Abstract
Identification of major glucan-associated proteins (GAPs) of the cell wall of a number of Candida albicans isolates susceptible or resistant to fluconazole (FLC) was addressed by direct sequencing of the protein bands resolved by unidimensional gel electrophoresis. Changes in the GAP compositions of the different strains grown in the presence of the drug were also investigated. In the FLC-susceptible strains, the major (more abundant) GAPs were enolase (46 kDa), two isoforms of phosphoglyceromutase (32 and 29 kDa), and two β-(1-3)-exoglucanases (44 and 34 kDa), one of which (the 34-kDa component) was glycosylated. When these strains were grown in the presence of FLC there were substantial decreases in the intensities of the two enzymes of the glycolytic pathway (enolase and the phosphoglyceromutases), which were apparently replaced by enhancement of the exoglucanase constituents, particularly the 44-kDa one. This GAP pattern closely mimicked that observed in the FLC-resistant strains whether they were grown in the presence or in the absence of the drug. Both the enolase and the exoglucanase constituents were detected in the culture supernatants of FLC-treated cells, together with substantial amounts of highly glycosylated, probably mannoprotein secretory material, suggesting that FLC may cause marked alterations of GAP incorporation into the cell wall. Altogether, we were able to identify all major GAP constituents and monitor their distributions in the cell wall of C. albicans during treatment with FLC. The near equivalence of the GAP profile for the FLC-susceptible strain grown in the presence of FLC to that for the FLC-resistant strain suggests that the effects of the drug on GAPs may be stably incorporated into the cell wall of the fungus upon acquisition of resistance.
The cell wall of the human opportunistic pathogen Candida albicans is a complex biochemical entity (organelle) mainly composed of neutral polysaccharides. These polysaccharides appear to be interspersed to constitute a three-dimensional network. Several proteins are embedded in this network, and these proteins are more or less tightly bound to the polysaccharides. Those which are released from the purified cell wall following digestion with endoglucanases are often referred to as glucan-associated proteins (GAPs) (4, 13, 18, 23). There is a great deal of interest in understanding the nature and function of these proteins in terms of their roles in cell wall construction, morphogenesis, and interaction with the host (4, 6, 9, 10, 18). Detailed knowledge of the compositions and structures of GAPs could also generate useful targets for novel anticandidal agents, facilitate a better understanding of the mechanisms of action of the existing antifungals, or even help with the identification of novel immunostimulating agents.
One of the areas which remains poorly investigated is the influence that current anticandidal drugs exert on GAP composition and function. Although most effective anticandidal drugs have direct or indirect activities on one or more steps of cell wall construction, it is not clear whether and to what extent the emergence of drug resistance or the use of suboptimal drug concentrations may affect cell wall proteins.
In previous studies we evaluated the relationship between a lipopeptide antimycotic and GAPs (2, 3). In the present study we have specifically addressed the relationship between fluconazole (FLC) and GAPs. To clarify this relationship, a major focus of the present study was on the precise identification of GAPs by amino acid sequence determination. We also assessed changes in the GAPs of strains of C. albicans with different susceptibilities to FLC both in the presence and in the absence of this clinically important antimycotic agent.
MATERIALS AND METHODS
Organisms and growth conditions.
C. albicans strains from type collections and recent clinical isolates of the fungus were used throughout the study. Namely, strains 3153 and CA2 were laboratory strains with different competences for germ tube formation (2). Strains AIDS 68 and AIDS 126 were originally isolated from AIDS patients with oropharyngeal candidiasis. Both were resistant to FLC at the time of isolation (see below). Strain CO23s was isolated from a subject with vulvovaginal candidiasis and was originally sensitive to FLC. Strain CO23s was made resistant to FLC (strain CO23r) by growth in stepwise-increasing FLC concentrations. As expected, strains CO23s and CO23r had identical electrophoretic karyotypes, as determined by pulse-field gel electrophoresis (5), thus demonstrating that there was no selection of a contaminating, FLC-resistant strain during the process of resistance acquisition.
Each strain was routinely maintained on Sabouraud dextrose agar medium (Difco, Detroit, Mich.) at 28°C. For experimental purposes, the strains were grown on yeast nitrogen base medium (Difco) supplemented with 0.1% glucose at 28°C. When necessary, FLC, which had previously been dissolved in distilled water at a concentration of 10 mg/ml, was added to the culture medium to the final concentrations indicated for each experiment. MICs were determined by methods of the NCCLS (24), and resistance to FLC was established as an MIC of >64 μg/ml.
FLC treatment and GAPs.
To assess the influence of FLC on the GAP composition, each FLC-susceptible strain was cultured in yeast nitrogen base medium at 28°C for 24 h by using an inoculum size of approximately 107 cells/ml to recover a sufficient amount of cell wall material for GAP analysis. Under these conditions, the final optical density of the control culture reached a value of 1.2 to 1.4. The drug was added at a concentration (5 μg/ml) to cause only a slight inhibition of growth (final optical density, about 1.0) For the resistant strains, FLC was added at a concentration 50 μg/ml and the other conditions were the same as those described above. In some experiments, following centrifugation of the culture at 1,500 × g for 10 min (at 4°C), the supernatants were collected and concentrated with a tangential flow filtration device (Mini-Ultrasette 10 K; Pall-Filtron, Barrington, Ill.). High-molecular-mass proteins were recovered by concentration (10:1) of the samples from the medium and were precipitated in absolute ethanol (1:3, by volume).
