Abstract
Rieske nonheme iron oxygenases form a large class of aromatic ring-hydroxylating dioxygenases found in microorganisms. These enzymes enable microorganisms to tolerate and even exclusively utilize aromatic compounds for growth, making them good candidates for use in synthesis of chiral intermediates and bioremediation. Studies of the chemical stability and thermostability of these enzymes thus become important. We report here the structure of free and substrate (indole)-bound forms of naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. The structure of the Rhodococcus enzyme reveals that, despite a ∼30% sequence identity between these naphthalene dioxygenases, their overall structures superpose very well with a root mean square deviation of less than 1.6 Å. The differences in the active site of the two enzymes are pronounced near the entrance; however, indole binds to the Rhodococcus enzyme in the same orientation as in the Pseudomonas enzyme. Circular dichroism spectroscopy experiments show that the Rhodococcus enzyme has higher thermostability than the naphthalene dioxygenase from Pseudomonas species. The Pseudomonas enzyme has an apparent melting temperature of 55°C while the Rhodococcus enzyme does not completely unfold even at 95°C. Both enzymes, however, show similar unfolding behavior in urea, and the Rhodococcus enzyme is only slightly more tolerant to unfolding by guanidine hydrochloride. Structure analysis suggests that the higher thermostability of the Rhodococcus enzyme may be attributed to a larger buried surface area and extra salt bridge networks between the α and β subunits in the Rhodococcus enzyme.
Members of the genus Rhodococcus are found in many environmental niches and have been demonstrated to catabolize many toxic xenobiotic compounds (16, 34, 53). Polycyclic aromatic hydrocarbons, such as naphthalene, are widespread in the environment and pose a notable health hazard due to their toxic, mutagenic, and carcinogenic properties. Naphthalene is released into the environment in complex mixtures of coal tar and coal tar products such as creosote (42). Many diverse groups of bacteria that degrade naphthalene are widely distributed in nature (6, 21). Rhodococcus sp. has been demonstrated to play a significant role in the degradation of these compounds at many contaminated sites (24). The first step in the biocatalytic degradation of these aromatic compounds is catalyzed by a class of enzymes called the Rieske nonheme iron oxygenases (ROs), of which naphthalene 1,2-dioxygenase (NDO) is the most studied (19). These enzymes catalyze cis-dihydroxylation reactions and require an oxygen molecule and two electrons that are not from the aromatic substrate. The electrons are produced by a reductase and transported to the dioxygenase by a ferredoxin (39). Besides bioremediation, the ability of these ROs to form enantiopure products and the potential to tailor their regio- and stereospecificities make them very useful in the chiral synthesis of precursor compounds (4).
Naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4 (NDO-P) (5, 26, 27, 54-56) has served as the structural and functional prototype for this copious class of dioxygenases which includes, among others, phthalate, biphenyl, benzoate, and toluene dioxygenases. While this work was inprogress, NDO-P and biphenyl dioxygenase (BPDO-R) from Rhodococcus sp. strain RHA1 (18) were the only terminal RO dioxygenases for which three-dimensional structures had been reported. Recently, structures of other terminal ROs, such as cumene dioxygenase (14), 2-oxoquinoline 8-monooxygenase (38), and nitrotoluene dioxygenase (17), have been determined. However, this work focuses on the NDO-P and BPDO-R structures in the discussions. What is now clear from the RO structures is that these enzymes have either an α3β3 or an α3 structure. All these ROs have been shown to have two metal centers. The metal centers include one Rieske-type iron-sulfur center and one mononuclear iron center. The mononuclear iron is located in the active site and binds oxygen. The residues that ligate the Rieske cluster (two cysteines and two histidines) as well as the 2-His-1-carboxylate facial triad residues that ligate the mononuclear iron center are conserved among ROs. Another conserved residue is an aspartic acid that bridges the ligands of the two metal centers across subunit interfaces. This aspartic acid has been shown to be important for the transfer of electrons between the Rieske iron cluster and the mononuclear iron in the neighboring subunit (47) and to play a part in conformational change at the mononuclear iron active site via hydrogen bonds upon reduction of the Rieske cluster (38).
