Abstract
The human mucosal surface is colonized by the indigenous microflora, which normally maintains an ecological balance among different species. Certain environmental or biological factors, however, may trigger disruption of this balance, leading to microbial diseases. In this study, we used two oral bacterial species, Streptococcus mutans and Streptococcus sanguinis (formerly S. sanguis), as a model to probe the possible mechanisms of competition/coexistence between different species which occupy the same ecological niche. We show that the two species engage in a multitude of antagonistic interactions temporally and spatially; occupation of a niche by one species precludes colonization by the other, while simultaneous colonization by both species results in coexistence. Environmental conditions, such as cell density, nutritional availability, and pH, play important roles in determining the outcome of these interactions. Genetic and biochemical analyses reveal that these interspecies interactions are possibly mediated through a well-regulated production of chemicals, such as bacteriocins (produced by S. mutans) and hydrogen peroxide (produced by S. sanguinis). Consistent with the phenotypic characteristics, production of bacteriocins and H2O2 are regulated by environmental conditions, as well as by juxtaposition of the two species. These sophisticated interspecies interactions could play an essential part in balancing competition/coexistence within multispecies microbial communities.
The human mucosal surface is colonized by large numbers of bacterial species, the so-called indigenous flora (14, 18, 36). In a homeostatic state, this indigenous flora plays an important role in protecting the host from invasions by exogenous pathogens; however, when the homeostasis is disrupted, it can cause diseases, such as dental caries, periodontal disease (35), vaginitis (10), and inflammatory bowel disease (23). Understanding the molecular mechanisms through which interspecies interactions can lead to homeostasis would shed new light on the development of novel measures to curb these “polymicrobial” diseases. In this study, we used two members of the dental biofilm, Streptococcus mutans and Streptococcus sanguinis (formerly S. sanguis) (22), as a model to investigate the interspecies interactions leading to competition and coexistence.
The dental biofilm is a good model system for studying interspecies interactions owing to its vast biodiversity (>500 bacterial species) (17, 26, 30), high cell density (∼1011 cells/g [wet weight]) (9), and easy accessibility (29). In addition, the oral cavity is an environment with constant cycles of feast and famine and fluctuations of pH due to food intake from the host. The high density and diversity of oral biofilm community members coupled with a limited food supply should create an environment that is conducive to fierce competition for available resources.
S. mutans is considered a major pathogen causing human dental caries (also known as tooth decay) (19). S. mutans normally exists as a regular member of the mature dental biofilm community; however, under certain conditions, it can become dominant to cause dental caries (21). S. sanguinis is also a member of the oral biofilm community (28). Except for reported associations with bacterial endocarditis (37), S. sanguinis is considered a benign, or even a beneficial, bacterium with regard to dental caries (2, 5). The antagonism between S. mutans and S. sanguinis at the ecological level has been known for many years. Epidemiological studies showed that early colonization and high levels of S. sanguinis in an infant's oral cavity correlate with significantly delayed colonization by S. mutans (5). Similarly, high levels of S. mutans in the oral cavity correlate with low levels of S. sanguinis (20). Early studies with germ-free rats also demonstrated a so-called “competitive exclusion” between S. mutans and S. sanguinis depending on the sequence of inoculation (25). Despite these interesting early findings, no further studies were conducted to understand the molecular mechanisms underlying these interspecies interactions. In this study, we developed several new cellular assays for more defined analyses of the competition and coexistence between S. mutans and S. sanguinis. Our results, obtained by using a combination of physiological, genetic, and biochemical approaches, led us to propose a possible molecular mechanism underlying these fascinating interspecies interactions.
MATERIALS AND METHODS
Bacterial strains, media, and enzymes.
S. mutans UA140 (32) and derivative strains constructed in this study are listed in Table 1. S. sanguinis ATCC 10556 was used for competition analysis. Other streptococcal species used in the initial screening were S. gordonii, S. pyogenes, S. oralis ATCC 10557, S. mitis ATCC 33399, S. mitis ATCC 903, S. pneumoniae, S. cristatus ATCC 49999, S. parasanguinis ATCC 15911, S. sanguinis NY101, and S. sobrinus OMZ176. All species were routinely grown in brain heart infusion (BHI) medium (Difco, Sparks, MD) or on BHI plates under anaerobic conditions (90% N2, 5% CO2, 5% H2) at 37°C unless otherwise indicated. Peptidase, hydrogen peroxide (30% [wt/wt]), and horseradish peroxidase were from Sigma (St. Louis, MO).
TABLE 1.
Bacterial strains used in this study
| Strain | Characteristics | Reference |
|---|---|---|
| S. sanguinis ATCC 10556 | Oral commensal | |
| S. mutans UA 140 | Wild-type MutI+IV+ | 32 |
| UA140I−IV+ | ΔmutC MutI−IV+ | This study |
| UA140I+IV− | ΔnlmAB MutI+IV− | This study |
| UA140I−IV− | ΔmutC ΔnlmAB MutI−IV− | This study |
| UA140::Φ(mutAp-luc) | Φ(mutAp-luc) MutI+IV+ | This study |
| UA140::Φ(mutAp-mrfp) | Φ(mutAp-mrfp) MutI+IV+ | 15 |
| UA140::Φ(ldhp-luc) | Φ(ldhp-luc) MutI+IV+ | 24 |
| UA140::Φ(ldhp-gfp) | Φ(ldhp-gfp) MutI+IV+ | This study |
| UA140::Φ(nlmAp-luc) | Φ(nlmAp-luc) MutI+IV+ | This study |
Competition assays on plates and in biofilms.
For competition assays on plates between S. mutans and S. sanguinis, 10 μl of an overnight culture of either species adjusted to an optical density at 600 nm (OD600) of ∼0.5 in 50% BHI was inoculated on half-strength BHI plates as the early colonizer. After an overnight incubation, 10 μl of the competing species at the same OD600 was inoculated beside the early colonizer as the later colonizer, or both species were inoculated at the same time beside each other. The plates were further incubated at 37°C anaerobically overnight before cell growth was inspected. For competition assays in biofilms, overnight cultures of S. mutans or S. sanguinis were diluted 1:100 in 50% BHI plus 0.1% sucrose and inoculated into a slide chamber. The cultures were incubated at room temperature for 3 h to allow cell attachment before the competing species was inoculated, or both species were inoculated at the same time. The biofilm was grown for 16 h at 37°C as a static culture. CellTracker Orange (Molecular Probes, Eugene, OR) was used to label all cells for 2 h before confocal microscopy. Confocal microscopy was performed as described previously (15).
