± 0.2 × 104) | 0.25 (± 0.05) | 17.4 (± 4) | 4.6 (± 0.6) | 84.2 (± 14) |
| A2B4262 | 4.2 × 103 (± 0.4 × 104) | 250 (± 50) | 0.67 (± 0.05) | 35.9 (± 3) | 9.7 (± 6) | 150.8 (± 35) |
| A2B4472 | 1.3 × 104 (± 0.2 × 104) | 220 (± 11) | 0.43 (± 0.06) | 63.1 (± 38) | 7.9 (± 1) | 1,600 (± 537) |
In the course of one supercoiling reaction cycle, DNA gyrase binds and hydrolyzes two molecules of ATP (23, 36). Gyrase is known to have an intrinsic rate of ATP hydrolysis, which is generally low but is known to vary significantly depending on the GyrB preparation (40); the ATPase activity is stimulated by the presence of DNA (21). The results of ATP hydrolysis experiments (Fig. 2) show that, under these conditions, the wild-type enzyme is stimulated ≈2-fold by the presence of DNA; A2B2426 shows an ATP hydrolysis rate similar to that of the wild-type enzyme but is not stimulated by DNA. A2B2447 shows a low intrinsic rate and is also not stimulated by DNA. In summary, the mutant proteins both show reduced supercoiling and relaxation activities and altered ATPase activities.
FIG. 2.

ATPase rates of wild-type and mutant GyrBs (100 nM) in the presence of excess GyrA and the absence (light gray) and presence (dark gray) of linear pBR322 (present at a concentration of 300 bp per tetramer).
GyrB(Asp426→Asn) and GyrB(Lys447→Glu) show resistance to both CFX and OXO in both supercoiling and relaxation reactions.
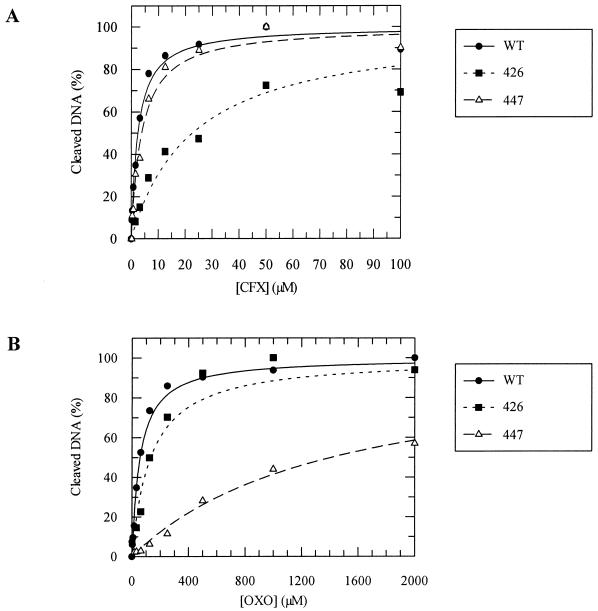
In order to assess the quinolone sensitivity of the supercoiling and relaxation reaction of the mutant enzymes, a concentration of enzyme known to result in submaximal supercoiling or relaxation was used, and CFX or OXO was titrated into the reaction (Fig. 3). The 50% inhibitory concentration (IC50) for each mutant and drug was recorded (Table 1). A2B2426 showed the expected resistance to both CFX and OXO. A2B2447, however, showed resistance to OXO and also a low level of resistance to CFX rather than the slight hypersensitivity to CFX in vivo reported before (45) and to several amphoteric drugs reported previously in vitro (47).
FIG. 3.
Supercoiling inhibition by quinolones. Inhibition by CFX (A) and OXO (B) of the supercoiling (s.c.) reactions of wild-type enzyme (WT), A2B2426, and A2B2447. CFX or OXO was titrated into supercoiling reactions containing less gyrase than is required for full supercoiling. Lines were fitted using a 1:1 ligand-binding curve. Derived IC50s are shown in Table 1.
A2B2447 binds DNA poorly.
Two methods were used to measure the binding of gyrase to DNA. The first method was a supercoiling time course. By taking time points during the course of a supercoiling reaction, intermediate stages in the reaction can be observed. Wild-type gyrase is a processive enzyme (25), meaning that few topological intermediates are observed between the relaxed starting material and the supercoiled product. Supercoiling time courses show that A2B2426 is at least as processive as the wild-type enzyme, suggesting that it binds DNA with a similar affinity. A2B2447, however, was found to be a more distributive enzyme, suggesting that it binds DNA less well (data not shown).
