Abstract
The genetic organization of the region coding for CTX-M-2 in Salmonella enterica serovar Infantis was determined by PCR mapping. This gene seems to have been mobilized from the Kluyvera ascorbata chromosome to a complex sulI-type integron, similar to In6 and In7.
After the initial isolation during May 1989 of a Salmonella enterica serovar Agona strain highly resistant to cefotaxime (A. Rossi, M. Woloj, G. Gutkind, et al., Abstr. 92nd Gen. Meet. Am. Soc. Microbiol. 1992, abstr. A135, 1992), dissemination of this resistance marker took place within the different salmonellae that were present in the three pediatric hospitals of Buenos Aires, Argentina, by the end of 1989 (H. Lopardo, N. Fernandez, M. Fernandez-Cobo, et al., Proc. 17th Congr. Chemother., abstr. 2088, 1991) (16). Among many outbreaks due to extended-spectrum-β-lactamase (ESBL)-producing enterobacteria reported in Argentina (2), CTX-M-2 (a cefotaximase) was present in almost 75% of all ESBLs detected in extended-spectrum-cephalosporin (ESC)-resistant strains, while PER-2 and other less frequent enzymes (SHV-5 and SHV-2) accounted for the remaining 25% (M. Quinteros, M. Mollerach, M. Radice, et al., 39th Int. Conf. Antimicrob. Agents Chemother., abstr. 893, 1999). At that time, it was assumed that the initial ESC-resistant salmonella outbreak was not due to the spread of a single salmonella clone but to dissemination of resistance markers within different salmonella serovars.
Between April 1997 and August 1998, a ESC-resistant serovar Infantis outbreak was detected in Hospital Gutierrez, Santa Fe, Argentina (C. Mayoral, P. Andres, J. Di Conza, et al., 9th Int. Congr. Infect. Dis., abstr. 82.014, 2000). Six strains were selected as representative of those causing the outbreak, and a wide antimicrobial susceptibility profile was carried out following NCCLS recommendations by agar dilution (10). All strains were resistant to cephalothin (MIC ≥ 512 μg/ml), cefuroxime (MIC ≥ 512 μg/ml), ceftriaxone (MIC ≥ 128 μg/ml), cefotaxime (MIC ≥ 64 μg/ml), and aztreonam (MIC ≥ 32 μg/ml). An 8- to 10-fold decrease in MICs was detected when these drugs were combined with lithium clavulanate (2 μg/ml). MICs for the following β-lactam drugs were as indicated: ceftazidime, 4 to 16 μg/ml; cefepime, 16 μg/ml (intermediate); and imipenem, ≤ 0.03 μg/ml (susceptible). Strains were also resistant to other antimicrobial agents, including gentamicin (MIC, 256 μg/ml), and nitrofurantoin (MIC, 256 to 512 μg/ml), remaining susceptible to amikacin (MIC, 4 to 8 μg/ml) and tetracycline (MIC, 0.5 to 2 μg/ml). Also, the isolates were resistant to kanamycin, tobramycin, netilmicin, and sulfonamides, tested by the disk diffusion method (11).
Crude extracts, obtained from late-log-phase cultures, were analyzed for their β-lactamase content by isoelectric focusing in broad-range (pH 3 to 10) polyacrylamide gels. All extracts displayed two β-lactam-hydrolyzing enzymes (with apparent pIs of 5.4 and 8.2) when revealed with an iodometric overlay system (16) using ampicillin (500 μg/ml) as the substrate, but only the pI 8.2 enzyme was detected when ceftriaxone (1,000 μg/ml) was the substrate (the same profile detected in salmonellae isolated in 1989). The suspected β-lactamases were TEM-1 (a pI 5.4 broad-spectrum enzyme) and CTX-M-2, the most-prevalent ESBL in Argentina and neighboring countries.
A 73-kb plasmid (pS21) was isolated by alkaline lysis extraction (3) from serovar Infantis S21. In order to search for the suspected β-lactamase-encoding genes, screening for blaTEM and blaCTX-M genes was performed by PCR amplification (17) using pS21 as template and the following specific primers: OT3 and OT4 (TEM-specific primers) (1) and bla1 and bla2 (CTX-M-specific primers) (14). A 0.85-kb fragment was obtained with OT3-OT4 primers, and a 0.9-kb fragment was obtained when bla1-bla2 were used. Both fragments were sequenced in both strands by the Big Dye terminator method (ABI PRISM 377); sequencing finally confirmed the presence of a blaTEM-1-like gene and blaCTX-M-2.