Luciferase and mutacin production assays.
Luciferase assays were performed as previously described (16). For planktonic culture, 100 μl of cell culture was used; for plate culture, cells were scraped from the plate and resuspended in 100 μl of BHI. The production of mutacin on the plate was measured by the antagonistic assay as described previously (32). Briefly, the plates were overlaid with the indicator strain S. sobrinus OMZ176 in a 1:5 dilution of an overnight culture in soft agar. After further incubation, the cleared zone was measured.
Assays for H2O2 production in liquid and on plate cultures.
The production of H2O2 by S. sanguinis in liquid culture was measured as described previously (27). To measure the effect of S. mutans on the H2O2 production of S. sanguinis, an overnight culture of S. sanguinis was diluted to ∼107 cells/ml (OD600, ∼0.025) and incubated anaerobically at 37°C. After two doubling times, the cells were washed twice with BHI and the OD600 was adjusted to 0.2. One milliliter of the cell suspension was transferred to a tube, and 1 ml of either BHI or S. mutans UA140::Φ(mutAp-luc) cell suspension (OD600 = ∼0.2) was added. The cells were further incubated either as a planktonic culture or as a cell pellet with medium (16,000 × g for 1.5 min) for 2 h before the H2O2 concentration was measured with the culture supernatant. For the determination of H2O2 production on the plate, 10 μl of peroxidase (64 μg) was added to a half-strength BHI plate containing 1 mg/ml leuco crystal violet. After the liquid was absorbed into the agar, 5 μl of S. sanguinis was inoculated at the same spot. After overnight incubation with a subsequent 2-h air exposure, the plate was inspected for the development of a purple color on and around the colony.
Construction of mutacin-defective strains.
S. mutans strain UA140 produces two major mutacins, the lantibiotic mutacin I and the nonlantibiotic mutacin IV (32). To study the role of each mutacin in interspecies competition, we constructed three derivative strains defective in either mutacin I (UA140I−IV+), mutacin IV (UA140I+IV−), or both (UA140I−IV−). To construct UA140I−IV+, the tetracycline (Tet) resistance gene tetM from Tn916 (7) was amplified by PCR and cloned into pCR2.1 cloning vector (Invitrogen). A DNA fragment encompassing 1 kb upstream and downstream of mutC (32) was amplified by PCR and cloned into pCR2.1 to form pCRBCD. To delete mutC, an inverse PCR was performed by using two primers, BR1 and DF1, both of which had a StuI restriction site incorporated at their 5′ ends. The tetM gene cassette was released from pCR2.1 by cutting with StuI restriction enzyme, whose recognition sequence was also incorporated into the primers for amplifying tetM, and inserted into pCRBCD at the same restriction site. The resulting plasmid was digested with PstI and SphI and transformed into UA140. The deletion construct was integrated into the chromosome by double-crossover homologous recombination. The transformants were selected on Tet plates (10 μg/ml). Ten independent transformants were randomly selected, tested, and confirmed for no production of mutacin I by the deferred-antagonism assay (8) using S. sobrinus OMZ176, which is sensitive only to mutacin I. The deletion construction in the mutacin I gene locus was further confirmed by PCR and genetic complementation. To construct UA140I+IV−, the same strategy was used, except that the kanamycin resistance gene aphIII (38) was used. The deletion mutation was also confirmed by PCR and genetic complementation. Since no indicator strain was sensitive only to mutacin IV, the defect in mutacin IV production was further confirmed by mutacin isolation under conditions in which mutacin I was not produced (32). Briefly, mutacin IV was isolated from the culture supernatant by extraction with an equal volume of chloroform from a wild-type strain and the mutacin IV-defective strain. For purification, the crude extract was applied to a Source 15RPC column and eluted with a gradient of buffers A (0.1% trifluoroacetic acid) and B (0.085% trifluoroacetic acid in 60% acetonitrile) using an LKB Purifier (Amersham Pharmacia Biotech, Piscataway, N.J.). The activity of the purified peptide was tested in an overlay assay with the indicator strain S. sanguinis NY101. Ten microliters each of different serial dilutions were spotted onto a BHI agar plate and, after they dried, overlaid with BHI soft agar (0.7% agar) containing the indicator strain S. sanguinis NY101. The activity was monitored by the occurrence of a cleared zone. To construct the double-mutant strain UA140I−IV−, chromosomal DNA was isolated from strain UA140I+IV− and transformed into strain UA140I−IV+. Ten independent transformants were selected on Tet-plus-kanamycin plates and tested, and the lack of mutacin production was confirmed by using the indicator strain NY101, which was sensitive to both mutacins (32). The mutants were further confirmed by PCR.
Construction of reporter strains.
The mutacin I promoter-luciferase reporter strain UA140::Φ(mutAp-luc) was constructed essentially as described previously (15), except that pFW5-luc (31) was used as the backbone plasmid. The lactate dehydrogenase (ldh) promoter-green fluorescent protein (gfp) reporter strain UA159::Φ(ldhp-gfp) was constructed in the same manner as described previously (16), except that the luciferase gene was replaced with gfp. Reporter strains were confirmed by PCR, as well as by spectinomycin resistance (800 μg/ml).
Inhibition assays with H2O2 and mutacin.
To assay the inhibition of S. mutans by H2O2, a fresh overnight culture of strain UA140 was diluted 25-fold in fresh BHI. After 2.5 doubling times, the culture was divided and treated with different concentrations of H2O2 (0.0005%, 0.0025%, and 0.005%). The growth inhibitions were monitored by following the OD600 at indicated time points (see Fig. 4A). The inhibition of S. sanguinis was tested with purified mutacin I and mutacin IV. Mutacins I and IV were purified as described above and earlier (33). The activities of the individual peptides were tested in an overlay assay as described above with the indicator strain S. sanguinis NY101.