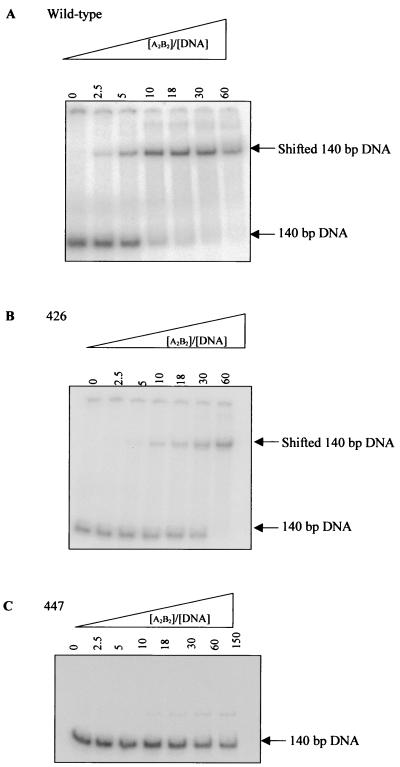
DNA binding was measured more directly using a gel retardation assay. In this assay, the binding of gyrase to a 140-bp radiolabeled DNA fragment was measured by virtue of the fact that DNA with enzyme bound migrates through a polyacrylamide gel more slowly than DNA without enzyme bound (21). The 140-bp fragment was chosen because this is similar to the length of DNA required for a single gyrase tetramer to bind (29). An example of the results obtained is shown in Fig. 4. It was found that A2B2447 does not bind to the 140-bp fragment under these conditions, up to an excess of enzyme over DNA of 150-fold. This is consistent with the supercoiling time course results, which show this mutant to be distributive rather than processive. The affinity of A2B2426 for DNA was ≈3-fold lower than the wild-type enzyme.
FIG. 4.
Gyrase-DNA binding assay. Gel retardation of 1 nmol of radiolabeled 140-bp DNA by increasing amounts of A2B2wt (A), A2B2426 (B), and A2B2447 (C) in the absence of drug and presence of excess GyrA. Apparent binding constants (Kdapps) were determined from phosphorimager analysis and were found to be 7.8 μM (A2B2wt) and 24.3 μM (A2B2426).
For A2B2wt and A2B2426, the presence of both CFX and OXO increased the affinity of the enzyme for the DNA, presumably by formation of a stable cleaved complex (14, 35); addition of quinolone to A2B2447 did not result in detectable DNA binding. These results suggest that the GyrB447 mutation affects the ability of the enzyme to bind DNA; this would account for the observed low DNA-stimulated ATPase rate and the low activity of this enzyme.
Affinity of CFX and OXO for mutant gyrase-DNA complexes.
The apparent affinity of quinolone drugs for gyrase-DNA complexes can be measured using the DNA cleavage reaction (4). Quinolones are known to bind to the gyrase-DNA complex and stabilize a form of the complex in which the DNA is cleaved across both strands; in the absence of drug, cleavage is not detectable. Therefore, measuring the amount of cleaved DNA produced at different quinolone concentrations (as shown in Fig. 5) is an indirect method of measuring the amount of quinolone bound. The results of such cleavage experiments (Table 2) suggest that the A2B2426-DNA complex has reduced affinity for both OXO and CFX, reflecting its resistance to both drugs. These results suggest that the resistance to quinolone drugs observed for A2B2426 is due to decreased drug binding. In the case of A2B2447, which is resistant to OXO and shows sensitivity to CFX similar to the wild-type, the IC50 for CFX is <2-fold that for the wild-type enzyme, whereas the IC50 for OXO is ≈25-fold higher, i.e., the wild-type-like sensitivity to CFX is reflected in nearly wild-type levels of CFX binding, whereas the resistance to OXO is reflected in reduced OXO binding.
FIG. 5.
Cleavage of 3.4 nM relaxed pBR322 in the presence of increasing amounts of CFX (A) or OXO (B) by wild-type enzyme (100 nM), A2B2426 (200 nM), and A2B2447 (200 nM). The amount of linear DNA as a percentage of the total was determined by densitometry (Syngene gel documentation system); results are normalized to 100% maximal cleavage. The results were fitted with a 1:1 ligand-binding curve. Derived IC50s are shown in Table 2.
TABLE 2.
Binding of quinolones to gyrase as measured by DNA cleavage and binding assaysa
| Quinolone | A2Bwt2
|
A2B4262
|
A2B4472
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| IC50 (μM) | Kdapp (μM) | Total bound (nM) | IC50 (μM) | Kdapp (μM) | Total bound (nM) | IC50 (μM) | Kdapp (μM) | Total bound (nM) | |
| CFX | 2.4 ± 0.2 | 4.2 ± 0.5 | 77 ± 2.4 | 22.6 ± 3.7 | 108 ± 0.0 | 26 ± 4.7 | 3.8 ± 0.3 | 18.3 ± 4.6 | 52 ± 3.5 |
| OXO | 56.5 ± 3.7 | 121 ± 30 | 33 ± 3.3 | 131.6 ± 64.8 | >500 | NDb | 1,413 ± 72.0 | >500 | ND |
Enzyme concentrations in DNA cleavage experiments (IC50s) were 100 nM for wild-type enzyme and 200 nM for mutants; enzyme-DNA complex concentrations in drug-binding experiments (Kdapps and stoichiometries) were 40 nM for wild-type enzyme and 34 nM for mutants.