The conjugative transfer of serovar Infantis' pS21 to recipient Escherichia coli CAG12177 (E. coli Genetic Stock Center, New Haven, Conn.) was performed by solid-medium mating, after several unsuccessful attempts using the liquid-medium assay (17). Selection was carried out on Luria-Bertani agar plates supplemented with tetracycline (20 μg/ml) and cefotaxime (30 μg/ml), and only three cefotaxime-resistant clones were obtained. The ESC resistance transfer was associated with cotransmission of aminoglycoside, sulfonamide, and nitrofurantoin resistance.
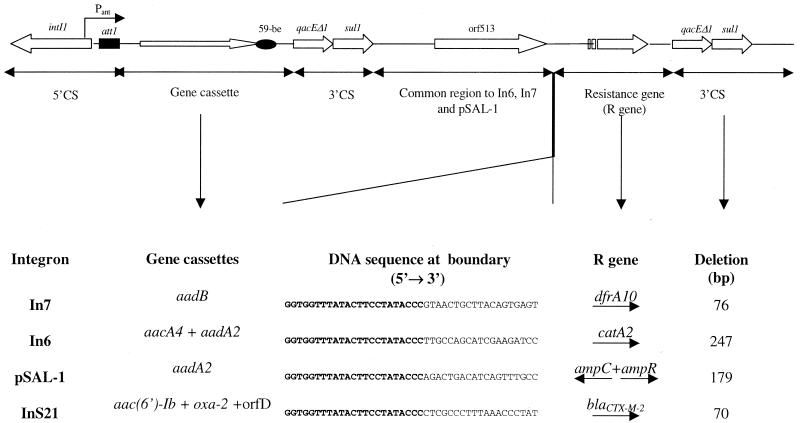
The sequence of a 497-bp region upstream of blaCTX-M-2 in pS21 was determined using oligonucleotide 14 (Table 1). Immediately upstream of blaCTX-M-2 there is a sequence of 266 bp 96% identical to the upstream region of the KluA-1-encoding gene (a chromosomal β-lactamase from Kluyvera ascorbata; EMBL accession number AJ272538). The distal 231-bp fragment has a 100% identity to the last 231-bp common region of In6 and In7 integrons (GenBank accession number L06418) (8). These unusual integrons contain the complete sequence of the 5′-conserved segment (5′-CS) and two copies of part of 3′-CS. Between both copies of 3′-CS there is a shared region (common region) and a variable one, containing different resistance genes, depending on the integron (Fig. 1) (8).
TABLE 1.
PCR primers for identification and characterization of integron and gene cassettes
| Primer no. | Primer name | Primer sequence (5′ → 3′) | Accession no. |
|---|---|---|---|
| 1 | 15 | ACCGCCAACTTTCAGCACAT | M95287 |
| 2 | 13 | GCGTTCGGTCAAGGTTCTGG | M95287 |
| 3 | 5′-CS | GGCATCCAAGCAGCAAGC | M73189 |
| 4 | 3′-CS | AAGCAGACTTGACCTGAT | M73189 |
| 5 | qacEΔ1F | ATCGCAATAGTTGGCGAAGT | X15370 |
| 6 | qacEΔ1B | CAAGCTTTTGCCCATGAAGC | X15370 |
| 7 | Sul1F | CTTCGATGAGAGCCGGCGGC | X12869 |
| 8 | Sul1B | GCAAGGCGGAAACCCGCGCC | X12869 |
| 9 | Oxa2-A | CCTGCATCGACATTCAAGATA | P05191 |
| 10 | Oxa2-B | CTCAACCCATCCTACCCACCA | P05191 |
| 11 | Bla1 | TTAATGATGACTCAGAGCATT | X92507 |
| 12 | Bla2 | GATACCTCGCTCCATTTATTGC | X92507 |
| 13 | Bla4 | TACCCAACCGGAGCAGAAGG | X92507 |
| 14 | Bla-up | GGCTTCCAGCTGCTGTTGCAC | This work |
| 15 | Bla-down | TTGACTGTCGACCCCAAATCC | This work |
| 16 | ORFend | CCGTTAAGCTCTTATGTGGG | L06418 |
FIG. 1.
Schematic drawing of the class 1 integrons In6, In7, pSAL-1, and InS21. In the sequence at the boundary, the boldface text corresponds to conserved nucleotides. Deletions correspond to the 5′ end of the second 3′-CS region. Arrows indicate the direction of transcription.
