Abstract
In a previous study, an analysis of 77 ampicillin-nonsusceptible (resistant plus intermediate categories) strains of Vibrio cholerae non-O1, non-O139, isolated from aquatic environment and diarrheal stool, showed that all of them produced a β-lactamase with a pI of 5.4. Hybridization or amplification by PCR with a probe for blaTEM or primers for blaCARB gene families was negative. In this work, an environmental ampicillin-resistant strain from this sample, ME11762, isolated from a waterway in the west region of Argentina, was studied. The nucleotide sequence of the structural gene of the β-lactamase was determined by bidirectional sequencing of a Sau3AI fragment belonging to this isolate. The gene encodes a new 288-amino-acid protein, designated CARB-7, that shares 88.5% homology with the CARB-6 enzyme; an overall 83.2% homology with PSE-4, PSE-1, CARB-3, and the Proteus mirabilis N29 enzymes; and 79% homology with CARB-4 enzyme. The gene for this β-lactamase could not be transferred to Escherichia coli by conjugation. The nucleotide sequence of the flanking regions of the blaCARB-7 gene showed the occurrence of three 123-bp V. cholerae repeated sequences, all of which were found outside the predicted open reading frame. The upstream fragment of the blaCARB-7 gene shared 93% identity with a locus situated inside V. cholerae's chromosome 2. These results strongly suggest the chromosomal location of the blaCARB-7 gene, making this the first communication of a β-lactamase gene located on the VCR island of the V. cholerae genome.
Non-O1 Vibrio cholerae strains are natural inhabitants of aquatic environments. Although this group of microorganisms does not produce choleric diseases, a large cholera-like outbreak in Bangladesh and India was found to be caused by a V. cholerae non-O1 strain, which was named O139 because of its new O serogroup. Since 1981, ampicillin (AMP)-resistant V. cholerae strains have been sporadically isolated. These strains harbored plasmid-mediated β-lactamases, such as TEM-1 (8) and SAR-1 (19). More recently, a nonconjugative β-lactamase belonging to the CARB group, named CARB-6, was characterized from a clinical strain of V. cholerae non-O1, non-O139 (5). Among the carbenicillin-inactivating β-lactamases, the CARB family is the main group, including PSE-4 (CARB-1), PSE-1 (CARB-2), CARB-3, CARB-4, CARB-6, and N29, a β-lactamase from Proteus mirabilis. In addition, a new group of carbenicillinases, the RTG group, was proposed, which includes RTG-1 (P. mirabilis GN79 enzyme) and RTG-2 (CARB-5). Although these two chromosomal enzymes showed a low identity with members of the CARB family (44%), they may be considered to be the ancestors of this enzymatic group (6).
Some structural genes for CARB enzymes were located on resistance cassettes as part of integrons (18) that so far had been characterized as genetic elements capable of capturing and rearranging resistance genes (24). Integrons from class 1 were already observed in V. cholerae (7, 9). In addition, V. cholerae strains have a large chromosomal region, which acts as an integron-like structure, capturing genes from plasmids or via phages (15). This super integron is restricted to a region of about 10% of V. cholerae chromosome 2 (11) and is characterized by many 123- to 126-bp sequences of imperfect symmetry with a high of identity, named V. cholerae repeats (VCR). VCR flank one single gene or in some cases pair of genes (4). Most of the 216 open reading frames (ORFs) described in the VCR region have not shown homology with any known sequence, and only three appear to be involved in drug resistance (chloramphenicol [CMP] acetyltransferase, fosfomycin resistance protein, and glutathione transferase) (11).