ND, could not be determined.
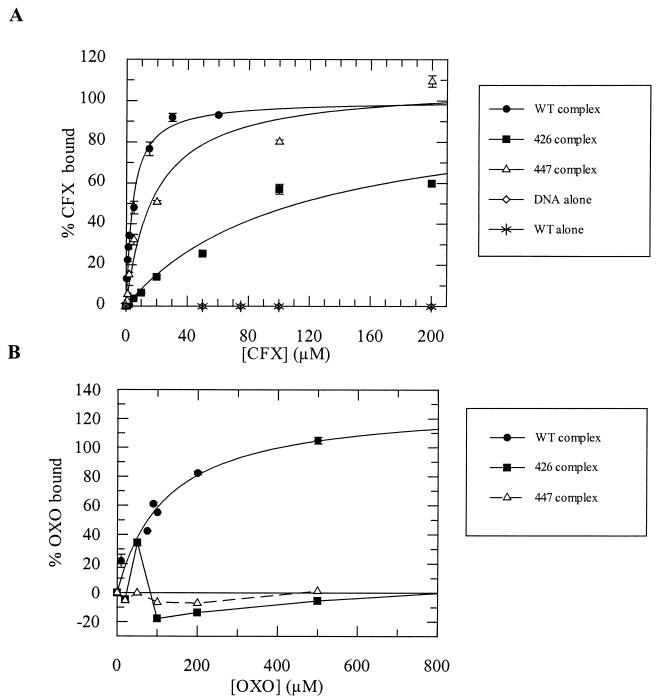
The binding of CFX and OXO to enzyme-DNA complexes was also measured directly using [3H]CFX. CFX was titrated into mixtures of gyrase-DNA complexes, which were spun through a G-50 Sephadex column in order to remove unbound drug, and the resulting bound drug was quantitated. A similar method has previously been used to quantitate enoxacin binding to the two mutants (47). OXO binding was measured indirectly, by a competition method in which complexes were formed with saturating amounts of tritiated CFX and unlabeled OXO was titrated into the mix to displace the labeled drug. The results of the binding experiment (Fig. 6, Table 2) show that A2B2426 binds both OXO and CFX with reduced affinity; Kdapp increased >4-fold and ≈25-fold, respectively, compared to the wild-type, whereas A2B2447 binds OXO with reduced affinity (Kdapp increased >4-fold) and CFX ≈4-fold less well than the wild-type. The stoichiometry of drug binding ranges from 0.8 to 2 drug molecules per complex. Given the dimeric form of the protein and the symmetrical nature of the binding site, it is likely that the actual stoichiometry is 2 (8).
FIG. 6.
Binding of CFX (A) and OXO (B) to 40 nM wild-type gyrase-DNA complexes and 34 nM mutant gyrase-DNA complexes, as measured by rapid gel filtration. Apparent dissociation constants are given in Table 2. Data were fitted with a 1:1 ligand-binding curve.
Resistant mutants do not show protection of the 47-kDa C-terminal region of GyrB from proteolytic degradation by trypsin in the presence of quinolones.
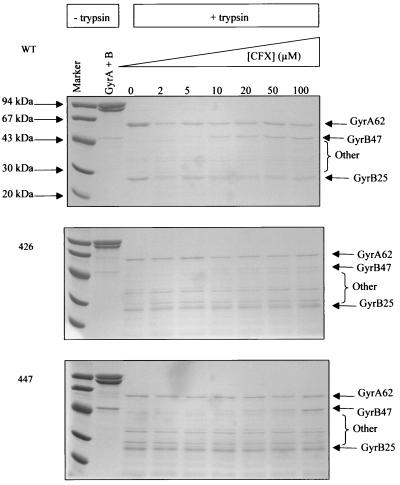
It has been shown that, in the presence of quinolone drugs and DNA, gyrase undergoes a conformational change that protects the 47-kDa C-terminal region of GyrB from proteolytic degradation by trypsin (18). This suggests that quinolones stabilize a particular conformation of the enzyme that is usually transient and involves a conformational change of the 47-kDa domain, protecting it from proteolysis. This characteristic protection did not occur with A2B2426 in the presence of CFX or OXO or with A2B2447 in the presence of OXO. It did, however, occur with A2B2447 in the presence of CFX, where it was present to a similar extent as in the wild-type sample (Fig. 7; OXO results not shown).