In order to determine if the integron is present as a single unit, pS21 was digested with different restriction endonucleases with no restriction sites within the intI1 gene (SacI, AccI, and HindIII, respectively). These digestions were subjected to Southern blot DNA-DNA hybridization (17), using a PCR fragment corresponding to a part of intI1 gene as a probe, obtained with primers 1 and 2 (Table 1). We observed a unique hybridization band in all reactions, of 5.5 kb (SacI), 8 kb (AccI), and 15 kb (HindIII), suggesting that pS21 carries only one allele of the class 1 integron (data not shown).
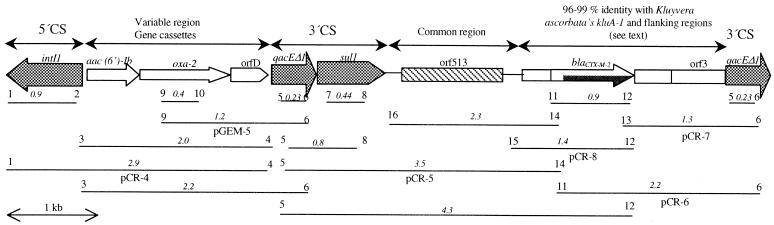
In order to determine if blaCTX-M-2 is part of an In6- or In7-like structure, the presence of the intI1, qacEΔ1, and sul1 class 1 integron-specific genes was confirmed using specific primers (18), while the location of the blaCTX-M-2 gene within the integron was assessed upon a strategy of different PCR amplifications, combining several primers (Table 1). Conditions for PCR amplifications were as follows: 10 ng of pS21 and 0.5 to 3 U of Taq DNA polymerase (Perkin-Elmer, Boston, Mass.); cycling parameters of 6 min at 94°C, 30 cycles of 1 min at 94°C (denaturation), 1 min at 50 to 65°C (annealing), and 1 to 2 min at 72°C (polymerization); and a final extension of 10 min at 72°C. The proposed scheme of the integron deduced from the results (InS21) is shown in Fig. 2, in which putative resistance genes in the pS21 structure are highlighted.
FIG. 2.
PCR mapping strategy and resulting structure of InS21. Numbers in italics correspond to the approximate sizes (in kilobases) of the PCR products. Numbers at both ends of each PCR product indicate the pair of primers used for the different PCR amplifications. Dashed boxes correspond to the common region, whereas striped boxes indicate the region (blaCTX-M-2 and ORF 3) having homology with kluA-1 and flanking regions. Arrows indicate the direction of transcription. Cloned inserts used for sequencing are indicated with the plasmid designation given to each construct.
In addition, different PCR products—obtained using primer combinations 1-4, 5-14, 11-6, 13-6, and 15-12—were cloned in pCR2.1 vector (Invitrogen Inc., Leek, The Netherlands), and a 1.2-kb fragment from primer combination 9-6 was cloned in pGEM-T vector (Promega, Madison, Wis.) (the resulting constructions were named pCR-4, pCR-5, pCR-6, pCR-7, pCR-8, and pGEM-5, respectively). These constructions (two clones for each fragment) were used for sequencing genes belonging to the proposed integron (InS21) shown in Fig. 2. It is noteworthy that blaTEM-1, cotransferred during conjugation of pS21, is not associated with InS21.
By sequencing the fragments cloned in pCR-4, pCR-5, and pGEM-5, the presence of several elements characteristic of class 1 integrons was confirmed: the 5′-CS included intI1 (a gene coding for the site-specific recombinase), attI (a cassette integration site), the promoters Pant and P2 (responsible for expression of the cassette genes) (4, 5); and the suitably oriented gene cassettes, with their corresponding 59-base elements (59-be) downstream of each cassette (described below) (9, 15). In this case, the 3′-CS extends from bp 2920 to bp 4932 (EMBL accession number AJ311891), including the qacEΔ1 and sulI genes. The length of this region varies in different integrons (4).
Briefly, the variable region of the integron (2 kb) contains the following cassettes, from 5′-CS to 3′-CS: an aac(6′)-Ib, oxa-2, and an open reading frame (ORF) (ORF D). The oxa-2 pseudogene contains two significant changes compared with the prototype sequence (EMBL accession number M95287): a Ser10Phe substitution (C29T) and a stop codon interrupting the ORF (A638G), which explains why this enzyme could not be detected after several attempts (data not shown). In addition, downstream of the 3′-CS region, a 2,153-bp fragment having 99% identity with the common region of In6/In7 (which also includes ORF 513, formerly ORF 341) (13, 20) was found.