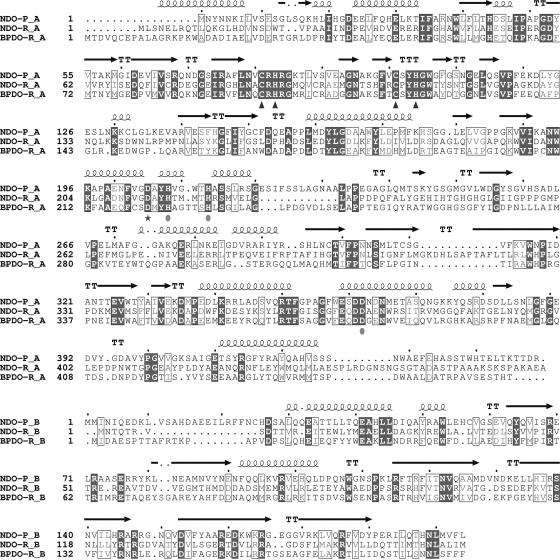
Purification and characterization of the NDO subunits NarAa and NarAb (gene products of α and β subunits, respectively) from Rhodococcus strain NCIMB12038 (NDO-R) revealed that the deduced amino acid sequences have only 33% and 29% identities with the corresponding subunits of NDO-P (33) (Fig. 1). In contrast, the α and β subunits of nitrobenzene dioxygenase (35) have 81% and 77% identity with the respective NDO-P subunits. This clearly indicates that NDO-R is a phylogenetically distant relative of such enzymes that diverged at a very early stage (41). Structures of various ROs are helping explain how the wide range in sequence identities (from greater than 80% to less than 30%) among these enzymes affects substrate preferences. In this work, we report the structures of the free and indole-bound forms of NDO-R and find that all of the differences in the active sites of NDO-R and NDO-P relate to residues near the distal ring of the substrate being dihydroxylated.
FIG. 1.
Multiple sequence alignment of the α and β subunits of NDO-P, NDO-R, and BPDO-R. Secondary structure information for NDO-R is shown above the aligned sequences. The shaded residues are completely conserved, and similar residues based on MultAlin (9) are boxed. The conserved residues that coordinate the mononuclear iron (solid circles) and the Rieske iron-sulfur cluster (triangles) are indicated below the alignment. The bridging aspartic acid is indicated by a star.
Studies of the chemical stability and thermostability of these enzymes become important in view of their potential use for bioremediation and chiral intermediate synthesis. Circular dichroism (CD) spectroscopy studies presented in this work indicate that NDO-R is more thermostable than NDO-P. Structural factors found most consistently to explain thermostability are increased number of stabilizing salt bridges and hydrogen bond interactions (32). Salt bridges in proteins have a stabilizing effect if the centroids of their charged side groups are within 5 Å; if the definition of a salt bridge is narrowed to 4 Å, 92% of the salt bridges become stabilizing (30). In this work, buried surface area, salt bridges, and hydrogen bonds for NDO-R and NDO-P are analyzed to explain the greater thermostability of NDO-R. In addition, the structure of another Rhodococcus sp. enzyme, BPDO-R, is also compared with the NDO structures. Based on these analyses, we predict that the thermostability of BPDO-R is higher than that of NDO-P and comparable to that of NDO-R.
MATERIALS AND METHODS
Purification and crystallization.
Naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038 was purified using published protocols (33). The purified protein was crystallized by the hanging drop method using 2-methyl-2,4-pentanediol (MPD) as a precipitant (37). Two different crystal forms were obtained under very similar conditions—one needle-shaped form that belonged to the orthorhombic crystal system and a plate-shaped form that belonged to the tetragonal crystal system. The highest-diffracting crystals (2-Å resolution) belong to the orthorhombic space group P212121 with a = 87.5 Å, b = 144 Å, c = 185.6 Å, α = β = γ = 90°. The tetragonal crystal form has a = b = 179.5 Å, c = 245.4 Å, α = β = γ = 90° and belongs to the space group P43212. However, the tetragonal crystals diffract to a resolution of only 3.0 Å and, therefore, will not be discussed for the free enzyme. Soaking experiments were carried out to prepare crystals of the protein-indole complex. Attempts at soaking the orthorhombic crystals with indole were unsuccessful. The tetragonal crystals were transferred to solutions of the reservoir buffer containing saturating concentrations of indole. Crystals were frozen after 24 h of soaking. These crystals diffract to a resolution of 3.0 Å. We report here the refined structure of the free enzyme in the orthorhombic form (structure 1) and the substrate (indole)-bound enzyme in the tetragonal form (structure 2).
Structure determination and refinement.
Data for the free enzyme structure were collected at the Argonne National Laboratory Structural Biology Center (SBC) Collaborative Access Team (CAT) beamline in the Advanced Photon Source. Data for the ligand-bound structure were collected at the Industrial Macromolecular Crystallography Association (IMCA) CAT beamline in the Advanced Photon Source. Crystallographic details are presented in Table 1. Self-rotation function studies clearly demonstrated the presence of a threefold axis. An analysis of the Matthews coefficient and the solvent content suggested that there was a single α3β3 molecule in the asymmetric unit in both the crystal forms. Molecular replacement attempts using an αβ model of NDO from Pseudomonas sp. in the programs AMORE (45) and BEAST (50) did not provide any clear solution. However, when a polyalanine model of the full hexamer was used as a model, AMORE readily produced a clear solution for both crystal forms. For the tetragonal crystal data, a translation function search was carried out in the different possible space groups for the Laue group P4/m2/m2/m and a clear solution was obtained in the space group P43212. Density modification, using DM (10), with noncrystallographic symmetry (NCS) averaging of the individual crystal forms clearly showed electron densities for the presence of the mononuclear iron and the Rieske iron-sulfur cluster in the expected positions. However, even in the better-diffracting orthorhombic crystal form, it was not possible to trace the entire chain. Multiple crystal averaging using DM was then carried out, resulting in excellent maps, and it was possible to trace almost the entire chain in the electron density map. All the side chains were modeled, and further refinement was carried out for the orthorhombic crystal form. Refinement was carried out using the program REFMAC (43), and model building was carried out using the program O (25). The structure has been refined with NCS restraints, and details on the quality of the structure are provided in Table 1.