FIG. 7.
Proteolysis with trypsin (2 μg/ml) of complexes of gyrase (0.3 mg/ml) with pBR322 (0.4 mg/ml) and increasing amounts of CFX for wild-type enzyme, A2B2426, and A2B2447. Samples were analyzed by SDS-polyacrylamide gel electrophoresis. The subunit and molecular weight of the protein fragment are shown, e.g., GyrA62 (18).
This result suggests that the quinolone drugs are unable to stabilize an altered conformation of enzyme if GyrB contains a quinolone-resistant mutation (as has been shown to be the case with resistance mutations in the QRDR of GyrA [18]). The exception is A2B2447 in the presence of CFX, where the mutant is only marginally resistant to the drug. We note also the presence of other tryptic fragments with the mutant enzymes in Fig. 7 that are not present with the wild-type enzyme. These may suggest that there are conformational changes in the mutant enzymes that are not directly related to quinolone binding.
DISCUSSION
The quinolone resistance mutations in GyrB are unusual in that they lie outside the GyrA QRDR in which most other quinolone resistance mutations are found. The mutants show two sets of features. First, they show altered activities, and second, they show altered quinolone sensitivities.
Enzyme activities.
The mutants show reduced supercoiling and relaxation activities. The reduced supercoiling activities (≈18-fold lower than the wild-type level for A2B2426 and ≈6-fold lower for A2B2447) are similar to those found previously (47). The relaxation activities were not previously investigated, and we find that they are also reduced (≈40-fold) in both cases, showing that the effect of the mutations is not specific to the ATP-dependent parts of the catalytic cycle.
The ATPase reaction of the mutants has not been previously investigated. We found that A2B2447 had reduced ATP hydrolysis activity compared to the wild-type protein in both the absence and presence of DNA. This reduced activity may account for the reduced supercoiling activity of this mutant, although it would not account for the reduced relaxation activity, as relaxation does not require ATP (14, 35). In contrast, A2B2426 has an ATPase rate equal to that of the DNA-stimulated wild-type rate irrespective of the presence of DNA, i.e., it is permanently switched on. This suggests that the coordination between the hydrolysis of ATP and the capture of a strand of DNA by the ATP-operated clamp of GyrB and subsequent steps in the mechanism have become uncoupled. This could lead to enzyme turnover in which the coupling between capture of a DNA segment by the ATP-operated clamp and subsequent steps in the catalytic pathway is lost. This would result in an increase in the number of enzyme turnovers in which supercoiling did not take place. Such inefficient supercoiling would explain the reduced activity of this mutant.
The DNA-binding ability of these mutants was investigated first by assessing the processivity of their supercoiling reactions. This experiment showed that A2B2426 was at least as processive as the wild-type enzyme, while A2B2447 was considerably more distributive, suggesting that it bound DNA less well than the wild-type or A2B2426. These results were confirmed in a DNA retardation assay in which the affinity of A2B2426 for 140-bp DNA was found to be 24.3 μM (compared to a wild-type value of 7.8 μM), while A2B2447 was found to bind DNA so poorly that an affinity could not be measured. The fact that supercoiling occurs with A2B2447 shows that this complex is able to bind DNA, but this binding is presumably too weak to be detected by gel retardation assays. This explains why the supercoiling and relaxation activities of A2B2447 were reduced and why it showed no stimulation in its rate of ATP hydrolysis in the presence of DNA.
In summary, the reduced supercoiling activity of A2B2426 is likely to be a result of uncoupling of ATP hydrolysis from subsequent catalytic events, while the reduced activity of A2B2447 is likely to be due to its reduced affinity for DNA. It is less easy to explain why the relaxation activities of the two mutants are more profoundly affected than supercoiling. One possibility is that the mutations at 426 and 447 also affect the cleavage reaction as they are close to residues in GyrB that are likely to be important in cleavage (20). However, the rate of supercoiling is not apparently determined by the rate of the cleavage reaction; ATP hydrolysis is thought to be the rate-limiting step in this case (1). The rate of DNA relaxation, a much slower reaction than supercoiling (31), is more likely to be dependent on the rate of the cleavage reaction (or a conformational change associated with it), such that mutations that affect cleavage will have a more profound effect on relaxation than supercoiling. The effects of the GyrB mutations at 426 and 447 on the activities of the enzyme could, in principle, affect drug resistance, for example, by disrupting the interaction between the gyrase subunits or reducing its ability to form a cleavage complex. These issues need to be borne in mind when considering the effects of the mutations on the susceptibility of the enzyme to quinolones.
Drug resistance.