Cells wall preparation and purification.
Clean cells walls of the different strains of C. albicans were obtained by centrifugation of the fungal cells at 1,500 × g for 10 min at 4°C, followed by suspension of the cellular pellet in 0.5 M Tris-HCl buffer (pH 6.8) containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and a protease inhibitor cocktail (0.05 ml/g [dry weight] of cells; Sigma Chemical Co., St. Louis, Mo.). Cells were broken by homogenization with 0.45-mm-diameter glass beads in a Bead-Beater apparatus (Biospect Products, Bartelsville, Okla.) that was kept in an ice bath, with cycles of 1 min of treatment and 1 min of rest in an ice bath for 20 min. The broken cells were centrifuged at 1,500 × g for 10 min, and the resulting pellet was washed 10 times with PMSF solution (1 mM), sonicated for 5 min at an amplitude of 28 at 9-s intervals, and washed again to yield the final cell wall material. Observations under an electron microscope confirmed the purity of the cell walls, as demonstrated elsewhere (2).
GAP extraction.
Clean cell walls were resuspended in 0.05 M Tris-1 M Tris-HCl-1 mM PMSF-50 mM dithiothreitol (DTT) (Tris-DTT) containing 20 U of purified β-(1-3)-glucanase (Zymolyase 100 T; Seigakaku, Tokyo, Japan) per ml. After overnight incubation at 37°C with mild agitation, the material was centrifuged at 1,500 × g for 10 min. The supernatant was collected and added to 3 volumes of absolute ethanol to precipitate the GAPs, which were stored at 4°C overnight.
Protein and carbohydrate concentration determinations.
A protein assay (Bio-Rad Laboratories, Richmond, Calif.) was used to estimate the amounts of proteins, with bovine serum albumin used as the standard. The carbohydrate content was assayed by the method of Dubois et al. (8).
SDS-polyacrylamide gel electrophoresis (PAGE) and Western (immunoblotting) assays.
Protein samples (7 μg) in sodium dodecyl sulfate (SDS) buffer were applied to 10% (wt/vol) polyacrylamide gels for electrophoresis. A mini vertical gel apparatus was used (Bio-Rad), and the samples were run at a constant voltage of 200 V for 50 min at room temperature. After electrophoresis, the gels were stained with Coomassie brilliant blue or silver to detect proteins bands, as described previously (3). Otherwise, the gels were prepared for transfer of the proteins to nitrocellulose (as specified below).
The apparent molecular masses of the separated proteins were determined with Kaleidoscope prestained standards (Bio-Rad), which contained myosin (>209 kDa), β-galactosidase (134 kDa), bovine serum albumin (64 kDa), carbonic anhydrase (40.6 kDa), soybean trypsin inhibitor (31.9 kDa), lysozyme (18.5 kDa), and aprotinin (7.2 kDa).
The samples were transferred to nitrocellulose paper with a mini transblot cell (Bio-Rad). Electrotransfer was done at room temperature for 1 h at a constant voltage of 100 V. The same prestained standards described above were used, and the transfer buffer was 20 mM Tris, 192 mM glycine, and 20% (vol/vol) methanol (pH 8.3).
Protein samples on nitrocellulose sheets were stained with antienolase antibody or concanavalin A (ConA)-digoxigenin, as follows. For immunoblotting, the transferred samples were treated for 1 h with 3% (wt/vol) bovine serum albumin in phosphate-buffered saline (PBS) and were then incubated at 4°C overnight with the antienolase serum (dilution, 1:200; see below) in 0.15% Tween 20 plus PBS. After several washings with PBS, a second antibody (alkaline phosphatase-conjugated anti-mouse immunoglobulin G [dilution, 1:10,000; Sigma]) was used. For ConA staining, the nitrocellulose sheets were incubated for at least 30 min in blocking solution (the reagents were from the Digoxigenin Detection kit [Boehringer Mannheim, Mannheim, Germany]) in TBS (50 mM Tris-HCl, 150 mM NaCl [pH 7.5]). They were incubated at 4°C overnight with ConA-digoxigenin in TBS plus 1 mM each MnCl2, MnCl2, and CaCl2. After three washes with TBS, a second antibody (alkaline phosphatase-conjugated antidigoxigenin [at a dilution of 0.75 U/ml]) was added. In both cases, the samples were then incubated with 5-bromo-4-chloro-3-indolylphosphate and 4-nitroblue tetrazolium chloride (Boehringer Mannheim) in alkaline phosphatase buffer (containing 100 mM Tris-HCl [pH 9.5], 100 mM NaCl, and 5 mM MgCl2) as the enzyme substrate.
Sequence determination.
Samples obtained by SDS-PAGE were electroblotted onto a ProBlot membrane (Busystem; Perkin-Elmer, Norwalk, Conn.). The pertinent bands were excised and analyzed with a Perkin-Elmer model AB 476A sequencer by previously published techniques (11, 14). Similarity searches in the SwissProt database were performed by use of the Basic Local Alignment Sequence Tools (BLAST) algorithm from the National Center for Biotechnology Information (Bethesda, Md.).
Serum with antienolase antibodies.
Serum with antienolase antibodies was kindly donated by R. La Valle (Department of Bacteriology and Medical Mycology, Istituto Superiore di Sanità, Rome, Italy). It was generated in mice immunized with recombinant C. albicans enolase, and the titer was >1:4,000, as determined by an in-house immunoassay (R. La Valle et al., unpublished data).
