Abstract
A screening assay to test the inducing capacity of macrolides by fusing the attenuator of the inducible erm(B) gene from Streptococcus pneumoniae HM28 with the gfpmut1 gene has been designed. Fluorescence was detected under UV light around disks impregnated with inducer macrolides (erythromycin or azithromycin) but not with noninducer ketolides. Induction could be quantified by fluorometry.
Resistance to macrolide-lincosamide-streptogramin B antibiotics defining the so-called MLS phenotype is common in streptococci and enterococci (11). It is mediated by dimethylation of adenine 2058 in the ribosomal 23S rRNA target, which reduces the affinity between the antibiotic and the ribosome (17). The genes that encode 23S rRNA methylases are designated erm (erythromycin resistance methylase). In streptococci and enterococci, MLS resistance is generally encoded by genes belonging to the erm(B) group (13). Expression of MLS resistance may be inducible or constitutive, depending upon a regulatory region preceding the gene (10, 18). In contrast to the pattern of inducer macrolides for the staphylococcal gene erm(C), which is limited to 14- and 15-member ring macrolides, erm(B) is inducible by most members of the MLS group (10). Recently, a new class of macrolides, the ketolides, which are derivatives of clarithromycin or erythromycin A characterized by a 3-keto function instead of the cladinose moiety and an 11- or 12-carbamate extension, has been shown to be active against most streptococci resistant to erythromycin (1, 3, 7, 8; C. Agouridas, Y. Benedetti, A. Denis, O. Le Martret, and J. F. Chantot, Abstr. 35th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F157, 1995). This activity was attributed, in part, to the lack of induction of MLS resistance by these antimicrobials (2, 3, 5, 19). We have fused the inducible attenuator of erm(B) with the green fluorescent protein reporter gene to provide a new fluorescence assay to easily detect the inducing capacity of macrolides and related antimicrobials.
Gene fusions.
We have amplified by PCR a 741-bp fragment from plasmid pAT505 composed of the entire gfpmut1 gene except the initiation codon (9) using oligonucleotides modified by insertion of SmaI and PstI restriction sites (underlined): GFP1 (5′-GGA GAT ATC CCC GGG GGT AAA GGA GAA G-3′) and GFP2 (5′-GCA TGC CTG CAGTTA TTT GTA CAA TT-3′). The PCR product was digested with SmaI and PstI, cloned in pUC18, and introduced into Escherichia coli DH10B cells by electrotransformation. A 384-bp fragment comprising the attenuator and the first 54 nucleotides of the erm(B) gene of Streptococcus pneumoniae HM28 inducibly resistant to MLS antibiotics (14) was amplified by PCR using oligonucleotides modified to include restriction sites EcoRI and SmaI (underlined): ermB1 (5′-CTT AGA AGA ATT C TT AAG AGT GTG-3′) and ermB2 (5′-TTA TTA TTT GCC CGG GTA CCT TTT C-3′). The amplification product was digested with the appropriate enzymes and cloned upstream from the truncated gfpmut1 gene in pUC18. The fusion was electrotransformed into E. coli DH10B and subcloned in the EcoRI and SphI restriction sites of the shuttle multicopy vector pAT28 (which confers spectinomycin resistance) (16) to generate plasmid pUV4. The hybrid plasmids were introduced by electrotransformation into Staphylococcus aureus RN4220. The transformants were plated onto media containing 180 μg of spectinomycin/ml and an inducing concentration of erythromycin (0.03 μg/ml). The fluorescent colonies were identified by UV (at 385 nm).
A constitutively expressed gfpmut1 gene was also constructed by fusion of the gfpmut1 gene with a 179-bp sequence upstream from the constitutive erm(B) gene of Enterococcus faecalis BM4110/pAMβ1 amplified by PCR with the ermB1 and ermB2 primers. The amplified fragment lacked the leader peptide sequence (12). The recombinant plasmid, pUV5, was electrotransformed into S. aureus RN4220 and used as a positive control.
MICs and induction experiments.