DNA cleavage experiments showed that the amount of CFX required for 50% maximal DNA cleavage (IC50) is as expected based on the sensitivity of supercoiling to CFX and OXO, i.e., A2B2447 shows a high level (25-fold) of resistance to OXO but a nearly wild-type level of sensitivity to CFX; A2B2426 shows resistance to both quinolones. The resistance of the mutants is more dramatic when measured in the cleavage assay than in the supercoiling assay; this is not surprising, as these two assays are measuring different aspects of the reaction, and this is a manifestation of the fact that quinolone inhibition is a complex multistep process. These results do not indicate whether the resistance shown by the mutants is due to a decrease in drug binding, as appears to be the case with GyrA mutants (3, 41, 47), or is due to another mechanism.
Previous attempts to quantitate the binding of radiolabeled enoxacin (another amphoteric quinolone similar to CFX) suggested that A2B2447 and the wild-type enzyme bind similar amounts of drug, whereas A2B2426 was shown to bind significantly less drug than the wild type (47). These experiments, however, gave a stoichiometry for drug binding of ≈0.6 drug molecule per complex, whereas the most likely number is 2 (8). The drug-binding experiments in this paper were carried out with both an amphoteric drug (CFX) and an acidic drug (OXO). The CFX binding results are similar to those found previously for enoxacin, with A2B2426 showing the least binding and A2B2447 showing slightly less binding than the wild type (Fig. 6). For OXO, the results show that both mutants are resistant to drug binding. These results support the idea that the effect of the mutations is to decrease drug binding.
The results of the proteolysis experiments show that the conformational change involving the 47-kDa C-terminal region of GyrB, associated with quinolone binding and the formation of a cleaved complex, does not occur when the resistant mutants are used in conjunction with the quinolones to which they are resistant. Given that conformational changes involving this region of GyrB are thought to be required for cleavage to occur (20) and the fact that GyrB quinolone-resistant mutants are also resistant in their cleavage reactions, it is possible that the conformational change seen in proteolysis is synonymous with the conformation stabilized by the quinolone drugs when they arrest cleavage. This would mean that the quinolone resistance mutations in GyrB act by disfavoring quinolone stabilization of the cleaved complex. Given that the conformational changes involved in cleavage involve the 47-kDa region and are likely to bring it close to the QRDR, it is possible that residues 426 and 447 have a direct effect on drug binding by constituting part of the quinolone-binding pocket.
Model for the quinolone-binding pocket.
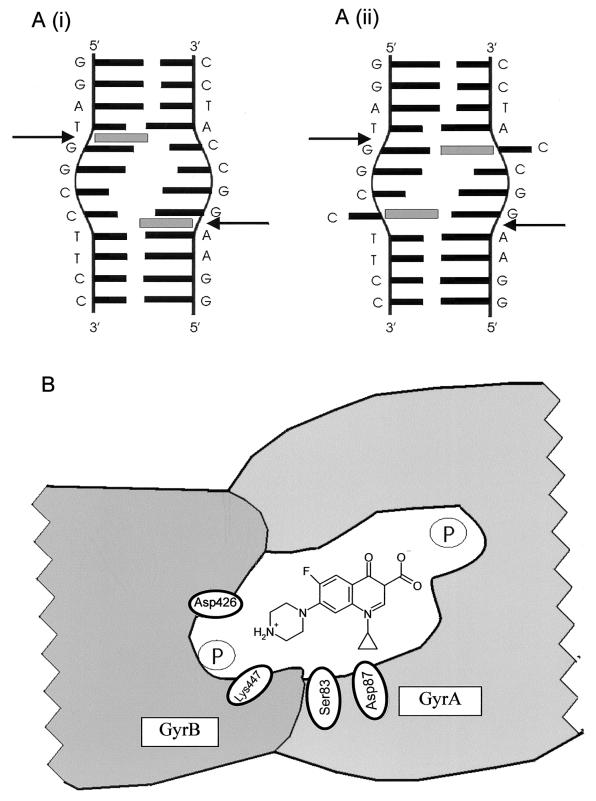
The model that we propose (Fig. 8) is that two ciprofloxacin molecules stack between DNA bases at the active site of gyrase, where the DNA is distorted (Fig. 8A). The exact site and mode of interaction are unclear. Here we present two possibilities. Ciprofloxacin may intercalate between the bases on either side of the scissile bond (Fig. 8Ai); this is supported by previous work which suggested that quinolones can cause DNA unwinding (37, 38). Alternatively, the quinolone could replace the cytosine opposite the scissile bond and hydrogen bond with the opposing guanine (Fig. 8Aii); to do this, the cytosine would have to be “flipped out.” Hydrogen bonding of quinolones to guanine has been suggested previously (33), and DNA cleavage assays have indicated a preference for G's around the cleavage site (7, 15).
FIG. 8.