In a previous work, we had performed susceptibility screening by disk diffusion methods of 669 isolates of V. cholerae non-O1, non-O139, which had been received at the National Institute of Infectious Diseases “Dr. C. G. Malbrán” between January 1993 and May 1998. Thirty percent of those isolates presented an AMP-nonsusceptible (resistant plus intermediate categories) phenotype. The analysis of a subset of 131 AMP-nonsusceptible isolates showed that 57% of them produced a β-lactamase with a pI of 5.4 with high activity against amino- and carboxy-penicillins. For this group, the MICs at which 50 and 90% of isolates tested were inhibited (in micrograms per milliliter) were, respectively, as follows: AMP, 64 and 256, and ticarcillin, 128 and 256. Interestingly, the codifying gene for this β-lactamase did not belong to the TEM family. Moreover, no PCR products were obtained when specific primers for the CARB-encoding genes were employed (L. Mange, H. Saka, A. Garutti, M. Galas, R. Melano, M. Rapoport, M. Caffer, and A. Rossi, 9th Int. Congr. Infect. Dis., abstr. 95.048, 2000).
In this work we analyzed an isolate belonging to that major group and we determined the nucleotide sequence of a new β-lactamase, named CARB-7, which in addition seems to be located on the VCR island.
MATERIALS AND METHODS
Bacterial strains and plasmids.
V. cholerae non-O1, non-O139 (ME11762) was isolated from water samples from the Capilla waterway, Mendoza, Argentina. Pesc37 is an AMP-susceptible V. cholerae non-O1, non-O139 strain included in the initial sample of 669 isolates. Nalidixic acid-resistant Escherichia coli ER1793 was used as the recipient in conjugation experiments. E. coli DH5α was used as the host for recombinant plasmids in cloning assays.
Plasmids pACYC184, encoding tetracycline and CMP resistance, and pBGS19, encoding kanamycin resistance (23), were used as cloning vectors.
Antimicrobial agents and susceptibility tests.
The following antimicrobial agents were used and were obtained as standard laboratory powders from the sources indicated: AMP, Bagó; cefoxitin, Merck Sharp & Dohme; cephalothin, John Wyeth; piperacillin, Richmond; sulbactam, Pfizer; ticarcillin, Schering Plough; clavulanic acid, SmithKline Beecham; nalidixic acid, Winthrop.
MICs were determined by the agar dilution technique under V. cholerae NCCLS specifications (17), adopting the criteria for Enterobacteriaceae for β-lactam agents other than AMP.
Conjugative assays.
Cells of both the donor and recipient strains were mixed on Luria-Bertani agar, and the mixture was incubated for 18 h at 25°C (3) or 35°C, as described previously (20). Mueller-Hinton agar supplemented with nalidixic acid (50 μg/ml) and AMP (8 μg/ml) was used to select the transconjugant cells.
DNA manipulations and PCR assays.
Genomic DNA and plasmids were purified by employing the Wizard Genomic DNA Purification Kit and the Wizard Minipreps DNA Purification System (Promega, Madison, Wis.), respectively. The restriction enzymes as well as the DNA ligase were purchased from Gibco BRL (Gaithersburg, Md.). The genomic DNA was partially digested with Sau3AI and ligated with the pACYC184 vector linearized with BamHI. The ligation mixture was electroporated into E. coli DH5α competent cells using the Gene Pulser II Electroporation System (Bio-Rad) according to the manufacturer's recommendations. Selection of recombinant strains was achieved in Mueller-Hinton agar supplemented with AMP (16 μg/ml) and CMP (50 μg/ml).
Amplifications were performed with a Biometra thermal cycler in a final volume of 50 μl containing 20 pmol of each primer (Gibco BRL), a 25 μM concentration of each deoxynucleoside triphosphate, 1.5 mM MgCl2, 2.5 U of Taq polymerase (Promega), and 1 μl of DNA of ME11762 as the template. After a denaturation step of 10 min at 94°C, the amplifications were performed over 25 cycles, each one of which consisted of 30 s of denaturation at 94°C, 45 s of annealing at 60°C, and 30 s of extension at 72°C, with a final extension step of 5 min at 72°C. Seminested PCR was performed under the same conditions, using forward PD (5′-CTCTGATGCATGCCTTGCTACT-3′) and reverse P3+ (5′-GCCTTATCAACGCGACTGTGAT-3′) primers in the first round of amplification and forward PD and reverse P2+ (5′-GGCGTTGTCGTATCCCTCAAAT-3′) primers in the second round. One microliter of amplimers obtained in the first round was employed as the template for the second round.
DNA sequencing and sequence analysis.