RESULTS
MIC determination.
In a preliminary set of experiments, the MICs of FLC for the C. albicans strains used throughout this study were determined by the method of NCCLS (24). The MICs for the two FLC-susceptible strains from a stock collection (non-germ-tube-forming strain CA2 and germ-tube-forming strain 3153) were <1 μg/ml, while the MICs for the two FLC-resistant strains (strains AIDS 126 and AIDS 68) were ≥400 μg/ml. The MICs for strains CO23s and CO23r were redetermined and were <1 and >128 μg/ml, respectively.
GAP detection and modulation in FLC-sensitive C. albicans strain.
We previously characterized some of the GAPs of C. albicans strains 3153 and CA2 and also demonstrated that enolase was a major constituent of the GAPs (2, 3). The following experiments were therefore performed to compare FLC-susceptible strains of C. albicans for the presence and distributions of GAPs, as well as the effect of FLC treatment on the presence and distributions of GAPs.
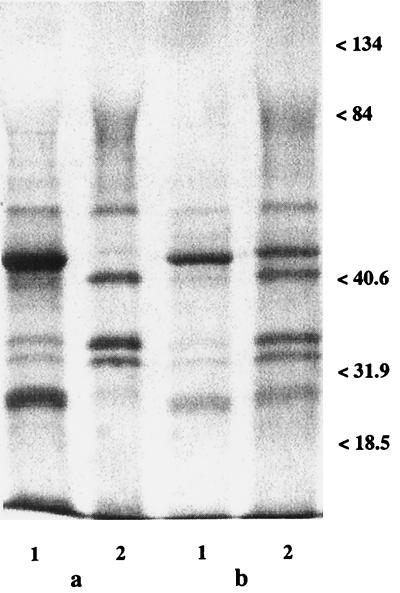
Figure 1 shows the GAP profiles of strains CA2 (Fig. 1a) and 3153 (Fig. 1b) grown in the absence (lanes 1) or in the presence (lanes 2) of FLC after staining with Coomassie brilliant blue. In both strains, the major GAPs of FLC-untreated cells were those with molecular masses of 46 and 29 kDa and a doublet of 34 and 32 kDa. Other components detectable at various intensities in the two strains and in repeated extractions were those of 22, 44, and 56 kDa. (The last protein was subsequently identified by sequence determination to be one of the zymolyase constituents used for extraction of the GAPs; thus, it will not be further dealt with here.)
FIG. 1.
Cell wall GAPs of C. albicans strains CA2 (a) and 3153 (b). Lanes 1, controls; lanes 2, samples after treatment with FLC (5 μg/ml). The samples (7 μg of protein) were run on an SDS-10% polyacrylamide gel and stained with Coomassie brilliant blue. Molecular mass standards are expressed on the right of each gel (in kilodaltons). For technical details, see Materials and Methods.
Figure 1 (lanes 2) also shows the remarkable changes in GAP profiles and contents in cells treated with FLC. There was a prominent increase in the intensities of the 34- and 32-kDa doublet and of the 44-kDa band, coupled with the substantial disappearance of the 46-kDa constituent and, at least in strain CA2, of the 29-kDa band. The levels of all other constituents were not greatly affected, and for this reason they were not investigated further. Since the GAP profiles and the changes in the GAP profiles associated with growth in the presence of FLC were similar in the two FLC-susceptible strains, further detailed investigations on the nature and identification of major cell wall GAPs were performed with only one of the strains (strain CA2).
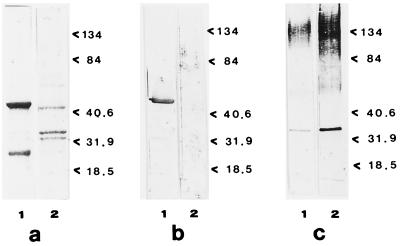
Figure 2a confirms the changes in the GAP profile in strain CA2 exerted by growth in the presence of FLC, as detected in an additional experiment. Figure 2b shows the strong reaction of the 46-kDa band, present in the cell walls of cells not treated with FLC, with the antienolase serum, confirming the previous identification of this protein band as enolase (2) (see below). This protein is also known to be nonglycosylated (6, 19, 27, 29); thus, no reactive 46-kDa band was observed in samples stained with ConA-digoxigenin (Fig. 2c). The latter treatment resulted in the appearance of the 34-kDa band and the pronounced glycosylation (which was particularly apparent in the FLC-treated cells) of high-molecular-mass complexes, present in the upper part of the gel in Fig. 2c.
FIG. 2.
Cell wall GAPs of FLC-sensitive strain C. albicans CA2 after staining of the gel with different strains. (a) Electrophoresis on an SDS-10% polyacrylamide gel stained with Coomassie brilliant blue; (b) transblotting onto nitrocellulose paper and reaction with antienolase immune serum (dilution, 1:100); (c) transblotting onto nitrocellulose paper and reaction with ConA-digoxigenin. In all cases, the gels were loaded with 7 μg of protein. Lanes 1, GAP patterns of control cells not treated with FLC; lanes 2, GAP patterns of FLC-treated cells. Molecular mass standards are expressed on the right of each gel (in kilodaltons). For technical details, see Materials and Methods.