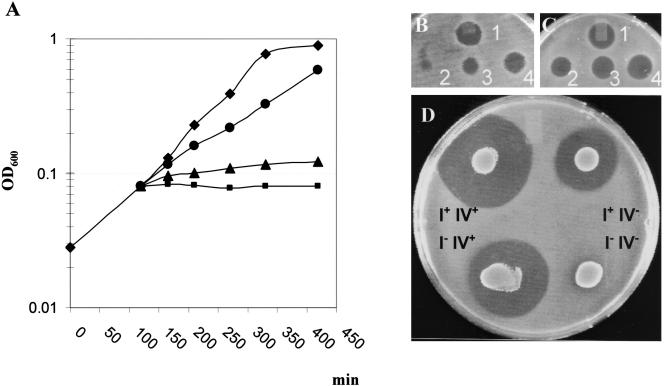
FIG. 4.
Effects of H2O2, mutacin I, and mutacin IV on the growth of S. mutans and S. sanguinis. (A) Growth inhibition of S. mutans UA140 treated with different concentrations of H2O2; ⧫, no H2O2; •, 0.0005% (142 μM) H2O2; ▴, 0.0025% (710 μM) H2O2; ▪, 0.005% (1.42 mM) H2O2. Experiments were repeated two times with similar results. Shown is a representative result of one experiment. (B and C) Inhibition of S. sanguinis with purified mutacin I and partially purified mutacin IV. Different dilutions of purified mutacin I (B) and partially purified mutacin IV (C) were spotted onto a BHI plate and overlaid with S. sanguinis. Each spot contained 10 μl of twofold serially diluted extract (i.e., no. 1, undiluted; no. 4, twofold diluted; no. 3, fourfold diluted, etc.). (D) Effects of mutations in mutacin I and mutacin IV genes on the growth of S. sanguinis. Overnight cultures of a mutacin I-defective (I−IV+), a mutacin IV-defective (I+IV−), a double-mutant (I−IV−), and a wild-type (I+IV+) strain of UA140 were spotted (10 μl) onto BHI plates and overlaid with S. sanguinis.
Expression of mutAp-luc and ldhp-luc under biofilm conditions.
Overnight cultures of strains UA140::Φ(ldhp-luc), UA140::Φ(mutAp-luc) and S. sanguinis were adjusted to an OD600 of 1. Ten microliters of strain UA140::Φ (ldhp-luc) or UA140::Φ(mutAp-luc) alone or mixed in a 1:1 ratio with S. sanguinis was spotted onto a BHI plate. After 6 h of incubation, the cells were scraped from the plate and the luciferase activity was determined. The activity was normalized by the cell counts of S. mutans after serial dilution.
RESULTS
Characterization of interspecies competition between S. mutans and other oral streptococcal species. To get a global view of how prevalent interspecies competition is between S. mutans and other oral streptococci, we analyzed the inhibitory spectrum of S. mutans strain UA140 against 11 streptococcal species, including members of the mitis, mutans, viridans, and pyogenic groups: S. gordonii ATCC 10558, S. oralis ATCC 10557, S. mitis ATCC 33399, S. mitis ATCC 903, S. pneumoniae, S. parasanguinis ATCC 15911, S. sanguinis ATCC 10556, S. sanguinis NY101, S. sobrinus OMZ176. S. cristatus ATCC 49999, and S. pyogenes. UA140 was inoculated onto BHI plates and grown for 24 h before the other species were inoculated nearby. As shown in Fig. 1, S. mutans could inhibit the growth of all tested strains; however, the growth inhibition was less severe against S. sobrinus, a member of the mutans group. Based on this result, S. sanguinis was chosen for further analysis because of its well-known history of antagonism toward S. mutans.
FIG. 1.
Inhibition of oral streptococcal species by S. mutans UA140. 1, S. gordonii; 2, S. pyogenes; 3, S. oralis; 4, S. mitis ATCC 33399; 5, S. mitis ATCC 903; 6, S. pneumoniae; 7, S. cristatus; 8, S. parasanguinis; 9, S. sanguinis ATCC 10556; 10, S. sanguinis NY101; 11, S. sobrinus.
Competition between S. mutans and S. sanguinis in time and space.
A simple competition assay was developed to test the antagonistic interactions between S. mutans and S. sanguinis. Overnight cultures of S. mutans UA140 and S. sanguinis ATCC 10556 were inoculated on half-strength BHI plates. Three tests were conducted: (i) S. mutans was inoculated first and allowed to grow overnight (as the early colonizer) before S. sanguinis was inoculated nearby (as the later colonizer), (ii) vice versa, and (iii) both species were inoculated at the same time. As shown in Fig. 2A, the early colonizer always inhibited the growth of the later colonizer regardless of the bacterial species (left and middle). This competitive exclusion was reduced to a negligible level when both species were inoculated at the same time (right). This suggested that the sequence of inoculation determined the competition outcome.
FIG. 2.
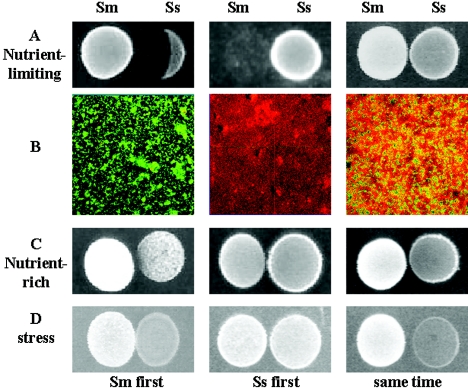
Competition assays between S. mutans and S. sanguinis. (A) Competition assay on half-strength BHI plate. (B) Confocal laser scanning microscopy analysis of competition in biofilms. Green cells, S. mutans (green fluorescent protein); red cells, S. sanguinis (Cell-tracker orange). The pictures were taken at ×100 magnification. (C) competition assays on “nutrient-rich” plate (BHI plus 1% sucrose, buffered to pH 7). (D) competition assays on “stress” plate (BHI at pH 5.5). (C and D) Left, S. mutans (Sm) was inoculated first; middle, S. sanguinis (Ss) was inoculated first; right Sm and Ss were inoculated at the same time.