TABLE 1.
Crystallographic data collection and refinement statistics
| Parameter | Free enzyme | Enzyme substrate complex |
|---|---|---|
| Unit cell parameters (Å) | a = 87.7, b = 144.2, c = 185.2 | a = b = 179.5, c = 245.5 |
| Space group | P212121 | P43212 |
| Images collected | 879 (SBC-CAT) | 180 (IMCA-CAT) |
| Resolution (Å) | 20.8-2.0 | 20.0-3.0 |
| No. of measured reflections | 449,811 | 461,815 |
| No. of unique reflections | 147,609 | 79,677 |
| Completeness (%)a | 92.9 (85.0) | 99.2 (98.6) |
| I/σ (I) | 7.4 (1.1) | 12.3 (5.4) |
| Rsym (%) | 7.6 (54.0) | 11.6 (26.5) |
| R factor | 19.1 (25.5) | 28.7 (44.0) |
| Rfree | 22.4 (26.5) | 30.0 (42.1) |
| Water molecules | 1,119 | 351 |
| RMS bond distance (Å) | 0.016 | 0.02 |
| RMS bond angles (°) | 1.5 | 1.4 |
Numbers in parentheses correspond to values in the highest-resolution shell.
All the nonprotein atoms, except the active site iron and iron-sulfur cluster, were removed from the refined native enzyme. This model was used to calculate the starting phases for the indole complex. Rigid body refinement was followed by a round of density modification, including threefold NCS averaging (10). In spite of poorer resolution, the maps clearly showed the position of the bound indole. Refinement was carried out in a similar fashion with high NCS restraints using the program REFMAC. The statistics are presented in Table 1.
CD spectroscopy.
CD spectra of NDO-R and NDO-P were recorded using an Aviv 62DS spectrometer with a Peltier temperature control system. Quartz cells (Starna) with a path length of 1 cm were used for temperature and guanidine hydrochloride-induced unfolding with a protein concentration of 0.15 mg/ml. The protein was in 50 mM Tris-Cl buffer at pH 7.8 containing 10% (vol/vol) ethanol and 10% (vol/vol) glycerol. For urea-induced unfolding, a 0.1-cm-path-length quartz cell was used and a protein concentration of 1.5 mg/ml. The CD spectra in the 320- to 216-nm range were recorded at 22°C, as was the CD signal at 222 nm for guanidine hydrochloride and urea unfolding. For temperature-induced unfolding, the CD signal at 222 nm was monitored for a 1°C/min temperature rise from 5 to 95°C. Baseline correction using the solution without the protein was made for every run. Multiple runs were conducted for every experiment, and negligible variation was found in the results. A representative curve for each experiment is shown in the figures.
Computational analyses.
Multiple sequence alignment of the sequences was carried out using ClustalW 1.82 (8) and annotation using ESPript (20). Structural analysis was carried out on the coordinates obtained from the Protein Data Bank (PDB) database (2) for NDO-P (PDB identification [id] 1EG9 and 1NDO) and BPDO-R (PDB id 1ULJ) and from structure 2 for NDO-R. The program WHAT IF (52) was used to determine all salt bridges (defined with a 5-Å-distance cutoff) in the three proteins. WHAT IF was also used to determine the number of optimal hydrogen bonds that could form within and across the subunits. WHAT IF utilizes the geometry of the donor and acceptor atoms to give each hydrogen bond a score between 0 and 1 to represent its quality. The scores are assigned based on the contribution of the hydrogen bond to the stability of the protein (23). These hydrogen bond values are reported in this study. The accessible surface area (Connolly surface) buried at the interfaces is calculated using a 1.4-Å probe in SYBYL (51). Cavity volume calculations were performed using VOIDOO (28). All structure figures have been generated using PyMOL (12).
Protein structure accession numbers.
The structures have been deposited at the RCSB (Protein Data Bank) with accession numbers 2B1X and 2B24.
RESULTS
CD spectroscopy studies of NDO-R and NDO-P unfolding.