We also investigated whether a similar pattern of FLC-induced changes in the GAP profiles were manifested in a recent clinical isolate of C. albicans (strain CO23s [see Materials and Methods]). As shown in Fig. 3, the most relevant change described above, i.e., the decrease in the intensities, if not the disappearance, of the 46- and 29-kDa bands, was also detectable in this strain, demonstrating that the effect of FLC on the GAP profile is not restricted to laboratory-maintained strains of C. albicans.
FIG. 3.
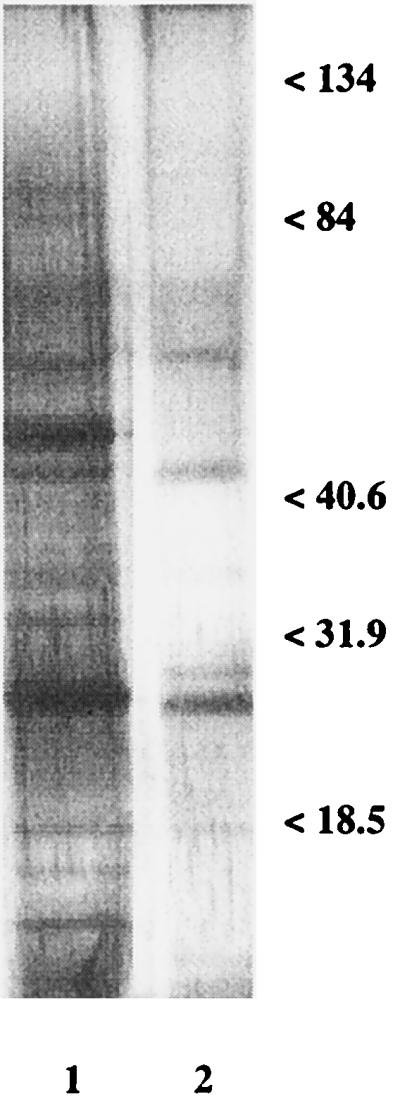
Cell wall GAPs of FLC-sensitive clinical strain CO23s of C. albicans. Lane 1, controls; lane 2, samples after treatment with FLC (1 μg/ml). Electrophoresis on an SDS-10% polyacrylamide gel loaded with 0.5 μg of protein and stained with silver. Molecular mass standards are expressed on the right (in kilodaltons). For technical details, see Materials and Methods and the legends to Fig. 1 and 2.
Treatment of the cell walls with SDS at 100°C for 5 min before zymolyase extraction did not show relevant differences in the SDS-PAGE profiles of the GAPs, suggesting that GAPs are intrinsic constituents of the cell wall (data no shown).
GAP identification.
In addition to enolase (2), we attempted to identify those main GAP constituents which apparently varied in their relative contents with FLC treatment. This was done by peptide sequencing of protease-digested bands excised from the polyacrylamide gel and automated Edman degradation. The sequenced peptides of the N-terminal moiety of each GAP are aligned with corresponding sequences from the SwissProt EMBL databases in Table 1. The sequence identities identified the 29- and 32-kDa proteins as two forms of the enzyme phosphoglyceromutase, while they identified the 34- and 44-kDa proteins as members of the β-(1-3)-glucanase family (7, 17, 22, 26). As shown by ConA staining (Fig. 2c), these two glucanases differed, in that the 34-kDa constituent, but not the 44-kDa constituent, was glycosylated. The identical 34-kDa constituent of Saccharomyces cerevisiae is indeed a glycoprotein with an N glycosylation site at amino acids 197 to 199 (7).
TABLE 1.
Comparison of peptide sequences of C. albicans GAPs with database protein sequences
| GAP size (kDa) | Sequencea | Protein identifiedb |
|---|---|---|
| 29 | Pro Lys Leu Val Leu Val Arg His Gly Gln Ser Glu Trp Asn Glu Lys Asn | S. cerevisiae phosphoglyceromutase (X06408) |
| 33 | Pro Lys Leu Val Leu Val Arg His Gly Gln Ser Glu Trp Asn Glu Lys Asn | S. cerevisiae phosphoglyceromutase (X06408) |
| 34 | Met Gly Asp Leu Ala Phe Asn Leu Gly Val Lys Asn Asp Asp | C. albicans β-(1-3)-glucanase (U12975) |
| 44 | Met Gly Asp Leu Ala Phe Asn Leu Gly Val Lys Asn Asp Asp | C. albicans β-(1-3)-glucanase (U12975) |
From the N terminus peptides 1 to 17 and 19 to 31 of the 29- and 33-kDa GAPs and the 34- and 44-kDa GAPs respectively.
From the SwissProt EMBL Database, with the accession numbers indicated in parentheses.
Cell wall protein profiles of FLC-resistant strains.