Since competitive exclusion could result from either nutritional deprivation by the growth of the early colonizer or production of inhibitory substances by the early colonizer, we decided to test the first possibility by performing the same competition assay described above but using different strains of the same species. We reasoned that nutritional deprivation would be more severe within the same species because the bacteria have the same nutrient requirements. We used another S. mutans strain, UA159 (1), in the competition assay with UA140 and another S. sanguinis strain, NY101, in the competition assay with ATCC 10556. No growth inhibition was observed in either competing pair regardless of the sequence of inoculation (data not shown). This result suggested that some diffusible substance produced by S. mutans and S. sanguinis rather than nutrient deprivation was responsible for the observed competitive exclusion.
To see if this competitive exclusion also occurred in space, such as in biofilms, we constructed an S. mutans green fluorescent protein (gfp) reporter strain, UA140::Φ(ldhp-gfp) (see Materials and Methods). UA140::Φ(ldhp-gfp) carries a gfp fusion to the lactate dehydrogenase (ldh) promoter on the chromosome. Since the ldh promoter is constitutively expressed (24), UA140::Φ(ldhp-gfp) cells continually exhibit green fluorescence throughout growth. This property made it easier to distinguish S. mutans from S. sanguinis, which was labeled with red fluorescence using a cell tracker dye (CellTracker Orange) 2 h prior to microscopy. UA140::Φ(ldhp-gfp) and S. sanguinis were then subjected to the previously described competition assays (see Materials and Methods). As shown in Fig. 2B, when S. mutans attached first, almost no S. sanguinis bacteria could attach and grow in the biofilm (left). The same was true for S. sanguinis when it attached first (middle). However, when both were inoculated at the same time, a mixed-species biofilm could form (right). This result was reminiscent of the observations made by Mikx et al. nearly 30 years ago in the germ-free-rat experiment (25), suggesting that the competition between S. mutans and S. sanguinis observed in this in vitro assay may also occur in vivo.
Environmental conditions modulate competition and coexistence between S. mutans and S. sanguinis.
Since the dental biofilm in nature is continually challenged by adverse conditions, such as cycles of feast and famine and fluctuations of pH, we were interested to see whether the competition between S. mutans and S. sanguinis was influenced by these environmental conditions. We performed a plate assay similar to that shown in Fig. 2A under three conditions: a “nutrient-rich” growth condition in which sucrose was added to BHI and the medium was buffered to pH 7.0 with phosphate buffer, a “stress” condition in which the pH of BHI was lowered to 5.5, and a “nutrient-limiting” condition in which BHI was diluted to half strength, as in Fig. 2A. As expected, the “nutrient-limiting” condition resulted in the same pattern of inhibition shown in Fig. 2A. Surprisingly, under “nutrient-rich” or “stress” conditions, there was negligible or no inhibition between the species regardless of the sequence of inoculation (Fig. 2C and D); the lesser growth of S. sanguinis under “stress” conditions is due to the growth inhibition of S. sanguinis by acidic pH. These results suggested that environmental conditions modulated competition/coexistence between bacterial species.
Investigation of possible inhibitory substances produced by S. mutans and S. sanguinis.
The results presented in Fig. 2A suggested that both S. mutans and S. sanguinis produced diffusible substances that inhibited the growth of the other species. To identify the possible inhibitory substances, we grew S. mutans and S. sanguinis on a half-strength BHI plate for 24 h and applied peroxidase (40 μg), peptidase (64 μg), or phosphate-buffered saline beside each colony for 10 min before the other species was inoculated at the same spot. The two enzymes (peptidase and peroxidase) were chosen based on previous knowledge that proteinaceous inhibitory substances (8) and H2O2 were produced by oral streptococci (34, 39). As shown in Fig. 3, addition of peroxidase abolished the inhibitory effect of S. sanguinis toward S. mutans (Fig. 3A, left), while addition of peptidase diminished the inhibitory effect of S. mutans toward S. sanguinis (Fig. 3B, middle). Given the fact that the inhibitory substance(s) produced by S. mutans is proteinaceous, one logical candidate would be a peptide antibiotic, e.g., bacteriocin, since S. mutans is known to produce multiple bacteriocins called mutacins (8). Strain UA140, used in this study, was known to produce two major mutacins, mutacin I and mutacin IV (32). To determine whether the mutacins were responsible for inhibiting the growth of S. sanguinis, we constructed a mutacin-defective isogenic strain, UA140I−IV−, in which the production of both mutacins was eliminated by inactivation of the mutacin-biosynthetic genes (see Materials and Methods). This mutant strain was tested in competition assays with S. sanguinis on the plate, as well as in the biofilm. As shown in Fig. 3C and D, UA140I−IV− could no longer inhibit the growth of S. sanguinis on the plate or in the biofilm even when it was inoculated first. Similar results were obtained with all 11 oral streptococci used in the initial screen (data not shown). To test which mutacin was responsible for the inhibitory effect, UA140 derivative strains defective in either mutacin I or mutacin IV were constructed (see Materials and Methods). Competition assays using these strains showed that they were still able to inhibit the growth of S. sanguinis (data not shown). These results demonstrate that both mutacins serve as inhibitory substances and that either mutacin is sufficient to inhibit the growth of S. sanguinis and other streptococcal strains.
FIG. 3.
Identification of inhibitory substances produced by S. mutans and S. sanguinis. (A) S. sanguinis (Ss) was inoculated first. (B) S. mutans (Sm) was inoculated first. After 24-h growth on half-strength BHI plates, 40 μg of peroxidase (left), 64 μg of peptidase (middle), or phosphate-buffered saline (right) was added beside the colony before the competing species was inoculated. (C) competition of the mutacin-defective strain UA140I−IV− with Ss on the plate (C) and in the biofilm (D) when Sm was inoculated first. Green cells, S. mutans (green fluorescent protein); red cells, S. sanguinis (Cell-tracker orange). The confocal micrograph was taken at ×100 magnification.