DNA sequencing was performed on both strands by the method of Sanger et al. (22), using the BigDye terminator methodology (Applied Biosystem/Perkin-Elmer, Foster City, Calif.) with universal (M13/pUC forward and reverse 23-base sequencing primers) and sequence-based primers which, besides P2+, P3+, and PD, consisted of the following: P1, 5′-GGGCAACCCGTATTGAGATCACA-3′; P2−, 5′-GCCTGCATTGGTCAGTTCATAGGT-3′; P3−, 5′-CCGAGGCTTCAATGGCAGAA-3′; and P4, 5′-TGCGCGAGGTTAGTGTTGTTG-3′. The sequences were analyzed in an ABI Prism 377 DNA sequencer (Applied Biosystem/Perkin-Elmer). Multiple nucleotide and protein sequence alignments were performed with the CLUSTAL facilities of the PCGENE software package (Intelligenetics, Inc.).
Nucleotide sequence accession number.
The CARB-7 β-lactamase sequence has been submitted to GenBank. Its accession number is AF409092
RESULTS
Antibiotic susceptibility.
Susceptibility testing of the ME11762 and Pesc37 isolates, respectively, determined the MICs (in micrograms per milliliter) of the following antibiotics: AMP, 256 and 0.5; AMP-sulbactam, 16 and 1; ticarcillin, 512 and 0.25; ticarcillin-clavulanic acid, 8 and 0.25; cephalothin, 2 and 0.25; piperacillin, 32 and 0.25; and cefoxitin, 8 and 1.
Conjugational transfer of resistance.
To analyze the mobility of the AMP resistance determinant, the ME11762 isolate was mated with nalidixic acid-resistant E. coli ER1793. After several attempts, including assays performed at 25 or 37°C, no transconjugants were obtained.
Cloning and sequencing of blaCARB-7 gene.
A Sau3AI-based library of the ME11762 isolate was prepared as indicated in Materials and Methods. E. coli DH5α electrotransformants harboring recombinant plasmids were screened on selective agar containing AMP plus CMP. Plasmids from several AMP- and CMP-resistant colonies were analyzed, and the one with the smallest insert (approximately 2.2 kb), named pGMPS1, was subjected to restriction analysis (Fig. 1A).
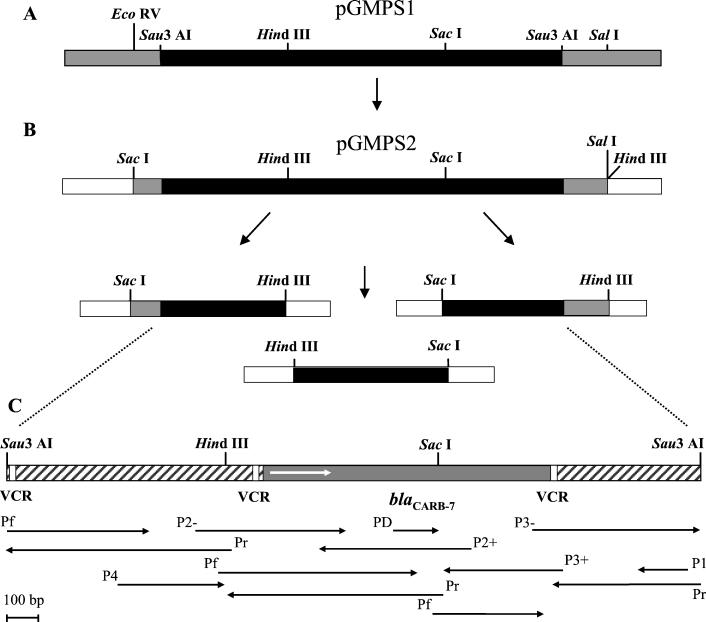
FIG. 1.