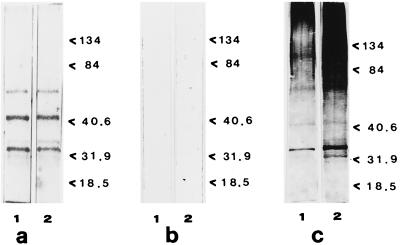
The presence and the distributions of the GAPs were also studied in the two FLC-resistant isolates of C. albicans (strains AIDS 68 and AIDS 126) as well as in strain CO23r, which was made resistant to FLC in the laboratory by growth in stepwise-increasing drug concentrations (see Materials and Methods). All the three FLC-resistant strains had similar GAP patterns, which were quite different from those for the susceptible strains but which were remarkably similar to those for the FLC-susceptible strains grown in the presence of FLC. Figure 4 shows the protein profile of strain AIDS 126, used as a representative of the other two FLC-resistant strains. As shown in Fig. 4a and b (lanes 1), the enolase was absent, while the 44-kDa band and the 32- and 34-kDa doublet bands were clearly evident. Also quite impressive was the remarkable amount of highly glycosylated high-molecular-mass material (Fig. 4c), again somewhat resembling the amount of this material detected in the FLC-susceptible strain after it was grown in the presence of FLC. As expected, the overall GAP profiles of these strains were not changed by the addition of FLC (Fig. 2a to c, lanes 2), with the exception of an apparent increase in the level of glycosylation of the protein bands, particularly in the high-molecular-mass region of the gel.
FIG. 4.
Cell wall GAPs of FLC-resistant strain AIDS 126 of C. albicans. Lanes 1, control; lanes 2, samples after FLC treatment. (a) SDS-10% polyacrylamide gel stained by Coomassie brilliant blue; (b) transblotting onto nitrocellulose paper and reaction with antienolase immune serum (dilution, 1:100); (c) transblotting onto nitrocellulose paper and reaction with ConA-digoxigenin. In all cases the gels were loaded with 7 μg of protein. Lanes 1, GAP patterns of control, FLC-untreated cells; lanes 2, GAP patterns of FLC-treated cells. Molecular mass standards are expressed on the right of each gel (in kilodaltons).
As a further control for protein identification, the bands with molecular masses of 32 and 34 kDa were again excised and sequenced. Sequence analysis confirmed their identities as phosphoglyceromutase and β-(1-3)-glucanase, respectively, as in the earlier determinations (see above).
Protein release.
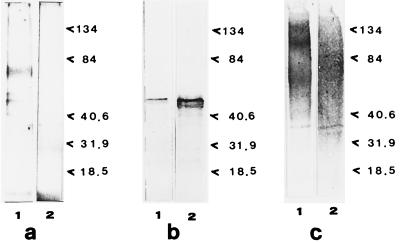
Since a main finding of the roles of GAP constituents observed in FLC-susceptible strains grown in the presence of FLC or in the FLC-resistant strain was the practical disappearance of the principal GAP, enolase, we wondered whether the disappearance of this GAP depended on the release of increased amounts of this protein due to the presence of the drug or to the acquisition of resistance to it. We therefore examined the supernatants of FLC-susceptible strains grown in the presence of FLC as well as the supernatant of the FLC-resistant strain. A protein reactive with the antienolase serum was detected in both supernatants. Figure 5 shows the results obtained with the FLC-resistant strain (strain AIDS 126). It is of interest that the presence of the drug caused an apparent further increase in the amount of enolase excreted into the supernatant (Fig. 5b, lanes 1 and 2). A substantial amount of glycosylated, polydisperse material was detected as a smear in the ConA-stained gels both in the presence and in the absence of the drug (Fig. 5c).
FIG. 5.
Proteins released by FLC-resistant strain AIDS 126 of C. albicans in the culture medium in the absence and in the presence of FLC. (a) SDS-10% polyacrylamide gel stained by use of the Silver Stain Plus kit; (b) transblotting onto nitrocellulose paper and reaction with antienolase immune serum; (c) transblotting onto nitrocellulose paper and reaction with ConA-digoxigenin. In all cases the gels were loaded with 1 μg of protein. Lanes 1, control supernatants of FLC-untreated cells; lanes 2, supernatants of FLC-treated cells. Molecular mass standards are expressed on the right of each gel (in kilodaltons). For other technical details, see Materials and Methods.
DISCUSSION
One of the most interesting aspects of the complex architecture of the C. albicans cell wall is the presence of several proteins and glycoproteins (generally mannosylated proteins) in it. Some of these proteins (e.g., enolase) are generally considered to be associated with cytoplasmic rather than cell wall functions, leading some investigators to question whether they are really present in the cell wall. The rather abundant secretion of enolase, as shown in the present study for the FLC-resistant strain and the inverse relationship between its apparent partition between the cell wall and the supernatants of these cells also provides further circumstantial evidence that enolase resides in the cell wall and is released into the external medium, as suggested previously (7, 24, 28). A detailed discussion of the genuine association of enolase and other “cytoplasmic” proteins with the fungal cell wall can be found in an excellent review by Chaffin et al. (6).
Our study had two principal aims: identifying the main (most abundant) GAP in the cell wall of C. albicans and studying the changes in GAP contents after treatment with a therapeutically important antifungal drug such as FLC. In this regard, we particularly addressed those proteins whose contents were seen to change remarkably following treatment of drug-susceptible strains or FLC-resistant strains with FLC.
As described in previous reports (9, 12), mannoproteins may be bound either noncovalently or covalently to both β-(1-3)- and β-(1-6)-glucans. The definition of GAPs given here would not distinguish between an association and true bonding; nonetheless, it is of interest that pretreatment of cell walls with SDS, which usually removes most of the noncovalently linked molecules, did not affect the protein pattern resulting from zymolyase digestion, indicating that at least some of these proteins could indeed be covalently bound to glucan. This raises the issue of the attachment of the protein to glucan through a glycosyl-phosphatidylinositol (GPI) anchor (15). However, this is certainly not the case for enolase, which was previously found to be cell wall associated and which lacks the signal for GPI in its nucleotide sequence (15). In addition, a signal peptide was also absent from enolase and other enzymes of the glycolytic pathway found in the C. albicans cell wall in the present study, also raising the issue of whether a nonclassical, nonsignal sequence-based secretory pathway is present in this and other fungi (6, 7).