By analyzing the constructions involving blaCTX-M-2 and their downstream sequences (pCR-6 and pCR-7 plasmids), we were able to confirm the presence of the following regions (from 5′ to 3′): a first region homologous to the downstream sequence of a variety of cefotaximases (including, once again, KluA-1) (EMBL accession number AJ272538); ORF 3 (homologous to that of K. ascorbata); and qacEΔ1, which corresponds to the first component of the second 3′-CS.
Taking into account the facts that blaCTX-M-2 and kluA-1 (the chromosomal β-lactamase from K. ascorbata) were 99% identical along the full length of the gene and that the upstream and downstream regions of the InS21 associated with blaCTX-M-2 have 96 and 99% identity, respectively, to the corresponding regions of kluA-1, it seems possible to support the argument that the hypothetical chromosomal origin of this plasmid-borne β-lactamase is K. ascorbata KluA-1 (or a similar enzyme), the possible precursor of CTX-M-2 subgroup β-lactamases (6, 12). The observed changes between KluA-1 and CTX-M-2 are Ile251Val and Val279Ile and three additional silent nucleotide changes: T336G, T357G, and G441T. The argument for this possible origin is especially reinforced if we consider that this enzyme, CTX-M-2, although not the first to be characterized, was the first of this family to be detected and was widely distributed in our region at a time when ESCs (and especially ceftriaxone) were overused. It is also noteworthy that blaCTX-M-2 maintains its own putative promoter (deduced by primer extension [data not shown]) in comparison with the KluA-1 sequence, and also that its upstream region conserves one of the proposed promoters for dfrA10, as reported by Parson et al. (13).
After further analysis of the proposed arrangement of genes in this new integron, InS21, we could confirm that blaCTX-M-2 is inserted in an In6-In7-like structure, where catA2 and dfrA10, respectively (13, 20), are replaced by the cefotaximase-encoding gene; all of them have the same orientation as the gene cassettes. On the other hand, even when InS21 has a similar genetic organization to that of pSAL-1 (22), in the latter, ampC-ampR (which replace blaCTX-M-2) are in the opposite orientation.
Another shared characteristic among these unusual integrons is a deletion (compared to the putative In1 precursor) at the 5′ end of the second 3′-CS region. The difference relies on the length of that deletion: 247 bp in In6 (20, 22), 179 bp in the pSAL-1 integron (22), 76 bp in In7 (20, 22), and 70 bp in InS21. Thus, deletions observed in In7 and InS21 have almost the same length (Fig. 3).
FIG. 3.
Sequence alignments of the second 3′-CS 5′ ends for the consensus sequence (19) of that region in In6, In7, pSAL-1, and the proposed InS21. Asterisks indicate sequence identity. The start codon for qacEΔ1 gene is underlined, and the sequence of ORF 3 in InS21 is double underlined.
The analysis of the sequence showed that blaCTX-M-2 does not have a typical 59-be, the main characteristic of which is the presence of two conserved regions (RH and LH motifs) (9, 15, 19). This finding is in agreement with the fact that previously reported In6-, In7-, and pSAL-1 integron-associated catA2, dfrA10, and ampC-ampR genes, respectively, do not have the 59-be either (13, 20, 22). In these cases, the insertion of these resistance genes between two copies of 3′-CS in the absence of a typical 59-be could not be explained (Fig. 2). However, ORF 513, associated with partial duplication of the 3′-CS, could encode a site-specific recombinase (21).
We might speculate, as already assumed by others (13, 20), that those genes could have been inserted as any other cassette genes, using a 59-be, with the subsequent loss of this element. Two indirect pieces of evidence may support the notion of the recombination-deletion event rather than a previously existing integron hypothesis. The first piece of evidence is the simultaneous isolation of an ESC-sensitive S. enterica, in which the 5′-CS and 3′-CS of the integron are already present, but lacking both catA2 and dfrA10 or any resistance-associated cassette (data not shown); the second piece of evidence is that the last 28 bp of the hypothetical primitive ORF 3 from K. ascorbata (absent in InS21) overlaps the 70-bp deletion in the second 3′-CS region. Assuming this hypothesis to be correct, it could be linked to the general model of genetic (resistance) islands (islets), in which formation of resistance islets might occur if the recombination sites, within mobile elements, had been lost (7).
As far as we know, there are no previous reports describing an integron-associated blaCTX-M gene. There is preliminary evidence for the presence of blaCTX-M-2 within the variable region of a class 1 integron, which is present in a high-molecular-weight plasmid of Morganella morganii, and a more-detailed analysis is being already done (P. Power, personal communication).