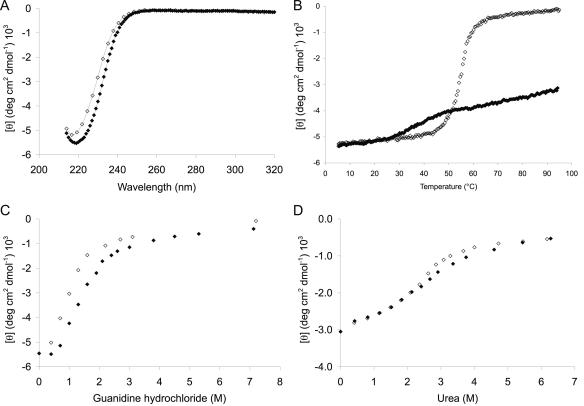
NDO-R and NDO-P demonstrate very different behavior during thermal unfolding. Both NDO-R and NDO-P undergo irreversible unfolding upon heating from 5 to 95°C. However, NDO-P precipitates out of solution as yellowish-white aggregates between 60 and 70°C, while NDO-R remains soluble even after heating to 95°C and does not completely recover its original folded state on cooling. This is clear from the reproducible shift of the ∼220-nm minima in the CD spectra of NDO-R at 22°C before heating to 95°C and after cooling to 5°C (Fig. 2A). The absence of a sharp transition in the CD signal on heating NDO-R to 95°C and the fact that the signal plateaus to an intermediate value instead of zero suggests that NDO-R undergoes partial unfolding in a noncooperative manner to a relatively stable intermediate (Fig. 2B). On the other hand, unfolding of NDO-P, though irreversible, is cooperative with an apparent melting temperature (Tm) of 55°C (Fig. 2B). It is clear that NDO-R is much more resilient than NDO-P to heat-induced unfolding.
FIG. 2.
Circular dichroism spectroscopy plots. (A) Spectra for NDO-R in the far-UV region at 25°C before (⧫) and after (⋄) heating of the sample to 95°C. (B) Temperature-induced unfolding curves for NDO-R (⧫) and NDO-P (⋄) at pH 7.8. Cooperative, though irreversible, unfolding of NDO-P suggests an apparent Tm of 55°C. NDO-R is only partially unfolded at 95°C. (C) Guanidine hydrochloride-induced unfolding curves for NDO-R (⧫) and NDO-P (⋄) at pH 7.8 and 25°C. (D) Urea-induced unfolding curves for NDO-R (⧫) and NDO-P (⋄) at pH 7.8 and 25°C.
NDO-R and NDO-P demonstrate similar unfolding behavior when urea and guanidine hydrochloride (GuHCl) are used as denaturants. With urea, both NDO-R and NDO-P follow almost identical unfolding curves with a relatively broad transition (Fig. 2D). Unfolding by GuHCl is cooperative for both the dioxygenases, with the NDO-R curve offset to an 0.5 M higher concentration of GuHCl (Fig. 2C).
Structure of free and indole-bound forms of NDO-R.
Two structures of NDO from Rhodococcus have been determined by X-ray crystallography. Structure 1, the native enzyme, has been refined to 2.0-Å resolution with noncrystallographic symmetry restraints to an R-factor of 19.1% and R-free of 22.4%. The final model contains α3β3 subunits with a total of 441 residues in each α subunit and 167 residues in each β subunit that could be modeled clearly in the electron density. Thirty amino acids at the C terminus of the α subunit and 12 amino acids at the N terminus of the β subunit were not visible in the electron density. The refined structure also has 1,119 water molecules in the asymmetric unit. Ninety-nine percent of the residues are in the allowed regions of the Ramachandran plot. The notable exception is serine 67 in the α subunit. This residue has strained phi-psi angles but is well defined in electron density. This serine is at the tip of the first turn between two β strands in the Rieske domain. Based on sequence alignment, this residue is a glycine in most of the other RO sequences.
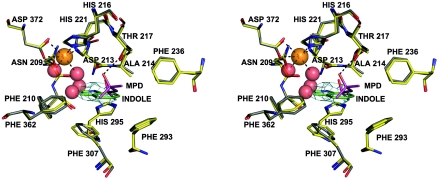
In structure 1, in contrast to the structures of NDO-P (5, 26, 27), there are two water molecules bound to the active site iron (Fig. 3). The aspartic acid (D372) coordinating the mononuclear iron is bound in a bidentate fashion (both OD1 and OD2 are 2.3 Å from the mononuclear iron). This results in an octahedral coordination for the active site iron similar to what has also been seen in the structure of reduced 2-oxoquinoline 8-monooxygenase (38). The octahedral coordination suggests that the active site iron, which is in a similar chemical environment as the NDO-P iron, is in the Fe3+ state (26).
FIG. 3.