Through automated Edman degradation we have been able to sequence and then identify all major protein constituents appearing in gels by one-dimensional gel electrophoresis with the exception of highly glycosylated, high-molecular-mass complexes. Besides enolase, which we (2) and others (9, 25, 26) have identified previously, the major GAP constituents were two isoforms of phosphoglyceromutase, corresponding to two Coomassie brilliant blue-stained bands of 29 and 32 kDa, and two forms of a β-(1-3)-glucanase, the 44- and 34-kDa bands, which differed in that only the latter was reactive with ConA and was therefore glycosylated.
Phosphoglyceromutase is the enzyme of the glycolytic pathway that converts 3-phosphoglycerate into 2-phosphoglycerate, thus just preceding the enolase step and following the phosphoglycerate kinase step in the glycolytic pathway. Interestingly, both phosphoglycerate kinase and glyceraldehyde-3-phosphodehydrogenase have been found in the C. albicans cell wall (1, 6). Pardo et al. (25) also recently identified by a proteomic approach phosphoglyceromutase as a protein secreted by the regeneration of C. albicans protoplasts. The presence of a substantial portion of the whole glycolytic pattern in the cell wall is intriguing. If the trivial explanation of contamination is excluded, the rather attractive hypothesis remains that these enzymes recognize unknown ligands or serve the same function in the cell wall that they serve in the cytoplasm, i.e., that of generating energy for local (e.g., in the cell wall itself) biosynthesis. In this context, the association of these enzymes with such a critical molecular complex for cell wall construction as the β-glucan is particularly interesting (15).
The sequences of the other two products found to be major GAPs in the present study, i.e., the 34- and 44-kDa constituents, exactly matched the amino acid sequence of the peptide from amino acids 19 to 31 of a BGL2 gene product of C. albicans present in the SwissProt EMBL database under accession number P43070 (A. D. Scadden and P. A. Sullivan) and is identified there as an exo-β-(1-3)-glucanase, an enzyme that catalyzes successive hydrolysis of one molecule of d-glucose from the nonreducing end of the polysaccharide. Both GAP constituents were indeed mature forms of the enzyme since the N-terminal moiety sequence started from amino acid 18, i.e., without the signal peptide, as expected from its extraction from the cell wall. The molecular mass of our glycosylated form (34 kDa) exactly corresponded to that of the peptide present in the SwissProt EMBL database. Molina et al. (21, 22) and Chambers et al. (7) also described a 44-kDa protein with exoglucanase activity. Our data suggest a precursor-product relationship between the two GAPs, and we hypothesize that the 34-kDa GAP derives from the 44-kDa protein after suitable processing and glycosylation.
The presence and quantities of cell wall proteins are regulated by several intracellular and environmental factors (6). We add here another factor which appears to exert a rather marked influence on the GAP pattern in the C. albicans cell wall, i.e., the antimycotic FLC. In drug-susceptible strains, FLC was indeed seen to provoke the substantial disappearance of enolase and one form of the phosphoglycerate mutase from the cell wall, with a shift to larger amounts of the two glucanases and an increased amount of polydisperse, high-molecular-mass, highly glycosylated material. Overall, there was a relative enrichment of the glycosylated constituents and enzymes involved in β-glucan metabolism. Since β-glucanases may also have transglycosylase activities (26, 30), the enrichment described above might indicate indirectly rather consistent rearrangements and modifications of the cell wall structure. This is in keeping with the previously reported influence of FLC on cell wall β-glucanases, coupled with decreased resistance to phagocytosis (12). The augmented susceptibility to glucanase digestion might also be due to the apparent increased proportion of cell wall endoglucanase, as shown here.
One interesting and intriguing observation that we have made in the present study is that the GAP patterns of FLC-susceptible strains in the presence of FLC are quite similar to those observed in drug-resistant strains, i.e., a relative diminution of glycolytic enzymes, in particular, the enolase, coupled with the relative enrichment in the enzymes involved with glucan metabolism. Two FLC-resistant strains used throughout this study (strains AIDS 68 and AIDS 126) were isolated from AIDS patients who were affected by recurrent oral candidiasis or recurrent vaginitis. Both subjects had undergone chronic treatment with FLC. This might have triggered the same changes in the GAP profile of the cell wall which are induced in vitro by the presence of growth in the presence of subinhibitory concentrations, as demonstrated here with strain CO23r.
It could be argued that the changes in the GAP profiles of FLC-resistant strains with respect to the GAP profiles of drug-susceptible strains were rather coincidental, being due to the different genetic backgrounds of the fungal isolates. This is unlikely, however, because a number of genetically unrelated strains, both strains from laboratory stocks and fresh clinical isolates, showed the same or highly comparable GAP profiles. Moreover, two independently isolated, originally FLC-resistant strains (strains AIDS 68 and AIDS 126) had the same GAP profile as one strain (CO23r) which was made resistant to the drug by culture in media with progressively increasing FLC concentrations. Strain CO23r had the same genotype as strain CO23s, which also had the same GAP profile and the same GAP response to FLC treatment as all other drug-susceptible strains did. Overall, the rather uniform pattern of changes in the GAPs in all strains evaluated in this study strongly argues against a purely coincidental event, while it substantially supports the genuine nature and consistency of the changes in GAPs in susceptible strains during FLC treatment and in the resistant strains.