Strategy used for sequencing the blaCARB-7 gene and the DNA flanking regions. (A) Restriction map of the Sau3AI fragment from the ME11762 strain (black box). Only the ending Sau3AI sites are indicated. Grey boxes indicate DNA of pACYC184 vector. (B) The insert of pGMPS1 was liberated by digestion with SalI and EcoRV and was subcloned into SmaI-SalI sites of pBGS19 vector (white boxes), rendering pGMPS2; grey boxes represent approximately 500 bp that belong to pACYC184 vector. Inserts of plasmids derived of pGMPS2 were sequenced. (C) blaCARB-7 gene and its flanking DNA regions. The sense, length, and primers used for each sequence reaction are indicated by the black arrows. The grey box represents the blaCARB-7 gene, the white arrow represent its translational orientation, the flanking fragments are depicted as hatched boxes, and VCR are depicted as open boxes. Restriction sites used for subcloning into pBGS19 vector are indicated. Pf and Pr, universal primers (forward and reverse, respectively).
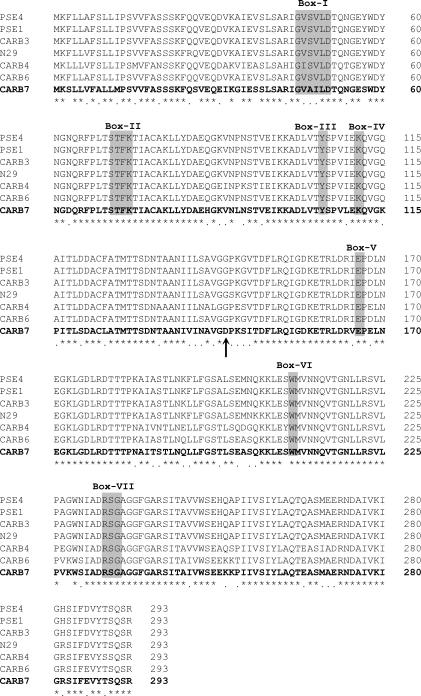
To sequence the gene encoding AMP resistance, a SalI-EcoRV fragment from pGMPS1 (about 2.7 kb, including 0.5 kb of the pACyC184 vector) was directionally subcloned into the SalI-SmaI sites of pBGS19, rendering plasmid pGMPS2 (Fig. 1B). From this recombinant plasmid, three SacI-HindIII fragments were subsequently obtained and sequenced (Fig. 1B). To overlap the inner ends of these fragments, a PCR product obtained with primers P2+ and P2− was also sequenced. In addition, the sequencing of this fragment directly amplified from the chromosome allowed us to discard putative rearrangements of the Sau3AI fragments produced in the construction of the DNA library. The cloned DNA region of ME11762 strain resulted in a sequence of 2,214 nucleotides containing a unique ORF of 867 nucleotides (Fig. 1C and 2). Neither putative ribosome-binding site nor promoter regions were found. The comparison of this ORF with GenBank sequences showed a high nucleotide identity (88.6%) with the CARB-6 gene and an overall 82.5% identity with other genes belonging to the CARB family. This ORF encodes a deduced protein of 288 amino acids which showed 88.5% identity with CARB-6 (differing in 31 amino acid positions) and an overall identity of 83.4% with other enzymes of the CARB family (Fig. 3 and Table 1). The new enzyme, named CARB-7, contains the common motifs found in serine β-lactamases: the active site of the enzyme contained a Ser-Thr-Phe-Lys tetrad at positions 70 to 73 according to the standard numbering scheme of Ambler et al. (1), all seven conserved amino acid boxes (12), and specific conserved residues (Fig. 3). Alignment of amino acid sequences of 28 class A enzymes belonging to different β-lactamase-groups (TEM, SHV, CARB, PER, and CTX-M) showed a conserved Gly residue at position 144 (data not shown). Interestingly, an Asp residue at this position was found in the deduced amino acid sequence of CARB-7. Therefore, to confirm this change we designed a specific sequence-based primer, PD, and carried out a seminested PCR assay using PD and P2+ primers in the second round of amplification. The sequence of both strands (191 bp) of the PD-P2+ PCR fragment confirmed the presence of a GAT codon coding for an Asp residue at position 144 of the CARB-7 enzyme.
FIG. 2.