Since the inhibitory substance(s) produced by S. sanguinis was sensitive to peroxidase (Fig. 3A), hydrogen peroxide (H2O2), became the likely candidate. To test this hypothesis, we used a leuco crystal violet assay (see Materials and Methods) to measure H2O2 production by S. sanguinis and found that under high-cell-density conditions, approximately 120 μM H2O2 was produced by S. sanguinis, which would be sufficient to affect the growth of S. mutans. Although a direct quantification of H2O2 production on the plate was not technically feasible, we did observe considerable H2O2 production by S. sanguinis grown on plates (see Fig. 5C). These data suggested that H2O2 produced by S. sanguinis could be one of the diffusible inhibitory substances responsible for preventing the growth of S. mutans.
FIG. 5.
Effects of growth conditions on mutacin I gene expression (A), mutacin production (B), and H2O2 production (C). Mutacin I gene expression (mutAp-luc) was measured as relative light units (RLU) per OD600 unit; mutacin production was measured by diameters of the inhibition zone against the indicator; H2O2 production by S. sanguinis was indicated by a purple color (see Materials and Methods). Cells were grown on different conditioned plates: 1, half-strength BHI; 2, BHI plus 1% sucrose, pH 7; 3, BHI, pH 5.5. Presented are representatives of at least two experiments performed on different days (the error bars indicate standard deviations).
To get more direct evidence that these compounds (mutacins and H2O2) indeed can carry out the inhibitory effects on S. sanguinis and S. mutans, respectively, we conducted direct growth inhibition studies. S. mutans was challenged with different concentrations of H2O2, and growth inhibition was measured (Fig. 4A). The lowest concentration that could inhibit the growth of S. mutans was 0.0005% (142 μM), which was in the same range as the H2O2 produced by S. sanguinis in the cell pellet (see Fig. 6B). Purified mutacin I and mutacin IV were both able to inhibit the growth of S. sanguinis in an overlay assay up to an eightfold dilution (Fig. 4B and C). In addition, we conducted overlay assays with the different mutacin mutants. These experiments showed that both mutacins are involved in the S. sanguinis growth inhibition and that the double mutant had a dramatically reduced ability to inhibit the growth of S. sanguinis (Fig. 4D). These results demonstrate the ability of H2O2 and mutacin to inhibit the growth of S. mutans and S. sanguinis, respectively.
FIG. 6.
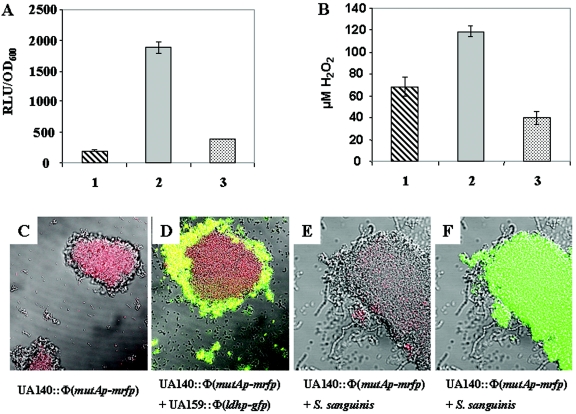
Effects of juxtaposition between S. mutans and S. sanguinis on mutacin gene expression and H2O2 production. (A) Mutacin I gene expression of strain UA140::Φ(mutAp-luc). (B) H2O2 production by S. sanguinis. Bars 1, single-species planktonic culture; bars 2, single-species pelleted culture; bars 3, mixed-species pelleted culture. The experiment was done three times on different days with similar results. Presented are representative data from one experiment done in duplicate. The error bars indicate standard deviations. (C) Mutacin I gene expression in strain UA140::Φ(mutAp-mrfp) single-strain culture. Red fluorescence indicates mutacin I gene expression. (D) Same experiment as in panel C, but mixed with strain UA159::Φ(ldhp-gfp). Red, UA140::Φ(mutAp-mrfp); green, UA159::Φ(ldhp-gfp); yellow, mixture of the two strains. (E) Same experiment as in panel C, but mixed with S. sanguinis. (F) Same experiment as in panel E with UA140::Φ(mutAp-mrfp) cells labeled with fluorescein isothiocyanate-conjugated anti-S. mutans monoclonal antibodies. Green cells, UA140::Φ(mutAp-mrfp); gray cells, S. sanguinis. The confocal micrographs were taken with fluorescent and differential interference contrast modes.
Mutacin gene expression and H2O2 production are regulated by growth conditions.
To determine the effect of mutacin and H2O2 production on the competition outcome between S. mutans and S. sanguinis, we studied the effect of medium conditions on the production of mutacin and H2O2. To quantify mutacin gene expression, we constructed reporter strains in which the promoterless firefly luciferase gene (luc) was fused to the mutacin I (mutA) and the mutacin IV (nlmA) promoters on the chromosome. The reporter strains were inoculated on the three conditioned plates as described in Fig. 2C and D. After 24 h of incubation, the cells were scraped from the plate and measured for luciferase activity and OD600. The spent plates were overlaid with an indicator strain to measure mutacin production. Both mutacin I (mutA) and mutacin IV (nlmA) promoters exhibited the same pattern of expression under these conditions; shown in Fig. 5A and B are the results of the mutacin I promoter expression (mutAp-luc) and mutacin I production. Compared to the “nutrient-limiting” plate (bar 1), mutacin I promoter expression was reduced ∼10-fold on both “nutrient-rich” (bar 2) and “stress” (bar 3) condition plates (Fig. 5A). Consequently, the inhibition zone on the “nutrient-rich” plate (Fig. 5B, bar 2) was reduced >5-fold compared to that on the “nutrient-limiting” plate (bar 1), and no inhibition zone was observed on the “stress” condition plate (bar 3).
The effect of environmental conditions on H2O2 production by S. sanguinis was measured on the plate by a modified peroxidase assay (see Materials and Methods). Darker color on and around the colony indicated the presence of larger amounts of H2O2. As shown in Fig. 5C, the amounts of H2O2 on the “nutrient-rich” (plate 2) and “stress” (plate 3) condition plates were conspicuously less than that on the “nutrient-limiting” (plate 1) plate. Taken together, these results correlated well with the phenotypic observations depicted in Fig. 2.