Finally, it is noteworthy that members of the family of integrons composed of In6, In7, the pSAL-1 integron, and the newly proposed InS21 have a similar organization and, possibly, derive from a common ancestor.
Nucleotide sequence accession numbers.
The complete integron sequence has been deposited into the EMBL database under accession number AJ311891.
Acknowledgments
We are grateful to Ana Amoroso and Marcela Radice for all their expert advice and helpful scientific discussions, Clara Mayoral for providing the clinical strains used in this study, and Andrea Nepote for her collaboration in the serotyping of the strains.
J.D.C. is a recipient of a FOMEC fellowship and was a recipient of a fellowship from the European Community, ALFA program, project ALR/B7-3011/94.04-5.0111.9. This work was supported in part by grants from the University of Buenos Aires (Buenos Aires, Argentina) (TB 039), CONICET (PID 4413), and SEPCYT (PICT 0693) to G.G. and grant PB97-1193 from the Ministerio de Ciencia y Tecnología, Spain, to J.A.A.
REFERENCES
- 1.Arlet, G., G. Brami, D. Décrè, A. Flippo, O. Gaillot, P. H. Lagrange, and A. Philippon. 1995. Molecular chracterisation by PCR-restriction fragment length polymorphism of TEM β-lactamases. FEMS Microbiol. Lett. 134:203-208. [DOI] [PubMed] [Google Scholar]
- 2.Bantar, C., A. Famiglietti, M. Goldberg, Antimicrobial Committee, et al. 2000. Three-year surveillance study of nosocomial bacterial resistance in Argentina. Int. J. Infect. Dis. 4:85-90. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, H. J., H. W. Stokes, and R. M. Hall. 1996. The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 178:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Decousser, J. W., L. Poirel, and P. Nordamann. 2001. Characterization of a chromosomally encoded extended-Spectrum class A β-lactamase from Kluyvera cryocrescens. Antimicrob. Agents Chemother. 45:3595-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hacker, J., and J. Kaper. 1999. The concept of pathogenicity islands. In J. Kaper and J. Hacker (ed.), Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, D.C.
- 8.Hall, R. M., and C. M. Collis. 1998. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updates 1:109-119. [DOI] [PubMed] [Google Scholar]
- 9.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 5th ed. M7-A5. National Committee for Clinical Laboratory Standards., Wayne, Pa.
- 11.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests; approved standard, 7th ed., vol 20. M2-A7. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 12.Oliver, A., J. C. Pérez-Díaz, T. M. Coque, F. Baquero, and R. Cantón. 2001. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-10) isolated in Spain. Antimicrob. Agents Chemother. 45:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parsons, Y., R. M. Hall, and H. W. Stokes. 1991. A new trimethoprim resistance gene, dhfrX, in the In7 integron of plasmid pDGO100. Antimicrob. Agents Chemother. 35:2436-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Power, P., M. Radice, C. Barberis, C. de Mier, M. Mollerach, M. Maltagliatti, C. Vay, A. Famiglietti, and G. Gutkind. 1999. Cefotaxime-hydrolyzing β-lactamase in Morganella morganii. Eur. J. Clin. Microbiol. Infect. Dis. 18:743-747. [DOI] [PubMed] [Google Scholar]
- 15.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 16.Rossi, A., H. Lopardo, M. Woloj, A. M. Picandet, M. Mariño, M. Galas, M. Radice, and G. Gutkind. 1995. Non-typhoid Salmonella spp. resistant to cefotaxime. J. Antimicrob. Chemother. 36:697-702. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd. ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1997. Characterization of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 157:177-181. [DOI] [PubMed] [Google Scholar]
- 19.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 20.Stokes, H. W., C. Tomaras, Y. Parsons, and R. M. Hall. 1993. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid 30:39-50. [DOI] [PubMed] [Google Scholar]
- 21.Valentine, C. R., M. J. Heinrich, S. L. Chissoe, and B. A. Roe. 1994. DNA sequence of direct repeats of the sulI gene of plasmid pSa. Plasmid 32:222-227. [DOI] [PubMed] [Google Scholar]
- 22.Verdet, C., G. Arlet, G. Barnaud, P. H. Lagrange, and A. Philippon. 2000. A novel integron in Salmonella enterica serovar Enteritidis, carrying the blaDHA-1 gene and its regulator gene ampR originated from Morganella morganii. Antimicrob. Agents Chemother. 44:222-225. [DOI] [PMC free article] [PubMed] [Google Scholar]