MICs of antibiotics were determined by agar dilution using Mueller-Hinton medium (Bio-Rad, Marnes-la-Coquette, France) supplemented with 5% horse blood (4). The following antibiotics were provided by their manufacturers: telithromycin, RU 69874, and the 2-fluoroketolides HMR 3562 and HMR 3787 (Aventis, Romainville, France) (6). RU 69874 is structurally similar to telithromycin except for a replacement of the 3-keto function by a l-cladinose sugar. Erythromycin, spectinomycin, and amoxicillin were from Sigma-Aldrich (St. Quentin Fallavier, France), and azithromycin was from Pfizer (Orsay, France). Induction of resistance by antibiotics was further assessed by growth curves of uninduced or induced cells (at a concentration equal to 1/10 the MIC of the macrolides) in the presence or absence of challenging (just subinhibitory) concentrations of antibiotic as described earlier (15). Bacterial growth was followed by CO2 production measured with the BacT/Alert 3D system (Organon Teknika Corp., Oklahoma City, Okla.). Growth curves were plotted and lag phases were calculated. All experiments were conducted twice.
Fluorescence induction assays.
Fluorescence induction was detected qualitatively by the disk diffusion method (4). Disks impregnated with 15 μg of the various macrolides and ketolides were placed on inoculated blood agar plates which were incubated for 24 h at 37°C and observed under UV light. For quantitative assays, overnight S. aureus RN4220/pUV4 or S. aureus RN4220/pUV5 cultures grown in Trypticase soy broth were used to inoculate fresh medium at a dilution of 1: 25. After 3 h of incubation at 37°C under aeration, cells were added to Trypticase soy broth at 106 CFU/ml, containing increasing concentrations of antibiotics, and incubated for 1 h at 37°C. The cultures were then washed three times with phosphate-buffered saline, and cells were resuspended in phosphate-buffered saline at 106 CFU/ml. The fluorescence intensity was determined by spectrofluorometry performed with a Hitachi model F-1200 fluorescence spectrophotometer (Hitachi Co., Tokyo, Japan) at an excitation wavelength of 460 nm and an emission wavelength of 510 nm.
MLS resistance phenotypes.
The MICs of and induction of resistance by erythromycin, telithromycin, and RU 69874 were determined for S. pneumoniae HM28 and E. faecalis BM4110/pAMβ1 (Table 1). Preinduced cells of S. pneumoniae HM28 challenged with erythromycin had a lag phase that was shortened, confirming that MLS resistance was inducible by this antimicrobial. As expected, expression of erythromycin resistance in E. faecalis BM4110/pAMβ1 was constitutive. Telithromycin did not appear to be an inducer for MLS resistance in S. pneumoniae HM28; in contrast, RU 69874 did reduce the lag phase, confirming the role of the l-cladinose residue in induction. Telithromycin was active only against the inducible strain S. pneumoniae HM28.
TABLE 1.
Induction of MLS resistance assessed by growth curve measurements against strains used in fusion experimentsc
| Strain | Antibiotic | MIC (μg/ml) | Lag phase time (in h) for
|
|
|---|---|---|---|---|
| Uninduced culture | Induced culture (inducing concen [μg/ml]) | |||
| S. pneumoniae HM28a | Erythromycin | 256 | 22.3 | 11.5 (25) |
| Telithromycin | 0.03 | 13.2 | 13.7 (0.003) | |
| RU69874 | 2 | 43.9 | 20.8 (0.2) | |
| E. faecalis BM4110/pAMβ1b | Erythromycin | 8,000 | 9 | 8.2 (800) |
| Telithromycin | 32 | 22.5 | 22.1 (3.2) | |
| RU69874 | 16 | 11.8 | 11.7 (1.6) | |
Challenge concentrations were 50, 0.006, and 0.4 μg/ml for erythromycin, telithromycin, and RU69874, respectively.
Challenge concentrations were 1,600, 6.4, and 3.2 μg/ml for erythromycin, telithromycin, and RU69874, respectively.
Results are means of two experiments.
Induction of fluorescence by macrolides.