Model of the quinolone-binding pocket. (A) Interaction of quinolone with DNA bound at the active site of gyrase. The DNA sequence is the preferred gyrase cleavage site from pBR322 (13). Sites of cleavage are shown by black arrows. The sequence surrounding the cleavage site is distorted (24). Drug is represented by a gray rectangle. A(i) shows the quinolone stacking between bases; A(ii) shows the quinolone replacing cytosine which is flipped out. (B) The quinolone-binding pocket in gyrase. GyrB is positioned close to the QRDR of GyrA, allowing residues 426 and 447 to interact with a bound quinolone molecule. The DNA axis is perpendicular to the plane of the diagram, and the approximate position of the sugar-phosphate backbone is shown by an encircled P. The quinolone (CFX) interacts with the distorted region of DNA at the active site. The bottom edge of the molecule can interact with residues of GyrA, including Ser83 and Asp87 (3). The C-7 group of the drug can interact with Asp426 and Lys447 in a way similar to that described previously (47). Lys447 may also interact with the phosphate backbone.
To explain the role of DNA gyrase, particularly GyrB, in the interaction with quinolones, we propose that, during a normal catalytic cycle, gyrase undergoes a large conformational change involving the 47-kDa subunit of GyrB. This brings the Rossman-like fold (Toprim [2]) region of GyrB near the active-site tyrosine and thus the QRDR of GyrA (17). This seems likely for a number of reasons. (i) In other type I and type II topoisomerase crystal structures, the Rossman fold lies close to the active site (27). (ii) It is known that in yeast topoisomerase II, the residue equivalent to 424 of GyrB, close to the quinolone resistance mutations, is absolutely required for DNA cleavage to occur (20). More recent work suggests that residues 498, 500, and 502 of GyrB also have a key role in DNA cleavage (28). For these residues to have a role in cleavage, it is likely that they are close to the active site. (iii) Additional evidence that such a conformational change is possible comes from comparing the two crystal structures solved for yeast topoisomerase II containing the region analogous to GyrB47 (5, 12). In the more recent structure, the 47-kDa regions have moved relative to the region of the protein equivalent to GyrA. The residues corresponding to GyrB426 and GyrB 447 thus move substantially closer to the active site.
Thus, in the model proposed (Fig. 8B), we envisage that, in order for cleavage to occur, the GyrB47 region moves close to the active site so that the Toprim fold, including residues 424, 498, 500, and 502, can participate in cleavage. For the wild-type protein in the presence of quinolone drug, this brings residues 426 and 447 close to the QRDR and the bound quinolone, resulting in additional stabilization of the quinolone-gyrase-DNA complex in the DNA-cleaved state. The proposed interaction with GyrA (Fig. 8B) is likely to involve residues on α-helix 4 of the N-terminal domain, specifically Ser83 and Asp87 (3, 24).
The reasons for the differential sensitivities of the GyrB mutants are presumably due to differences in the details of interactions between the amino acid residues and specific regions of the quinolone molecule. Thus, it is possible that the interaction of quinolones with gyrase may involve the QRDR of GyrA and residues 426 and 447 of GyrB in a manner similar to that first suggested by Nakamura and colleagues (26); we propose to refer to this model as the Nakamura model. However, we would also propose that residue 447 has an additional interaction with the DNA backbone. It is thought that the two DNA strands at the active site of gyrase are pulled apart to some extent in order for the phosphates to approach the active-site tyrosine (24). It is possible that GyrB447 helps to stabilize the DNA in the distorted form. This model suggests a binding pocket that includes residues of GyrA, GyrB, and the DNA molecule, all of which interact with the quinolone molecule. It is likely that a definitive description of the gyrase-quinolone-DNA complex will require the application of X-ray crystallography.
Acknowledgments
J.H. was supported by a studentship from the Medical Research Council. This work was supported by grants from the BBSRC and the Wellcome Trust.
We thank Faye Barnard, Karl Drlica, and Christian Noble for comments and Alison Howells for DNA and protein.