Nucleotide sequence of the 2,214-bp fragment of pGMPS2 containing the CARB-7 β-lactamase coding region. The deduced amino acid sequence of the ORF proposed to encode CARB-7 β-lactamase is represented in one-letter code. VCR on the flanking sequences of blaCARB-7 gene are underlined. The fragment showing high homology with the V. cholerae chromosome is indicated by the grey box (GenBank accession number AE004376 [nucleotides 7944 to 8706]). Restriction sites used for subcloning of blaCARB-7 gene are double underlined.
FIG. 3.
Alignment of the deduced amino acid sequence of CARB-7 with those of the closely related class A β-lactamases PSE-4 (CARB-1), PSE-1 (CARB-2), CARB-3, CARB-4, P. mirabilis N29, and CARB-6. The shadowed boxes (I to VII) correspond to amino acid boxes conserved in all penicillin-recognizing enzymes (12). The arrow indicates amino acid position 139, with the change G→D. Asterisks indicate identical residues. Dots indicate conserved amino acids. Sequences are numbered as described by Ambler et al. (1).
TABLE 1.
Amino acid identity of CARB-7 with several related class A β-lactamases
| β-Lactamase | % Identity with:
|
|||||
|---|---|---|---|---|---|---|
| PSE-4a | PSE-1b | CARB-3 | N29 | CARB-6 | CARB-4 | |
| PSE-4 | ||||||
| PSE-1 | 99.7 | |||||
| CARB-3 | 99.3 | 99.7 | ||||
| N29 | 97.9 | 98.2 | 97.9 | |||
| CARB-6 | 93.8 | 94.1 | 94.4 | 93.1 | ||
| CARB-4 | 85.1 | 85.1 | 85.4 | 84.7 | 85.1 | |
| CARB-7 | 83.0 | 83.3 | 83.7 | 83.0 | 88.5 | 78.8 |
Also designated CARB-1.
Also designated CARB-2.
Analysis of flanking regions of the blaCARB-7 gene.
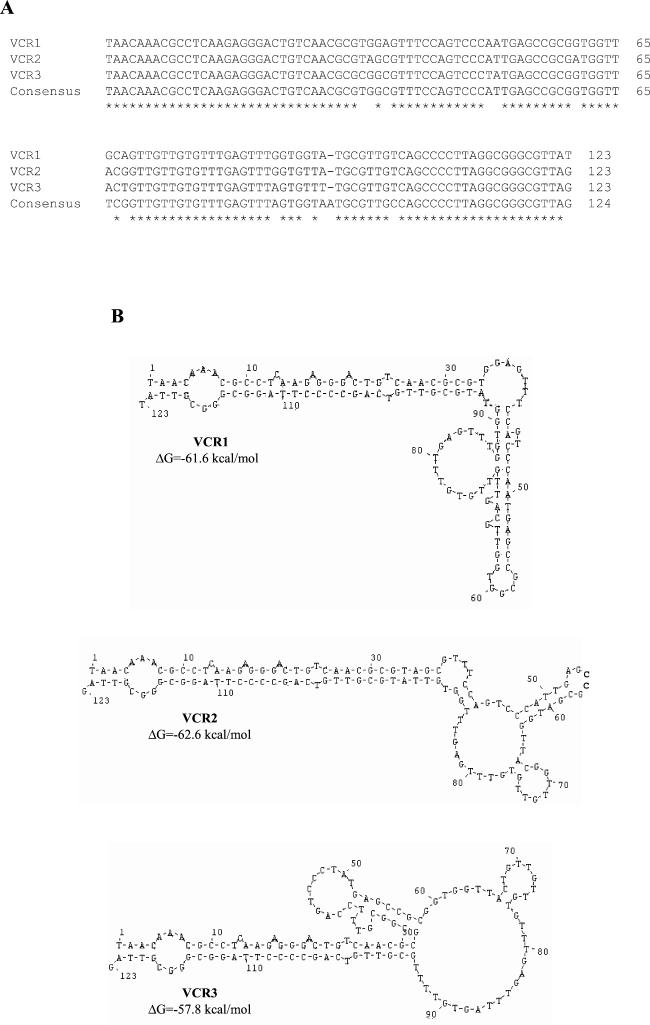
The analysis of the nucleotide sequence of the flanking regions of the blaCARB-7 gene (887 nucleotides upstream and 460 nucleotides downstream) showed the occurrence of three 123-bp repeat sequences, two of them located on the upstream fragment and the other on the downstream fragment (Fig. 1C and 2). The three repeated sequences share an overall 97% identity with the reported consensus sequence of the VCR from V. cholerae (Fig. 4). As expected from this high degree of identity, the predicted secondary structure for these three repeated sequences showed stem-loop conformations having free-energy stability values in the range of −62.6 to −57.8 kcal/mol, which clearly resembles that from the previously published VCR consensus sequence (−58.6 kcal/mol) (Fig. 4) (4). The three VCR are in the same orientation relative to one another, and all of them are found outside of the predicted ORF. Both flanking regions of the blaCARB-7 gene were compared with GenBank sequences. As is indicated in Fig. 2, 762 nucleotides belonging to the upstream fragment share 93% identity with locus VCA0424 (The Institute for Genomic Research), which is included within section 33 of V. cholerae chromosome 2 (GenBank AE004376, nucleotides 7944 to 8706). The downstream fragment mainly showed high homology with other VCR sequences.