Mutacin gene expression and H2O2 production are both inhibited by juxtaposition between S. mutans and S. sanguinis.
The results presented in Fig. 2A and B demonstrated that despite competitive exclusion between S. mutans and S. sanguinis, they can coexist under certain circumstances, such as when both species are inoculated at the same time (right). To determine whether close cell-cell proximity between the two species could result in mutual inhibition of inhibitory-substance production by the competing species, we developed a mixed-culture pelleting assay that would create an environment for cell-cell contact but without complications of extensive cell growth. Overnight cultures were diluted and grown to early log phase (OD600, ∼0.1), and the two species were mixed in a 1:1 ratio and centrifuged. The mixed cultures were incubated for 2 h as cell pellets before luciferase activity, H2O2 production, and OD600 were measured. As controls, single-species cultures of S. mutans and S. sanguinis in planktonic and pelleted conditions were used. Since mutacin I and IV promoters behaved similarly, only the results of mutacin I promoter expression are presented here (Fig. 6). In the single-species culture, mutacin I gene expression increased 10-fold in the cell pellet (bar 2) compared with the planktonic culture (bar 1) (Fig. 6A). Similarly, H2O2 production by S. sanguinis increased twofold in the cell pellet (bar 2) compared with the planktonic culture (bar 1) (Fig. 6B). These results suggested that high cell density enhanced mutacin gene expression by S. mutans and H2O2 production by S. sanguinis. Surprisingly, in the mixed-species cell pellet, mutacin gene expression by S. mutans was reduced fivefold (Fig. 6A, bar 3) and H2O2 production by S. sanguinis was reduced threefold (Fig. 6B, bar 3) compared with their respective single-species cell pellets.
To further confirm that this inhibition of mutacin gene expression by juxtaposition with S. sanguinis happens only between different species, not within the same species, we performed the same pelleting assay with two S. mutans strains carrying different fluorescent protein reporters. UA140::Φ(mutAp-mrfp) (15) carries a red fluorescent protein fused to the mutacin I promoter, and UA159::Φ(ldhp-gfp) carries a green fluorescent protein fused to the ldh promoter (16). Pelleting assays were performed with either UA140::Φ(mutAp-mrfp) alone (Fig. 6C), UA140::Φ(mutAp-mrfp) plus UA159::Φ(ldhp-gfp) (Fig. 6D), or UA140::Φ(mutAp-mrfp) plus S. sanguinis (Fig. 6E). After 2 h of incubation, the cell pellet was analyzed by confocal microscopy. UA140::Φ(mutAp-mrfp) cells alone or in a mixture with UA159::Φ(ldhp-gfp) exhibited bright-red fluorescence, indicating a high level of mutacin I gene expression; in contrast, the same UA140::Φ(mutAp-mrfp) cells displayed almost no fluorescence in the mixed culture with S. sanguinis (Fig. 6E). To exclude the possibility that the diminished fluorescence of UA140::Φ(mutAp-mrfp) in the mixed-species culture was due to fewer S. mutans cells in the cell aggregates, a fluorescein isothiocyanate-conjugated monoclonal antibody specific to S. mutans (1) was used to label cells in the mixed-species cell aggregates. As shown in Fig. 6F, similar amounts of UA140::Φ(mutAp-mrfp) cells existed in the mixed-species cell aggregates and in the single-species cell aggregates.
To test whether the reduced mutacin gene expression and H2O2 production in the mixed-species cell pellet was due to inhibition of cell growth, cells in the single-species and mixed-species cell pellets were plated at the beginning and the end of the experiment. No difference was observed between the single-species and mixed-species cultures, suggesting that the reduced mutacin gene expression and H2O2 production in the mixed-species cell pellet was not due to inhibition of cell growth of either species during the 2-h coculturing period (data not shown). To test further whether live cells were required to exert this inhibitory effect, S. mutans cells were mixed with UV-killed S. sanguinis cells or vice versa, and pelleting assays were performed. Mutacin gene expression or H2O2 production was not inhibited when dead cells of the other species were present (data not shown).
Since it could be argued that the pelleting assay created an artificial high-cell-density environment, which may not represent the natural dental biofilm situation, we did another experiment under biofilm conditions. We inoculated UA140::Φ(mutAp-luc) as a single-species culture and as a mixed-species culture with S. sanguinis in a 1:1 ratio on a BHI plate and incubated the cells for 6 h. Under this biofilm condition, both bacterial species could grow on a surface with an air interface, as could be found in the dental biofilm. The cells were scraped from the plate, and luciferase activity was determined. After normalization with the number of viable cells, we found a 15-fold reduction of the luciferase activity in the mixed-species culture compared to UA140::Φ(mutAp-luc) alone (Fig. 7A). As a control, UA140::Φ(ldhp-luc) was used to monitor the expression of the housekeeping gene ldh (lactate dehydrogenase), which would reflect the metabolic status of the cells (24). As shown in Fig. 7B, the expression of the ldh gene remained the same in the mixed-species biofilm as in the single-species biofilm. The increase in the change from 5-fold (in the pellet) to 15-fold (on the plate biofilm) could be explained by the longer incubation time of S. mutans in the presence of S. sanguinis. The longer incubation time was necessary to yield visible cell growth on the plate. This result further confirmed the observations made in the pelleting assays (Fig. 5), suggesting that in the dental biofilm, the presence of S. sanguinis could inhibit mutA gene expression of S. mutans.
FIG. 7.
Relative mutacin I (mutAp-luc) and lactate-dehydrogense (ldhp-luc) gene expression in single- and mixed-species surface biofilms. Overnight cultures of all strains were adjusted to an OD600 of 1. Ten microliters of strain UA140::Φ(mutAp-luc) or UA140::Φ(ldhp-luc) alone (bars 1 and 3) or mixed in a 1:1 ratio with S. sanguinis (bars 2 and 4) were spotted onto a BHI plate. After 6 h of incubation, the cells were scraped from the plate and the luciferase activity was determined. The activity was normalized by the cell counts of S. mutans after serial dilution. Experiments were repeated twice with similar results. Shown is a representative result of one experiment done with triplicate samples. (A) Expression of the mutacin I (mutA) gene. (B) Expression of the ldh gene.