A second intriguing aspect of our observations is that enolase and other GAPs underwent changes similar to those described here when C. albicans was grown in the presence of subinhibitory doses of cilofungin (3) or FK 463 (kindly donated by I. T. Murato, Fujisawa, GmBH, Tokyo, Japan) (L. Angiolella et al., unpublished data), two antimycotics that are quite distinct from FLC in terms of their mechanisms of action and that more directly affect β-glucan metabolism (2, 3). These observations suggest that the modulated expression of GAP constituents might not depend on a specific action of the antibiotic but, rather, might reflect some general metabolic change that is indirectly provoked by exposure to the antibiotic as a cell-stressing agent. In support of this speculation, it must be noted that some changes, such as an apparent increase in the levels of enolase secretion and in the amounts of highly glycosylated, high-molecular-mass cell wall material, were also caused by FLC in the FLC-resistant strain.
Overall, the change in the GAP profile from a prevalently “glycolytic” one (rich in enolase and phosphoglyceromutase) to a prevalently “glucanase” one may be envisaged as a sort of general reaction to an inhibitor, a reaction that may be stably incorporated into a resistant strain as a sort of antistress response because it offers the cell some selective advantage (31). For instance, glycolytic enzymes are highly antigenic (6, 16, 20, 27); thus, their decreased level of expression in the cell wall might render the cells less antigenic, therefore helping the cell to evade the host's immune responses. It would be of interest to examine whether other generic stresses, such as heat or nutritional shock, could provoke the same kinds of changes in the GAP profile shown by both FLC and cilofungin. At any rate, high levels of glycosylation and the relative enrichment of β-glucan-modeling enzymes suggest that antibiotics have profound influences on the metabolism and structure of the cell wall of such an important human opportunistic pathogen as C. albicans. The full identification of the major GAPs achieved here might also be useful in the creation of strategies for the targeting of new antimicrobials to these sensitive cell wall enzymes.
Acknowledgments
We are grateful to Lajean Chaffin (Texas Tech University Health Science Center, Lubbock) for critically reading the manuscript and providing useful suggestions. The donation of antienolase serum by R. La Valle is also kindly acknowledged. Thanks are also due to A. Botzios and G. Mandarino, who helped in the preparation of the manuscript.
This work was supported in part by a grant (contract 50 B/C) from the National AIDS Program of Italy.
REFERENCES
- 1.Alloush, H. M., J. J. Lòpez-Ribot, B. J. Masten, and W. L. Chaffin. 1997. 3-Phosphoglycerate kinase: a glycolytic enzyme present in the cell wall of Candida albicans. Microbiology 143:321-330. [DOI] [PubMed] [Google Scholar]
- 2.Angiolella, L., M. Facchin, A. Stringaro, B. Maras, N. Simonetti, and A. Cassone. 1996. Identification of a glucan-associated enolase as a main cell wall proteins of Candida albicans and an indirect target of lipopeptide antimycotics. J. Infect. Dis. 173:684-690. [DOI] [PubMed] [Google Scholar]
- 3.Angiolella, L., N. Simonetti, and A. Cassone. 1994. The lipoptide antimycotic, cilofungin, modulates the incorporation of glucan-associated proteins into cell wall of Candida albicans. J. Antimicrob. Chemother. 33:1137-1146. [DOI] [PubMed] [Google Scholar]
- 4.Cassone, A. 1986. Cell wall of pathogenic yeast and implications for antimycotic therapy. Drugs Exp. Clin. Res. 12:635-643. [PubMed] [Google Scholar]
- 5.Cassone, A., F. De Bernardis, E. Pontieri, G. Carruba, C. Girmenia, P. Martino, M. Fernández-Rodríguez, G. Quindós, and J. Ponton. 1995. Biotype diversità of Candida parapsilosis and its relationship to the clinical source and experimental pathogenicity. J. Infect. Dis. 171:967-975. [DOI] [PubMed] [Google Scholar]
- 6.Chaffin, W. L., J. L. Lopez-Ribot, M. Casanova, D. Gozaldo, and J. P. Martinez. 1998. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microb. Mol. Biol. Rev. 62:130-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers, R. S., M. J. Broughton, R. D. Cannon, A. Carne, G. W. Emerson, and P. A. Sullivan. 1993. An exo-β-(1,3)-glucanase of Candida albicans: purification of the enzyme and molecular cloning of the gene. J. Gen. Microbiol. 139:325-334. [DOI] [PubMed] [Google Scholar]
- 8.Dubois, M., K. A. Giles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350-356. [Google Scholar]
- 9.Edwards, S. R., R. Bradley, and W. L. Chaffin. 1999. Enolase is present in the cell wall of Saccharomyces cerevisiae. FEMS Microbiol. Lett. 177:211-216. [DOI] [PubMed] [Google Scholar]
- 10.Elorza, M. V., A. Murgui, and R. Sentandreu. 1985. Dimorphism in Candida albicans: contribution of mannoproteins to the architecture and mycelial walls. J. Gen. Microbiol. 131:2209-2216. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez, J., L. Andrews, and S. M. Mische. 1994. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Anal. Biochem. 218:112-117. [DOI] [PubMed] [Google Scholar]