FIG. 4.
Nucleotide sequence of the 123-bp element. (A) Multiple alignment of nucleotide sequence of the three VCR found on the flanking regions of the blaCARB-7 gene with a proposed VCR consensus sequence (4). Identical bases are indicated by an asterisk. The alignment was generated with the multiple-alignment program CLUSTAL. (B) Proposed secondary structure for each VCR sequence, determined with DNASIS software (version 2.5).
DISCUSSION
Both V. cholerae O1 and non-O1 are naturally susceptible in vitro to broad-spectrum penicillins and cephalosporins. However, strains producing β-lactamases such as SAR-1 (pI 4.9), TEM-1 (pI 5.4), or CARB-6 (pI 5.35), which conferred resistance to amino- and carboxy-penicillins, were previously described (5, 8, 19). We have reported here the sequence of a new β-lactamase gene isolated from a V. cholerae non-O1, non-O139 strain, which shares an overall 85.5% identity with the genes coding for several CARB enzymes. Moreover, from all the sequences currently comprised in the GenBank, only the CARB genes showed a relevant homology with the encoding gene for the new β-lactamase. In addition, the susceptibility profile conferred by the new β-lactamase suggests a strong activity against amino- and carboxy-penicillins, which resembled the phenotype conferred by PSE-1 (CARB-2) or PSE-4 (CARB-1) β-lactamases in Pseudomonas aeruginosa (14). Therefore, we propose that the new enzyme is a member of the CARB group and name it CARB-7.
Despite the fact that both the nucleotide and deduced amino acid sequences for CARB-7 showed high similarity with the those of the CARB enzymes, two relevant differences must be pointed out. First, as could be expected from having the lowest overall similarity percentage observed with other CARB genes, there is a poor homology between the blaCARB-7 gene and the reverse specific primer (OC-2) reported by Arlet and Philippon (2) for the identification of members of the blaCARB gene family. This fact can explain the lack of amplification in the PCR assays carried out in our preliminary work for the molecular identification of β-lactamases (Mange et al., 9th ICID). It points out the need to design new primers that can detect this new member of the CARB family. Second, the CARB group includes enzymes belonging to the molecular class A β-lactamases, for which several conserved residues or motifs have been identified. Comparing these conserved motifs on the deduced sequence for CARB-7 with those of β-lactamases from the major families classified into class A, such as TEM, SHV, CARB, CTX-M, and PER enzymes, we observed a change, Gly to Asp, at position 144 (Ambler classification scheme) of the CARB-7 enzyme. This is, to our knowledge, the first report of this amino acidic variant. It would be located far from the active site, as can be inferred from the crystallographic structure reported for the TEM-1 β-lactamase (25). The strong activity against amino- and carboxy-penicillins displayed by CARB-7 appears to support the assumption that Asp144 is not involved in the enzymatic activity. Strikingly, the two Gly residues at positions 143 and 144 are highly conserved among the Ambler class A enzymes. Therefore, this nonconservative change of a nonpolar residue to an acidic one at position 144 of CARB-7 could imply a conformational change in the secondary structure of the enzyme, which could additionally have evolutionary significance.