DISCUSSION
A unique feature of the oral biofilm is its high density and diversity of microbial species (9). This high cell density dictates close cell-cell contact within the same species or between different species, which results in inevitable intra- and interspecies interactions. Cooperative interactions among oral bacteria have been well studied. These include coaggregation to facilitate a cell's attachment to the tooth surface (4), nutritional complementation to enable cell growth in saliva (11), and metabolic cooperation between two species (6). These cooperative interactions probably have played very important roles in the development of the dental biofilm; however, antagonistic interactions among different species may be equally important given the conditions in the oral cavity. For example, Xie et al. reported inhibition of Porphyromonas gingivalis fimbrial gene expression by Streptococcus cristatus mediated by a 59-kDa surface protein (40). In this study, we initiated a systematic investigation of the molecular mechanisms of interspecies competition between S. mutans and S. sanguinis.
Competition assays on the plate and in the biofilm demonstrated a mutual exclusion between the two species depending on the sequence of inoculation (Fig. 2). This competitive exclusion turned out to be a result of the production of inhibitory substances by the two competing species. Interestingly, when both species were inoculated at the same time, negligible or no competition was observed. Further investigation revealed that when both species were juxtaposed to each other, both mutacin and H2O2 production were inhibited (Fig. 6). Since this mutual inhibition was observed only when live cells of the competing species were present in the cell pellet, we speculate that interspecies communications are involved. Further investigations are under way to elucidate the interspecies communication pathways and the molecular signals involved.
Another interesting finding from the competition assays was that on a BHI plate supplemented with sucrose and buffered to pH 7 with phosphate buffer, or on a BHI plate adjusted to pH 5.5 with HCl, competitive exclusion was not observed regardless of which species was inoculated first (Fig. 2C and D). We considered the former condition as “nutrient rich” because sucrose appeared to be the preferred carbohydrate for both species and buffering the medium to pH 7 would prevent inhibition of cell growth by the acids produced during the fermentation of sucrose. Indeed, cells growing on this plate always achieved higher cell mass than cells growing on a regular BHI plate (data not shown). The BHI plate with pH 5.5 was considered a stress condition because both bacterial species grew more slowly on this plate than on regular BHI plates, although S. mutans showed more acid tolerance than S. sanguinis (3, 12). How did “nutrient-rich” growth and “stress” conditions suppress competition between the two species? Further studies (Fig. 5) demonstrated that this was achieved at least partially through inhibition of mutacin and H2O2 production.
What is the ecological meaning of this environmental modulation of interspecies competition? From a cell's economic point of view, we speculate that it is related to the balance between the cost and benefit of producing mutacins and H2O2. Biosynthesis of mutacin I and IV is an expensive process. For mutacin I, at least 11 gene products are required for producing a functional mutacin molecule (32), and for mutacin IV, at least five gene products would be required. Although the exact mechanism of H2O2 production by S. sanguinis is not known, it also would require energy (13). In this case, mutacin or H2O2 production may become a double-edged sword. In a multispecies community, mutacin or H2O2 production may give the producer a competitive edge, while it may also slow down the growth of the producers due to the extra energy expenditure. Therefore, it would make perfect ecological sense that when excess nutrient is present, mutacin or H2O2 production is shut down to allow more energy to be used for cell growth and species proliferation. Similarly, under stress conditions where cell survival becomes more important than colony expansion, mutacin or H2O2 production is also shut off to focus energy expenditure on maintaining the essential cellular functions. Only under conditions where cells have enough energy to compete but not enough food for optimal growth is mutacin or H2O2 production activated for competition. These well-regulated strategies may be necessary for the survival and perpetuation of a species in a multispecies community under natural conditions and may be even more so in the oral cavity, where cycles of feast and famine and fluctuations of pH are daily routines. It is also worth noting that mutacin and H2O2 production is rather prevalent in clinical isolates of S. mutans and S. sanguinis (and other members of the mitis group streptococci), respectively (8, 34, 39). So is the competition between S. mutans and other oral streptococcal species, as shown in Fig. 1. Therefore, the molecular mechanisms underlying the competition and coexistence between the two species reported in this study may represent a general mechanism underlying interspecies interactions in the dental biofilm.
Acknowledgments
This work was supported in part by NIH grants U01-DE15018 to W.S. and R01-DE014757 to F.Q., NIDCR T32 Training Grant DE007296 to J.M., and Delta Dental grant WDS78956 to W.S.
REFERENCES
- 1.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, G. R., S. V. Sutton, and R. E. Marquis. 1986. Acid tolerance, proton permeabilities, and membrane ATPases of oral streptococci. Infect. Immun. 53:331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blehert, D. S., R. J. Palmer, Jr., J. B. Xavier, J. S. Almeida, and P. E. Kolenbrander. 2003. Autoinducer 2 production by Streptococcus gordonii DL1 and the biofilm phenotype of a luxS mutant are influenced by nutritional conditions. J. Bacteriol. 185:4851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caufield, P. W., A. P. Dasanayake, Y. Li, Y. Pan, J. Hsu, and J. M. Hardin. 2000. Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect. Immun. 68:4018-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egland, P. G., R. J. Palmer, Jr., and P. E. Kolenbrander. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. USA 101:16917-16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flannagan, S. E., L. A. Zitzow, Y. A. Su, and D. B. Clewell. 1994. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid 32:350-354. [DOI] [PubMed] [Google Scholar]
- 8.Hamada, S., and T. Ooshima. 1975. Production and properties of bacteriocins (mutacins) from Streptococcus mutans. Arch. Oral Biol. 20:641-648. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton, I. A. 2000. Ecological basis for dental caries, p. 219-275. In H. K. Kuramitsu and R. P. Ellen (ed.), Oral bacterial ecology. Horizon Scientific Press, Wymondham, Norfolk, England.