Stereo view of the active site of NDO-R. MPD from the buffer is found bound in the active site of the native enzyme (structure 1), and indole is bound in the substrate-soaked crystal structure (structure 2). Structure 1 is in gray and structure 2 in yellow. Four water molecules found in the cavity of the active site in structure 1 are shown as red spheres. Two of these waters form part of the octahedral coordination of the iron (dashed lines). A hydrogen bond between MPD and T217 (dashed line) suggests that polar groups on the nonhydroxylating ring of the substrates may interact with T217. A 2FoFc omit map for indole contoured at 0.8 sigma is shown in cyan.
Structure 2, an enzyme-substrate complex with indole bound, has been refined to a resolution of 3.0 Å. The bound indole had a temperature factor of 72 Å2, and a 2FoFc omit map contoured at 0.8 sigma is shown in Fig. 3. This structure contains 351 water molecules and has a final R-factor and R-free of 28.7% and 30.0%, respectively. This structure is discussed only in the context of indole binding at the active site. The root mean square deviation (RMSD) of the Cα atoms is 0.3 Å between the free enzyme and the indole-bound enzyme structures, suggesting very little change in the overall structure of the protein with respect to the native structure.
The overall structure of the enzyme is very similar to NDO-P and BPDO-R in spite of low (∼30%) sequence identity. A superposition of the Cα atoms of NDO-R and NDO-P gives an RMSD of 1.58 Å for 359 Cα atoms of the α subunit and 1.51 Å for 157 Cα atoms of the β subunit. BPDO-R could be superimposed on NDO-R, resulting in an RMSD of 1.73 for 370 Cα atoms of the α subunit and 0.93 Å for 161 Cα atoms of the β subunit.
Structural basis for higher thermostability of NDO-R (and proposed thermostability of BPDO-R).
Upon hexamer formation, the surface area of the individual monomers rendered inaccessible to the solvent is significantly higher for NDO-R (13,200 Å2 or 18%) and BPDO-R (13,500 Å2 or 19.3%) than for NDO-P (9,700 Å2 or 12%). The differences between NDO-R/BPDO-R and NDO-P in percent surface area buried are reflected in the differences in the volume of the central cavities. These central cavities are formed around the threefold axis of the hexamer when the subunits oligomerize. The biggest cavity in the hexamer is formed by the α subunits and the next biggest by the β subunits. The volumes of these central cavities are 6,320 Å3 and 447 Å3 in NDO-P, 4,055 Å3 and 496 Å3 in NDO-R, and 3,966 Å3 and 70 Å3 in BPDO-R, respectively. Salt bridges and hydrogen bonds in networks across subunits (discussed below in this section) are found in a greater number in the central cavity region for NDO-R and BPDO-R than for NDO-P. These extra interactions block off the cavities much nearer to the cavity openings for NDO-R and BPDO-R, leading to a greater reduction in accessible surface area and central cavity volume compared to NDO-P.
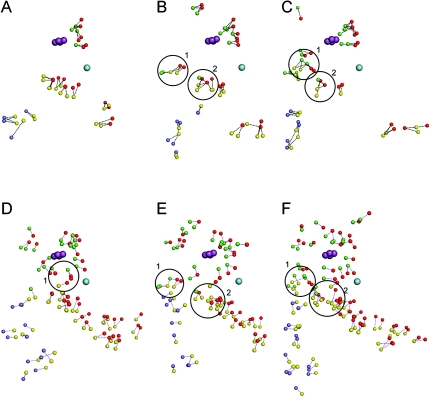
Stability analysis based only on the number of intrasubunit salt bridges and hydrogen bonds is inconclusive; either the numbers for these interactions are comparable or a decrease of the number of one type of interaction is compensated for by an increase in the other (Table 2). A visual inspection of the intrasubunit interactions in the structures of the three enzymes is also difficult to interpret. However, an analysis of the α—α, β—β, and α—β interfaces of these enzymes is quite revealing. The intersubunit salt bridges and hydrogen bonds are not only more numerous in NDO-R and BPDO-R (Table 2) but also form a greater number of networks (i.e., more than two interacting residues) than NDO-P. The distribution of these interface interactions can be seen in Fig. 4 for the three enzymes. Salt bridge and hydrogen bond networks are believed to contribute much more to the stability of a protein than similar interactions occurring only in pairs (31, 44, 57). NDO-R has five and BPDO-R has six salt bridge networks at the interfaces with three to five residues interacting in each network, whereas NDO-P has just one network with three interacting residues. While all the interface salt bridges in NDO-P are formed between only two subunits, two interface networks in NDO-R and BPDO-R are formed by residues from three subunits (circled in Fig. 4B and C). Residues in this case belong to the αi, βi, and αi + 1 subunits (where i + 1 is the neighboring subunit of i in the α3 and β3 homotrimers) and are located in the same region in both NDO-R and BPDO-R. In order of αi, βi, and αi + 1, the residues forming the first network are E374—R632—R97 in NDO-R and E380—R139—R107 in BPDO-R. These residues are also conserved in the sequence alignment. The intersubunit residues in the second network are R381—D634—R196 in NDO-R and E384—E141—R104 in BPDO-R. Of these, the βi residues line up in the sequence alignment.