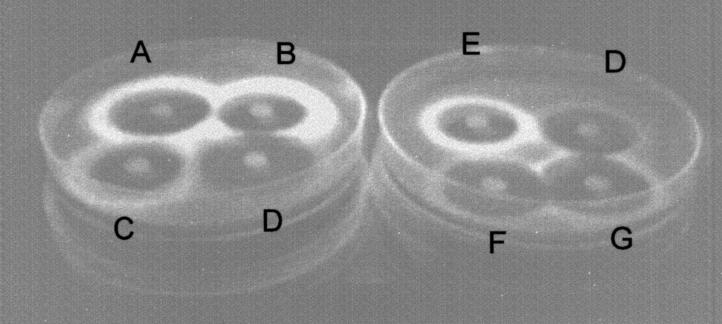
The ability of erythromycin and ketolides to induce fluorescence was studied with fusion constructs. After exposure to UV light, agar plates spread with S. aureus RN4220/pUV4 exhibited fluorescence localized at the border of inhibition zones for disks containing erythromycin and spiramycin. Fluorescence was barely visible with telithromycin and HMR 3787; however, HMR 3562 induced a weak fluorescence (Fig. 1). Fluorescence was strongly expressed in the presence of RU 69874. The fluorescence of S. aureus RN4220/pUV5 was expressed in the absence of antibiotic and was not enhanced in the presence of erythromycin or spiramycin (data not shown).
FIG. 1.
Fluorescence induced by various antibiotics in S. aureus RN4220/pUV4. A bacterial suspension was spread onto blood agar plates, and disks containing macrolides were placed on the surface of the plates. After an incubation for 24 h at 37°C, plates were examined under UV light. A, erythromycin; B, azithromycin; C, spiramycin; D, telithromycin; E, RU 69874; G, HMR 3562; and F, HMR 3787.
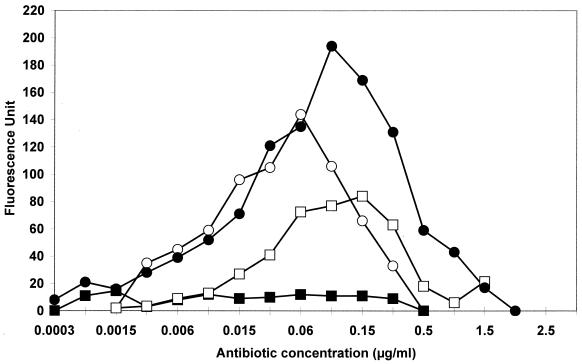
Fluorescence expressed in the presence of erythromycin, azithromycin, telithromycin, RU 69874, and two fluoroketolides (HMR 3562 and HMR 3787) was quantified by spectrofluorometry (Fig. 2). For noninduced S. aureus RN4220/pUV4, no basal fluorescence could be detected, while cells of S. aureus RN4220/pUV5 expressed a fluorescence equal to approximately 90 U, irrespective of the presence or absence of a macrolide (data not shown). Azithromycin and erythromycin were strong inducers with fluorescence peaks reaching 194 U at 0.09 μg/ml and 144 U at 0.06 μg/ml, respectively. Fluorescence was detected at 0.02 to 0.1 times the MIC of the macrolides for S. aureus RN4220/pUV4, reaching a peak at nearly one-quarter to one-half the MIC and then decreasing rapidly at higher concentrations. By contrast, telithromycin (Fig. 2) and the 2-fluoroketolides HMR 3562 and HMR 3787 (data not shown) induced a very weak fluorescence. The fluorescence maxima were 14.6, 18.6, and 21.4 U for telithromycin, HMR 3562, and HMR 3787, respectively. As expected, RU 69874 induced marked fluorescence.
FIG. 2.
Fluorometric detection of fluorescence induced by azithromycin (closed circles), erythromycin (open circles), RU 69874 (open squares), and telithromycin (closed squares) in S. aureus RN4220/pUV4. Fluorescence was measured in arbitrary fluorescence units.
Therefore, the pattern of induction by the antibiotics was similar, as determined by fluorescence or growth curve experiments. However, the fluorescence assay was convenient and easy to use, since it did not require substrates or cofactors. Additionally, the reporter system also delineates viable cells (A. Lefort, M. Arthur, C. Vignes-Colombeix, C. Vissuzaine, D. Henin, C. Carbon, and P. Courvalin, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 668, 2000). This assay can be used to study qualitatively or quantitatively the relationship between structure and activity of the macrolides, ketolides, or any inducer of methylase production.
Acknowledgments
We thank Patrice Courvalin and Stanley Falkow for the gift of plasmid pAT505 containing gfpmut1.
This work was supported by grants from Aventis and the Fondation de la Recherche Médicale.