Although initially the CARB enzymes were assumed to be confined to P. aeruginosa strains (10), some of these β-lactamases were isolated from enterobacteria (16), Alcaligenes xylosoxidans (CARB-2) (13), and from an isolate of V. cholerae non-O1, non-O139 (CARB-6) (5). In this study, CARB-7 was isolated from a V. cholerae non-O1, non-O139 strain from a sample of 77 AMP-resistant isolates that were producers of a β-lactamase with a pI of 5.4. The blaCARB-7 gene was not transferred to the E. coli ER1793 strain, regardless of the fact that several attempts employing different mating temperatures were made. This fact was previously reported for others CARB enzymes (5) and could be interpreted as either a location of the CARB gene on a nonconjugative plasmid or on the chromosome. This last possibility is strongly supported by two facts. First, three 123-bp repeats were detected on the flanking sequence of the blaCARB-7 gene showing a 97% overall similarity with the consensus sequence for VCR (4), which are restricted to a region of about 10% of V. cholerae's chromosome 2 (11). Second, the upstream fragment of the blaCARB-7 gene shared 93% identity with locus VCA0424 (The Institute for Genomic Research), situated inside section 33 of V. cholerae's chromosome 2. The assumption of a chromosomal location for the blaCARB-7 gene supports the notion that the VCR island functions as a “mega-integron.” To date, only in vitro works have demonstrated that intl4 V. cholerae integrase is active for site-specific cassette recombination (21), without in vivo proof of gene capture. Therefore, this would be the first evidence of in vivo capture of a gene by this system.
All the VCR-associated genes described to date are transcribed from promoter sequences located immediately upstream of the ORFs (C. Clark, L. Purins, P. Kaewrakon, and P. Manning, Letter, Mol. Microbiol. 26:1137-1138, 1997). However, putative ribosome-binding site or promoter regions (regarding E. coli consensus sequences) were not found in the upstream region of the blaCARB-7 gene. These facts suggest that the transcription may occur through a promoter located more distantly of the blaCARB-7 gene.
Most of the 216 ORFs situated on the VCR island have no homology to other sequences, although three genes were identified that may be involved in resistance to drugs other than β-lactam antibiotics (11). Therefore, this is the first report of a β-lactamase within the VCR region.
Acknowledgments
We thank Mariana Miranda for her assistance in technical work. We thank N. Binsztein for providing the V. cholerae non-O1, non-O139 collection and for reading the manuscript and thank M. I. Caffer for serotyping of the strains.
Footnotes
Dedicated to the memory of our dear Ali.
REFERENCES
- 1.Ambler, R. P., A. F. W. Coulson, J. M. Frere, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tirabi, and S. G. Waley. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276:269-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arlet, G., and A. Philippon. 1989. Construction by polymerase chain reaction and intragenic DNA probes for three main types of transferable β-lactamases (TEM, SHV, CARB). FEMS Microbiol. Lett. 82:19-26. [DOI] [PubMed] [Google Scholar]
- 3.Barja, J. L., Y. Santos, I. Huq, R. Colwell, and A. E. Toranzo. 1990. Plasmids and factors associated with virulence in environmental isolates of Vibrio cholerae non-O1 in Bangladesh. J. Med. Microbiol. 33:107-114. [DOI] [PubMed] [Google Scholar]