- 10.Hammill, H. A. 1989. Normal vaginal flora in relation to vaginitis. Obstet. Gynecol. Clin. N. Am. 16:329-336. [PubMed] [Google Scholar]
- 11.Hansen, M. C., R. J. Palmer, Jr., C. Udsen, D. C. White, and S. Molin. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383-1391. [DOI] [PubMed] [Google Scholar]
- 12.Harper, D. S., and W. J. Loesche. 1984. Growth and acid tolerance of human dental plaque bacteria. Arch. Oral Biol. 29:843-848. [DOI] [PubMed] [Google Scholar]
- 13.Higuchi, M., M. Shimada, Y. Yamamoto, T. Hayashi, T. Koga, and Y. Kamio. 1993. Identification of two distinct NADH oxidases corresponding to H2O2-forming oxidase and H2O-forming oxidase induced in Streptococcus mutans. J. Gen. Microbiol. 139:2343-2351. [DOI] [PubMed] [Google Scholar]
- 14.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 15.Kreth, J., J. Merritt, C. Bordador, W. Shi, and F. Qi. 2004. Transcriptional analysis of mutacin I (mutA) gene expression in planktonic and biofilm cells of Streptococcus mutans using fluorescent protein and glucuronidase reporters. Oral Microbiol. Immunol. 19:252-256. [DOI] [PubMed] [Google Scholar]
- 16.Kreth, J., J. Merritt, W. Shi, and F. Qi. 2005. Coordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighboring species. Mol. Microbiol. 57:392-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsen, B., and G. R. Monif. 2001. Understanding the bacterial flora of the female genital tract. Clin. Infect. Dis. 32:e69-e77. [DOI] [PubMed] [Google Scholar]
- 19.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loesche, W. J., J. Rowan, L. H. Straffon, and P. J. Loos. 1975. Association of Streptococcus mutans with human dental decay. Infect. Immun. 11:1252-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh, P. D. 2003. Are dental diseases examples of ecological catastrophes? Microbiology 149:279-294. [DOI] [PubMed] [Google Scholar]
- 22.Marsh, P. D. 2004. Dental plaque as a microbial biofilm. Caries Res. 38:204-211. [DOI] [PubMed] [Google Scholar]
- 23.Marteau, P., F. Daniel, P. Seksik, and R. Jian. 2004. Inflammatory bowel disease: what is new? Endoscopy 36:130-136. [DOI] [PubMed] [Google Scholar]
- 24.Merritt, J., J. Kreth, F. Qi, R. Sullivan, and W. Shi. 2005. Non-disruptive, real-time analyses of the metabolic status and viability of Streptococcus mutans cells in response to antimicrobial treatments. J. Microbiol. Methods 61:161-170. [DOI] [PubMed] [Google Scholar]
- 25.Mikx, F. H. M., J. S. Vanderhoeven, A. J. M. Plasschaert, and K. G. Konig. 1976. Establishment and symbiosis of Actinomyces viscosus, Streptococcus sanguis and Streptococcus mutans in germfree Osborne-Mendel rats. Caries Res. 10:123-132. [DOI] [PubMed] [Google Scholar]
- 26.Moore, W. E., and L. V. Moore. 1994. The bacteria of periodontal diseases. Periodontol. 2000 5:66-77. [DOI] [PubMed] [Google Scholar]
- 27.Mottola, H. A., B. E. Simpson, and G. Gorin. 1970. Absorptiometric determination of hydrogen peroxide in submicrogram amounts with leuco cristal violet and peroxidase as catalyst. Anal.l Chem. 42:410-411. [Google Scholar]
- 28.Nyvad, B., and M. Kilian. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:369-380. [DOI] [PubMed] [Google Scholar]
- 29.Palmer, R. J., Jr., R. Wu, S. Gordon, C. G. Bloomquist, W. F. Liljemark, M. Kilian, and P. E. Kolenbrander. 2001. Retrieval of biofilms from the oral cavity. Methods Enzymol. 337:393-403. [DOI] [PubMed] [Google Scholar]
- 30.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Podbielski, A., B. Spellerberg, M. Woischnik, B. Pohl, and R. Lutticken. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137-147. [DOI] [PubMed] [Google Scholar]
- 32.Qi, F., P. Chen, and P. W. Caufield. 2001. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl. Environ. Microbiol. 67:15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi, F., P. Chen, and P. W. Caufield. 2000. Purification and biochemical characterization of mutacin I from the group I strain of Streptococcus mutans, CH43, and genetic analysis of mutacin I biosynthesis genes. Appl. Environ. Microbiol. 66:3221-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryan, C. S., and I. Kleinberg. 1995. Bacteria in human mouths involved in the production and utilization of hydrogen peroxide. Arch. Oral Biol. 40:753-763. [DOI] [PubMed] [Google Scholar]
- 35.Sbordone, L., and C. Bortolaia. 2003. Oral microbial biofilms and plaque-related diseases: microbial communities and their role in the shift from oral health to disease. Clin. Oral Investig. 7:181-188. [DOI] [PubMed] [Google Scholar]
- 36.Simon, G. L., and S. L. Gorbach. 1986. The human intestinal microflora. Dig. Dis. Sci. 31:147S-162S. [DOI] [PubMed] [Google Scholar]
- 37.Stinson, M. W., S. Alder, and S. Kumar. 2003. Invasion and killing of human endothelial cells by viridans group streptococci. Infect. Immun. 71:2365-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trieu-Cuot, P., and P. Courvalin. 1983. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene 23:331-341. [DOI] [PubMed] [Google Scholar]
- 39.Uehara, Y., K. Kikuchi, T. Nakamura, H. Nakama, K. Agematsu, Y. Kawakami, N. Maruchi, and K. Totsuka. 2001. H2O2 produced by viridans group streptococci may contribute to inhibition of methicillin-resistant Staphylococcus aureus colonization of oral cavities in newborns. Clin. Infect. Dis. 32:1408-1413. [DOI] [PubMed] [Google Scholar]
- 40.Xie, H., G. S. Cook, J. W. Costerton, G. Bruce, T. M. Rose, and R. J. Lamont. 2000. Intergeneric communication in dental plaque biofilms. J. Bacteriol. 182:7067-7069. [DOI] [PMC free article] [PubMed] [Google Scholar]