TABLE 2.
Intra- and intersubunit optimal hydrogen bond values and numbers of salt bridges and salt bridge residues calculated using WHAT IF
| Interaction location | Hydrogen bond value
|
No. of salt bridges/no. of salt bridge residues
|
||||
|---|---|---|---|---|---|---|
| NDO-P | NDO-R | BPDO-R | NDO-P | NDO-R | BPDO-R | |
| Intrasubunit | ||||||
| αi | 276.8 | 223.3 | 260.4 | 77/39 | 104/49 | 73/35 |
| βi | 119.1 | 74.7 | 109.9 | 58/25 | 52/22 | 54/21 |
| Total | 395.9 | 298.0 | 370.3 | 135/64 | 156/71 | 127/56 |
| Homodimer interface | ||||||
| αi − αi+ 1 | 9.2 | 11.4 | 13.3 | 8/4 | 19/7 | 18/8 |
| βi − βi+ 1 | 5.1 | 4.2 | 11.1 | 6/3 | 10/5 | 13/4 |
| Total | 14.3 | 15.6 | 24.4 | 14/7 | 29/12 | 31/12 |
| Heterodimer interface | ||||||
| αi − βi | 18.9 | 12.7 | 16.6 | 21/7 | 20/8 | 12/5 |
| αi − βi+ 1 | 0 | 2.3 | 3.3 | 0/0 | 2/1 | 9/3 |
| αi − βi+ 2 | ||||||
| Total | 18.9 | 15.0 | 19.9 | 21/7 | 22/9 | 21/8 |
FIG. 4.
Interface salt bridges and hydrogen bonds for NDO-P (A and D), NDO-R (B and E), and BPDO-R (C and F), respectively. The small spheres represent nitrogen and oxygen atoms of subunits αi (red), βi (yellow), αi + 1 (green), and βi + 1 (blue), which interact to form salt bridges and hydrogen bonds (dashed lines).The mononuclear iron belonging to αi is shown as a sphere in cyan, and the Rieske cluster belonging to αi + 1 is shown as spheres in purple. NDO-R and BPDO-R have two salt bridge networks (circled areas 1 and 2 in panels B and C) which involve conserved residues from three subunits. NDO-P has one hydrogen bond network (circled area 1 in panel D) which involves residues from three subunits; NDO-R and BPDO-R have two hydrogen bond networks (circled areas 1 and 2 in panels E and F) which involve residues from three subunits. NDO-P is missing network 2 in panel D, the 8- to 10-residue network, which is present in the other two enzymes. NDO-R and BPDO-R in general have larger networks and more widespread interacting pairs.
There are two hydrogen bond networks across interfaces in NDO-R and BPDO-R that involve residues from three subunits, αi, βi, αi + 1, whereas NDO-P has one such network (circled in Fig. 4D to F). The hydrogen bond network circled as “1” for NDO-R and BPDO-R has 8 to 10 residues involved in interactions, and this network is missing in NDO-P.
These data suggest that, under identical perturbant conditions, NDO-P is more likely to dissociate and unfold than NDO-R and BPDO-R.
DISCUSSION
Thermal and chemical stability.
NDO-R appears to be marginally more stable than NDO-P with respect to chemical denaturants such as urea and GuHCl, but its resilience towards thermal unfolding is much more pronounced. This is not an exclusive case; while increased chemical stability of a protein generally goes hand in hand with increased thermostability, there are examples where chemical stability of a protein with a high Tm, such as thermophilic esterase from Archaeoglobus fulgidus, aspartate aminotransferase from Sulfolobus solfataricus, or glucoamylase from Aspergillus awamori, may be similar to or less than that of their lower-Tm homologs/mutants (1, 7, 13). Unfolding behaviors have also been found to vary greatly between closely related proteins. For example, in the case of serum albumin from different mammals with greater than 70% sequence homology, some were shown to unfold through at least one stable intermediate, while the others showed a single transition from the folded to the unfolded state (29).
While the difference in protein thermostability can often be explained by a comparison of the related structures (for example, the extra salt bridge networks for NDO-R in this work), differential stability towards chemical perturbants or comparison of thermal and chemical stability has been more difficult to interpret using sequence and structures (3, 11, 22).