- 12.Hazen, K. C., G. Mandell, E. Coleman, and G. Wu. 2000. Influence of fluconazole at sub-inhibitory concentrations on cell surface hydrophobicity and phagocytosis of Candida albicans. FEMS Microbiol. Lett. 183:89-94. [DOI] [PubMed] [Google Scholar]
- 13.Herrero, H., P. Sanz, and R. Sentandreu. 1987. Cell wall proteins liberated by zymolase from several ascomycetous and imperfect yeasts. J. Gen. Microbiol. 133:2895-2903. [Google Scholar]
- 14.Iwamatsu, A. 1992. S-carboxymethylation of proteins transferred onto polyvinylidene difluoride membranes followed by in situ protease digestion and amino acid microsequencing. Electrophoresis 13:142-147. [DOI] [PubMed] [Google Scholar]
- 15.Kapteyn, J. C., H. Van Den Ende, and F. M. Klis. 1999. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta 1426:373-393. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Ribot, J. L., R. K. McAtee, W. R. Kinkpatrick, R. La Valle, and T. F. Patterson. 1999. Low levels of antigenic variability in fluconazole-susceptible and -resistant Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Clin. Diagn. Lab. Immunol. 6:665-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luna-Arias, J. P., E. Andaluz, J. C. Ridruejo, I. Olivero, and J. Larriba. 1991. The major exoglucanase from Candida albicans: a non-glycosylated secretory monomer related to its counterpart from Saccharomyces cerevisiae. Yeast 7:833-841. [DOI] [PubMed] [Google Scholar]
- 18.Marcilla, A., M. V. Elorza, S. Mormeneo, H. Rico, and R. Sentandreu. 1991. Candida albicans mycelial wall structure: supramolecular complexes released by zymolyase, chitinase and β-mercaptoethanol. Arch. Microbiol. 155:312-319. [DOI] [PubMed] [Google Scholar]
- 19.Mason, A. B., H. R. Buckley, and J. A. Gorman. 1993. Molecular cloning and characterization of the Candida albicans enolase gene. J. Bacteriol. 175:2632-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthews, R. C., J. P. Burnie, and S. Tabaqchali. 1987. Isolation of immunodominant antigens from sera of patients with systemic candidiasis and characterization of serological response to Candida albicans. J. Clin. Microbiol. 25:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molina, M., R. Cenamor, and C. Nombela. 1987. Exo-1,3-β-glucanase activity in Candida albicans: effect of the yeast to mycelium transition. J. Gen. Microbiol. 133:609-617. [DOI] [PubMed] [Google Scholar]
- 22.Molina, M., R. Cenamor, M. Sanchez, and C. Nombela. 1989. Purification and some properties of Candida albicans exo-1,3-beta-glucanase. J. Gen. Microbiol. 135:309-314. [DOI] [PubMed] [Google Scholar]
- 23.Murgui, A., M. V. Elorza, and R. Sentandreu. 1986. Tunicamycin and papulocandin B inhibit incorporation of specific mannoproteins into cell wall of Candida albicans regenerating protoplasts. Biochim. Biophys. Acta 884:550-558. [DOI] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 1977. Reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard. NCCLS document M27-A. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 25.Pardo, M., M. Ward, A. Pitarch, M. Sanchez, C. Nombela, and W. C. Blackstock. 2000. Cross species identification of the novel Candida albicans immunogenic proteins by combination of two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Electrophoresis 21:2651-2659. [DOI] [PubMed] [Google Scholar]
- 26.Stubbs, H. J., D. J. Brasch, G. W. Emerson, and P. A. Sullivan. 1999. Hydrolase and transferase activities of the β-1,3-exoglucanase of Candida albicans. Eur. J. Biochem. 263:889-895. [DOI] [PubMed] [Google Scholar]
- 27.Sundstrom, P., J. Jensen, and E. Balish. 1994. Humoral and cellular immune response to enolase after alimentary tract colonization or intravenous immunization with Candida albicans. J. Infect. Dis. 170:390-395. [DOI] [PubMed] [Google Scholar]
- 28.Sundstrom, P., and G. R. Aliaga. 1994. A subset of proteins found in culture supernatants of Candida albicans includes the abundant, immunodominant, glycolytic enzyme enolase. J. Infect. Dis. 169:452-456. [DOI] [PubMed] [Google Scholar]
- 29.Sundstrom, P., and G. R. Aliaga. 1992. Molecular cloning of cDNA and analysis of protein secondary structure of Candida albicans enolase, an abundant, immunodominant glycolytic enzyme. J. Bacteriol. 174:6789-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasquez de Aldana, C., J. Correa, P. San Segundo, A. Bueno, A. R. Nebreda, E. Mendez, and F. del Rey. 1991. Nucleotide sequence of the exo-1,3-β-glucanase encoding gene, EXG1, of the yeast Saccharomyces cerevisiae. Gene 97:173-182. [DOI] [PubMed] [Google Scholar]
- 31.Wu, T., K. Wright, S. F. Hurst, and C. J. Morrison. 2000. Enhanced extracellular production of aspartyl proteinase, a virulence factor, by Candida albicans isolates following growth in subinhibitory concentrations of fluconazole. Antimicrob. Agents Chemother. 44:1200-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]