REFERENCES
- 1.Ali, J. A., A. P. Jackson, A. J. Howells, and A. Maxwell. 1993. The 43-kDa N-terminal fragment of the gyrase B protein hydrolyses ATP and binds coumarin drugs. Biochemistry 32:2717-2724. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L., D. D. Leipe, and E. V. Koonin. 1998. Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 26:4205-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnard, F. M., and A. Maxwell. 2001. Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser83 and Asp87. Antimicrob. Agents Chemother. 45:1994-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett, J. F., J. A. Sutcliffe, and T. D. Gootz. 1990. In vitro assays used to measure the activity of topoisomerases. Antimicrob. Agents Chemother. 34:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger, J. M., S. J. Gamblin, S. C. Harrison, and J. C. Wang. 1996. Structure at 2.7 Å resolution of a 92K yeast DNA topoisomerase II fragment. Nature 379:225-232. [DOI] [PubMed] [Google Scholar]
- 6.Champoux, J. J. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369-413. [DOI] [PubMed] [Google Scholar]
- 7.Cove, M. E., A. P. Tingey, and A. Maxwell. 1997. DNA gyrase can cleave short DNA fragments in the presence of quinolone drugs. Nucleic Acids Res. 25:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Critchlow, S. E., and A. Maxwell. 1996. DNA cleavage is not required for the binding of quinolone drugs to the DNA gyrase-DNA complex. Biochemistry 35:7387-7393. [DOI] [PubMed] [Google Scholar]
- 9.Cullen, M. E., A. W. Wyke, R. Kuroda, and L. M. Fisher. 1989. Cloning and characterization of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob. Agents Chemother. 33:886-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drlica, K. 1999. Mechanisms of fluoroquinolone action. Curr. Opin. Microbiol. 2:504-508. [DOI] [PubMed] [Google Scholar]
- 11.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fass, D., C. E. Bogden, and J. M. Berger. 1999. Quaternary changes in topoisomerase II may direct orthogonal movements of two DNA strands. Nat. Struct. Biol. 6:322-326. [DOI] [PubMed] [Google Scholar]
- 13.Fisher, L. M., K. Mizuuchi, M. H. O'Dea, H. Ohmori, and M. Gellert. 1981. Site-specific interaction of DNA gyrase with DNA. Proc. Natl. Acad. Sci. USA 78:4165-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gellert, M., K. Mizuuchi, M. H. O'Dea, T. Itoh, and J. Tomizawa. 1977. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. USA 74:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gmünder, H., K. Kuratli, and W. Keck. 1997. In the presence of subunit A inhibitors DNA gyrase cleaves DNA fragments as short as 20 bp at specific sites. Nucleic Acids Res. 25:604-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallett, P., A. J. Grimshaw, D. B. Wigley, and A. Maxwell. 1990. Cloning of the DNA gyrase genes under tac promoter control: overproduction of the gyrase A and B proteins. Gene 93:139-142. [DOI] [PubMed] [Google Scholar]
- 17.Heddle, J. G., F. M. Barnard, L. M. Wentzell, and A. Maxwell. 2000. The interaction of drugs with DNA gyrase: a model for the molecular basis of quinolone action. Nucleosides Nucleotides Nucleic Acids 19:1249-1264. [DOI] [PubMed] [Google Scholar]
- 18.Kampranis, S. C., and A. Maxwell. 1998. Conformational changes in DNA gyrase revealed by limited proteolysis. J. Biol. Chem. 273:22606-22614. [DOI] [PubMed] [Google Scholar]
- 19.Kuhlmann, J., A. Dalhoff, and H.-J. Zeiler (ed.). 1998. Quinolone antibacterials, vol. 127. Springer-Verlag, Berlin, Germany.
- 20.Liu, Q., and J. C. Wang. 1999. Similarity in the catalysis of DNA breakage and rejoining by type IA and IIA DNA topoisomerases. Proc. Natl. Acad. Sci. USA 96:881-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maxwell, A., and M. Gellert. 1984. The DNA dependence of the ATPase activity of DNA gyrase. J. Biol. Chem. 259:14472-14480. [PubMed] [Google Scholar]
- 22.Maxwell, A., and A. J. Howells. 1999. Overexpression and purification of bacterial DNA gyrase, p. 135-144. In M.-A. Bjornsti and N. Osheroff (ed.), Protocols for DNA topoisomerases. I. DNA topology and enzyme purification. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 23.Maxwell, A., D. C. Rau, and M. Gellert. 1986. Mechanistic studies of DNA gyrase, p. 137-146. In R. H. Sarma and M. H. Sarma (ed.), Biomolecular stereodynamics. III. Proceedings of the Fourth Conversation in the Discipline of Biomolecular Stereodynamics. Adenine Press, Albany, N.Y.
- 24.Morais Cabral, J. H., A. P. Jackson, C. V. Smith, N. Shikotra, A. Maxwell, and R. C. Liddington. 1997. Structure of the DNA breakage-reunion domain of DNA gyrase. Nature 388:903-906. [DOI] [PubMed] [Google Scholar]
- 25.Morrison, A., N. P. Higgins, and N. R. Cozzarelli. 1980. Interaction between DNA gyrase and its cleavage site on DNA. J. Biol. Chem. 255:2211-2219. [PubMed] [Google Scholar]
- 26.Nakamura, S., H. Yoshida, M. Bogaki, M. Nakamura, and T. Kojima. 1993. Quinolone resistance mutations in DNA gyrase, p. 135-143. In T. Andoh, H. Ikeda, and M. Oguro (ed.), Molecular biology of DNA topoisomerases and its application to chemotherapy. CRC Press, Inc., Boca Raton, Fla.