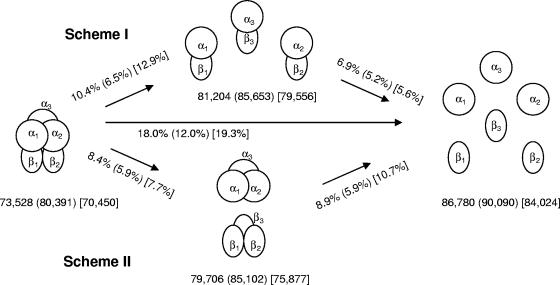
Dissociation schemes for the hexamer of NDO-P, NDO-R, and BPDO-R.
The β subunit (in ROs) has no direct interaction with the active site but has been known to affect substrate specificity in toluate, toluene, and biphenyl dioxygenases from gram-negative bacteria (49). The β subunit in NDO-P, NDO-R, and BPDO-R, however, has no known effect on enzyme specificity and is believed to provide only structural stability to the enzyme. Indeed, many dioxygenases, such as those of the phthalate family, lack the β subunit altogether (19). The recently solved structure of the oxygenase component of 2-oxoquinoline 8-monooxygenase (38) also lacks the β subunit. In the folding of oligomeric proteins, subunit association may occur during the monomer folding process or after monomer folding is complete (15). It is likely that oligomeric proteins like NDO unfold in a similar manner with subunit dissociation preceding complete unfolding of the monomers. Two schemes of subunit dissociation are presented in Fig. 5, where the α3β3 hexamers dissociate into three αβ heterodimers (scheme I) or into α3 and β3 homotrimers (scheme II) before disassembling to monomers and unfolding completely. Scheme II is interesting because a stable α3 homotrimer could potentially retain catalytic properties and be an active enzyme. For NDO-R and BPDO-R, scheme II is supported by the fact that the total hydrogen bond value and the number of salt bridges at the homodimer interfaces are greater than at the heterodimer interfaces (Table 2). Additionally, the percentage of interface buried surface area exposed to solvent upon dissociation is also appreciably smaller in scheme II for NDO-R and BPDO-R (Fig. 5). In the case of NDO-P, the total hydrogen bond value is smaller for the homodimer interfaces while the number of salt bridges is the same. Furthermore, the percentage of interface buried surface area exposed to the solvent upon dissociation of the NDO-P hexamer is only marginally smaller in scheme II. While not conclusive, these analyses indicate that the dissociation of NDO-R is similar to BPDO-R and likely to favor the “head” (α3) and “stem” (β3) separation of the mushroom-shaped hexamer during unfolding. Further studies could show if the stable intermediate formed upon thermal unfolding is indeed a functional “head.” Neither of the two simplistic schemes considered here seems significantly favored over the other for the dissociation of the NDO-P hexamer.
FIG. 5.
Cartoon showing likely schemes of dissociation of the α3β3 hexamer to trimers, dimers, and monomers. The accessible surface area calculated using a 1.4-Å probe with SYBYL (Tripos) is shown in Å2. The percent increase in accessible surface area upon exposure of the interface buried surface area to the solvent is given next to the arrows. All the numbers are in the order NDO-R (NDO-P) [BPDO-R].
Comparison of NDO-P, NDO-R, and BPDO-R structures.
Insertions and deletions in the exposed loop regions account for a number of differences between the two NDO structures. The most significant differences occur on the outside of the mushroom. The longer N terminus of NDO-R folds into an alpha helix, while it is a meandering loop in NDO-P. The C-terminal alpha helix present in NDO-P is absent in NDO-R. The other differences occur in extensions outside of the core domain made of β strands. The regions are amino acids 182 to 188, 228 to 239, 255 to 293, and 310 to 316 of NDO-R.
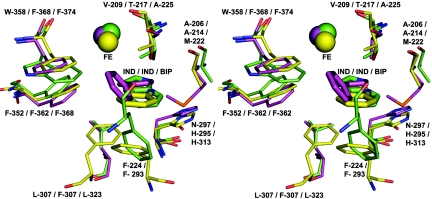
Data on substrate specificity profile for NDO-R are not available due to difficulties in purification of the other components of this dioxygenase system as well as nonexpression in other hosts such as Escherichia coli. In the absence of biochemical data, the changes seen in some of the loops for NDO-R and NDO-P cannot be rationalized. However, one effect of the difference in the loops is the change in the substrate cavity. It has been reported in the structure of NDO-P that two loops cover the active site and form a lid with residues 253 to 265 and 223 to 240 (27). The corresponding loops, 255 to 294 and 228 to 239 in NDO-R, are substantially different, and the entrance to the active site cavity is larger. The loops that cover the entrance to the active site are part of the nonconserved regions, and the effect of the differences in this region on substrate selectivity is not known. Some of the residues present in the variable loops, such as F293, contribute to the active site pocket that binds to the substrate. F293 of NDO-R is very similarly placed to F224 in the structure of NDO-P (Fig. 6). F224 in NDO-P belongs to the region described as the lid that covers the top of the active site. Table 3 lists the residues that are nearly equivalent in their interaction with indole as the substrate in the two structures. Also listed in Table 3 are structurally equivalent residues of BPDO-R. Of these residues, the ones that seem likely to affect substrate specificity and binding are shown in Fig. 6.