REFERENCES
- 1.Agouridas, C., A. Bonnefoy, and J. F. Chantot. 1997. Antibacterial activity of RU64004 (HMR 3004), a novel ketolide derivative active against respiratory pathogens. Antimicrob. Agents Chemother. 41:2149-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnefoy, A., A. M Girard, C. Agouridas, and J. F. Chantot. 1997. Ketolides lack inducibility properties of MLS(B) resistance phenotype. J. Antimicrob. Chemother. 40:85-90. [DOI] [PubMed] [Google Scholar]
- 3.Bryskier, A. 2000. Ketolides—telithromycin, an example of a new class of antibacterial agents. Clin. Microbiol. Infect. 6:661-669. [DOI] [PubMed] [Google Scholar]
- 4.Comité de l'Antibiogramme de la Société Française de Microbiologie. 1996. Technical recommendations for in vitro susceptibility testing. Clin. Microbiol. Infect. 2(Suppl. 1):11-25. [Google Scholar]
- 5.Denis, A., C. Agouridas, J. M. Auger, Y. Benedetti, A. Bonnefoy, F. Bretin, J. F. Chantot, A. Dussarat, C. Fromentin, S. G. D'Ambrieres, S. Lachaud, P. Laurin, O. Le Martret, V. Loyau, N. Tessot, J. M. Pejac, and S. Perron. 1999. Synthesis and antibacterial activity of HMR 3647 a new ketolide highly potent against erythromycin-resistant and susceptible pathogens. Bioorg. Med. Chem. Lett. 9:3075-3080. [DOI] [PubMed] [Google Scholar]
- 6.Denis, A., F. Bretin, C. Fromentin, A. Bonnet, G. Piltan, A. Bonnefoy, and C. Agouridas. 2000. Synthesis and antibacterial activity of 2-halogeno, 2-methyl and 2,3 enol-ether ketolides. Bioorg. Med. Chem. Lett. 10:2019-2022. [DOI] [PubMed] [Google Scholar]
- 7.Douthwaite, S., L. H. Hansen, and P. Mauvais. 2000. Macrolide-ketolide inhibition of MLS-resistant ribosomes is improved by alternative drug interaction with domain II of 23S rRNA. Mol. Microbiol. 36:183-193. [DOI] [PubMed] [Google Scholar]
- 8.Goldman, R. C., and S. K. Kadam. 1989. Binding of novel macrolide structures to macrolides-lincosamides-streptogramins B-resistant ribosomes inhibits protein synthesis and bacterial growth. Antimicrob. Agents Chemother. 33:1058-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grillot-Courvalin, C., S. Goussard, F. Huetz, D. M. Ojcius, and P. Courvalin. 1998. Functional gene transfer from intracellular bacteria to mammalian cells. Nat. Biotechnol. 16:862-866. [DOI] [PubMed] [Google Scholar]
- 10.Horinouchi, S., W. H. Byeon, and B. Weisblum. 1983. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J. Bacteriol. 154:1252-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leclercq, R., and P. Courvalin. 1991. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin, B., G. Alloing, V. Méjean, and J. P. Claverys. 1987. Constitutive expression of erythromycin resistance mediated by ermAM determinant of plasmid pAMβ1 results from deletion of 5′ leader peptide sequences. Plasmid 18:250-253. [DOI] [PubMed] [Google Scholar]
- 13.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppala. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosato, A., H. Vicarini, A. Bonnefoy, J. F. Chantot, and R. Leclercq. 1998. A new ketolide, HMR 3004, active against streptococci inducibly resistant to erythromycin. Antimicrob. Agents Chemother. 42:1392-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosato, A., H. Vicarini, and R. Leclercq. 1999. Inducible or constitutive expression of resistance in clinical isolates of streptococci and enterococci cross-resistant to erythromycin and lincomycin. J. Antimicrob. Chemother. 43:559-562. [DOI] [PubMed] [Google Scholar]
- 16.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in Gram-positive bacteria. Nucleic Acids Res. 18:4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weisblum, B. 1995. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 39:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong, P., Z. Cao, R. Hammond, Y. Chen, J. Beyer, V. D. Shortridge, L. Y. Phan, S. Pratt, J. Capobianco, K. A. Reich, R. K. Flamm, Y. S. Or, and L. Katz. 1999. Induction of ribosome methylation in MLS-resistant Streptococcus pneumoniae by macrolides and ketolides. Microb. Drug Resist. 5:183-188. [DOI] [PubMed] [Google Scholar]