- 27.Nichols, M. D., K. DeAngelis, J. L. Keck, and J. M. Berger. 1999. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo 11. EMBO J. 18:6177-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noble, C. G., and A. Maxwell. 2002. The role of GyrB in the DNA cleavage-religation reaction of DNA gyrase: a two-metal-ion mechanism. J. Mol. Biol. 318:361-371. [DOI] [PubMed] [Google Scholar]
- 29.Orphanides, G., and A. Maxwell. 1994. Evidence for a conformational change in the DNA gyrase-DNA complex from hydroxyl radical footprinting. Nucleic Acids Res. 22:1567-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterson, U., and T. Schemke. 1998. The chemistry of the quinolones: chemistry in the periphery of the quinolones, p. 63-118. In J. Kuhlmann, A. Dalhoff, and H.-J. Zeiler (ed.), Quinolone antibacterials, vol. 127. Springer, Berlin, Germany. [Google Scholar]
- 31.Reece, R. J., and A. Maxwell. 1991. DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 26:335-375. [DOI] [PubMed] [Google Scholar]
- 32.Reece, R. J., and A. Maxwell. 1989. Tryptic fragments of the Escherichia coli DNA gyrase A protein. J. Biol. Chem. 264:19648-19653. [PubMed] [Google Scholar]
- 33.Shen, L. L., J. Baranowski, and A. G. Pernet. 1989. Mechanism of inhibition of DNA gyrase by quinolone antibacterials: specificity and cooperativity of drug binding to DNA. Biochemistry 28:3879-3885. [DOI] [PubMed] [Google Scholar]
- 34.Shen, L. L., and A. G. Pernet. 1985. Mechanism of inhibition of DNA gyrase by analogues of nalidixic acid: the target of the drugs is DNA. Proc. Natl. Acad. Sci. USA 82:307-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugino, A., C. L. Peebles, K. N. Kruezer, and N. R. Cozzarelli. 1977. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. USA 74:4767-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura, J. K., A. D. Bates, and M. Gellert. 1992. Slow interaction of 5′-adenylyl-β,γ-imidodiphosphate with Escherichia coli DNA gyrase. J. Biol. Chem. 267:9214-9222. [PubMed] [Google Scholar]
- 37.Tornaletti, S., and A. M. Pedrini. 1988. DNA unwinding induced by nalidixic acid binding to DNA. Biochem. Pharmacol. 37:1881-1882. [DOI] [PubMed] [Google Scholar]
- 38.Tornaletti, S., and A. M. Pedrini. 1988. Studies on the interaction of 4-quinolones with DNA by DNA unwinding experiments. Biochim. Biophys. Acta 949:279-287. [DOI] [PubMed] [Google Scholar]
- 39.Wigley, D. B., G. J. Davies, E. J. Dodson, A. Maxwell, and G. Dodson. 1991. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351:624-629. [DOI] [PubMed] [Google Scholar]
- 40.Williams, N. L., A. J. Howells, and A. Maxwell. 2001. Locking the ATP-operated clamp of DNA gyrase: probing the mechanism of strand passage. J. Mol. Biol. 306:969-984. [DOI] [PubMed] [Google Scholar]
- 41.Willmott, C. J. R., and A. Maxwell. 1993. A single point mutation in the DNA gyrase A protein greatly reduces the binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob. Agents Chemother. 37:126-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamagishi, J., Y. Furutani, S. Inoue, T. Ohue, S. Nakamura, and M. Shimizu. 1981. New nalidixic acid resistance mutations related to deoxyribonucleic acid gyrase activity. J. Bacteriol. 148:450-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamagishi, J., H. Yoshida, M. Yamyoshi, and S. Nakamura. 1986. Nalidixic acid-resistant mutations of the gyrB gene of Escherichia coli. Mol. Gen. Genet. 204:367-373. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida, H., M. Bogaki, M. Nakamura, L. M. Yamanaka, and S. Nakamura. 1991. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother. 35:1647-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida, H., T. Kojima, J. Yamagishi, and S. Nakamura. 1988. Quinolone-resistant mutations of the gyrA gene of Escherichia coli. Mol. Gen. Genet. 211:1-7. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida, H., M. Nakamura, M. Bogaki, H. Ito, T. Kojima, H. Hattori, and S. Nakamura. 1993. Mechanism of action of quinolones against Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 37:839-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao, X., J. Domagala, and K. Drlica. 1997. DNA topoisomerase targets of the fluoroquinolones: a strategy for avoiding bacterial resistance. Proc. Natl. Acad. Sci. USA 94:13991-13996. [DOI] [PMC free article] [PubMed] [Google Scholar]