- 4.Barker, A., C. Clark, and P. Manning. 1994. Identification of VCR, a repeated sequence associated with a locus encoding a hemagglutinin in Vibrio cholerae O1. J. Bacteriol. 176:5450-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choury, D., G. Aubert, M. Szajnert, K. Azibi, M. Delpech, and G. Paul. 1999. Characterization and nucleotide sequence of CARB-6, a new carbenicillin-hydrolyzing β-lactamase from Vibrio cholerae. Antimicrob. Agents Chemother. 43:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choury, D., M.-F. Szajnert, M.-L. Joly-Gillou, K. Azibi, M. Delpech, and G. Paul. 2000. Nucleotide sequence of the blaRTG-2 (CARB-5) gene and phylogeny of a new group of carbenicillinases. Antimicrob. Agents Chemother. 44:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalsgaard, A., A. Forslund, O. Serichantalergs, and D. Sandvang. 2000. Distribution and content of class 1 integrons in different Vibrio cholerae O-serotype strains isolated in Thailand. Antimicrob. Agents Chemother. 44:1315-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupont, M., M. Jouvenot, G. Couetdic, and Y. Michel-Briand. 1985. Development of plasmid-mediated resistance in Vibrio cholerae during treatment with trimethoprim-sulfamethoxazole. Antimicrob. Agents Chemoter. 27:280-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falbo, V., A. Carattoli, F. Tosini, C. Pezzella, A. Dionisi, and I. Luzzi. 1999. Antibiotic resistance conferred by a conjugative plasmid and a class I integron in Vibrio cholerae O1 El Tor strains isolated in Albania and Italy. Antimicrob. Agents Chemother. 43:693-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hedges, R., and M. Matthew. 1979. Acquisition by Escherichia coli of plasmid-borne β-lactamases normally confined to Pseudomonas spp. Plasmid 2:269-278. [DOI] [PubMed] [Google Scholar]
- 11.Heidelberg, J., J. Eisen, W. Nelson, R. Clayton, M. Gwinn, R. Dodson, D. Haft, E. Hickey, J. Peterson, L. Umayam, S. Gill, K. Nelson, T. Read, H. Tettelin, D. Richardson, M. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. Fleishmann, W. Nierman, O. White, S. Salzberg, H. Smith, R. Colwell, J. Mekalanos, J. Venter, and C. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joris, B., J. Ghuysen, G. Dive, A. Renard, O. Dideberg, P. Charlier, J. Frère, J. Kelly, J. Boyington, P. Moews, and J. Knox. 1988. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem. J. 250:313-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Labia, R., M. Guionie, M. Barthélémy, and A. Philippon. 1981. Properties of three carbenicillin-hydrolyzing β-lactamases (CARB) from Pseudomonas aeruginosa: identification of a new enzyme. J. Antimicrob. Chemother. 7:49-56. [DOI] [PubMed] [Google Scholar]
- 14.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazel, D., B. Dychinco, V. Webb, and J. Davies. 1998. A distinctive class of integron in the Vibrio cholerae genome. Science 280:605-608. [DOI] [PubMed] [Google Scholar]
- 16.Medeiros, A., R. Hedges, and G. Jacoby. 1982. Spread of a “Pseudomonas-specific”β-lactamase to plasmids of enterobacteria. J. Bacteriol. 149:700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed., vol. 20, no. 2. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 18.Recchia, G., and R. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 19.Reid, A., and S. Amyes. 1986. Plasmid penicillin resistance in Vibrio cholerae: identification of new β-lactamase SAR-1. Antimicrob. Agents Chemother. 30:245-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice, L. B., and R. A. Bonomo. 1996. Genetic and biochemical mechanisms of bacterial resistance to antimicrobial agents, p. 453-501. In M. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. Williams & Wilkins, Baltimore, Md.
- 21.Rowe-Magnus, D., A. Guerout, P. Ploncard, B. Dychinco, J. Davies, and D. Mazel. 2001. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc. Natl. Acad. Sci. 98:652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanger, S., S. Micklen, and A. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spratt, B. G., P. J. Hedge, S. te Heesch, A. Edelman, and J. K. Broome-Smith. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 41:337-341. [DOI] [PubMed] [Google Scholar]
- 24.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669-1683. [DOI] [PubMed] [Google Scholar]
- 25.Strynadka, N. C. J., H. Adachi, S. E. Jensen, K. Johns, A. Sielecki, C. Betzel, K. Sutoh, and M. N. G. James. 1992. Molecular structure of the acyl-enzyme intermediate in β-lactam hydrolysis at 1.7 Å resolution. Nature (London) 359:700-705. [DOI] [PubMed] [Google Scholar]