FIG. 6.
View of the active site of NDO-P (from PDB id 1EG9) in green, NDO-R (structure 2) in yellow, and BPDO-R (from PDB id 1ULJ) in purple. The figure illustrates the positions of the bound substrate, the active site mononuclear iron, and the side chains of the residues that interact (similarly or differently) with the bound substrate as discussed in the text.
TABLE 3.
Residues in the active site of NDO-R (structure 2) which interact with indole and residues in NDO-P (1EG9.pdb) and BPDO-R (1ULJ.pdb) that are in similar spatial orientations
| NDO-R residue | Residue in similar spatial orientation
|
|
|---|---|---|
| NDO-P | BPDO-R | |
| N209 | N201 | Q217 |
| F210 | F202 | F218 |
| D213 | D205 | D221 |
| A214 | A206 | M222 |
| H216 | H208 | H224 |
| T217 | V209 | A225 |
| F293 | F224 | —a |
| H295 | N297 | H313 |
| F307 | L307 | L323 |
| F362 | F352 | F368 |
| F368 | W358 | F374 |
—, no corresponding residue.
One of the differences in the active site is the alteration of a V209 in NDO-P to T217 in NDO-R. In BPDO-R, the corresponding residue is A225. Extensive mutagenesis studies have been carried out on NDO-P to alter the regio- and stereospecificity of product formation (46, 48). None of these changes correspond to a mutation in V209. This residue in NDO-R interacts with the phenyl ring that is farther away from the iron of the substrate. In structure 1, a bound MPD molecule (from the precipitant used to crystallize the protein) at the active site is stabilized by a hydrogen bond between OG1 of T217 and O2 of MPD (Fig. 3). The presence of a polar group here would suggest a difference in the substrate selection profile of NDO-R. Structure 2, however, suggests that the substrates could bind in a similar fashion. Indole binds in a very similar position in both NDO-P (5) and NDO-R in the binary complex (Fig. 3). While the second ring is stabilized primarily by hydrophobic interactions in NDO-P, it is quite possible that there is a hydrogen bond between the OG1 atom of the threonine and the aromatic ring of the substrate. The distance between the OG1 and the plane of the ring is approximately 3.7 Å. This is very close to the minimum distance for a hydrogen bond interaction between a donor and an aromatic ring calculated by Levitt and Perutz (36). One can argue that the effect of the valine-to-threonine alteration changes the mode of stabilization of the ring, with little effect on the product specificity profile.
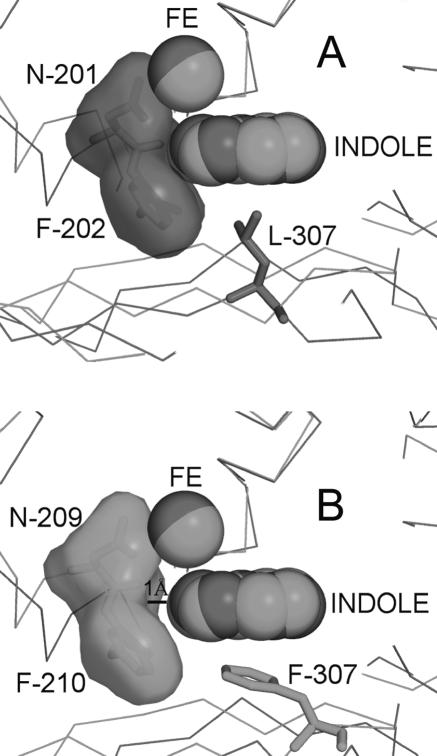
Other residues that are different in the active site of NDO-R include F307, F368, and H295. F307 forms a stacking interaction (π-π) with indole in NDO-R while the equivalent residues in NDO-P (L307) and BPDO-R (L323) have hydrophobic interactions. In NDO-R, there is a gap of 1 Å between the “head” of the bound indole and the F210 at the end of the cavity. In the case of NDO-P, indole is in van der Waals contact with F202 at the end of the cavity (Fig. 7). Docking of indole in the active sites of NDO-P and NDO-R using AutoDock 3.0.5 (40) (data not presented) reproduced the experimental pose of indole binding for NDO-P; however, indole docked ∼1 Å deeper into the active site of NDO-R. AutoDock does not implicitly account for π interactions, and docking in the case of NDO-R possibly failed due to this π-π interaction being unaccounted for. It is likely that the stacking interaction with F307 in NDO-R is at least partially responsible for holding the substrate in position.
FIG. 7.
NDO-P has an active site cavity (A) that is 1 Å shallower than NDO-R (B). However, indole is still held in the same place next to the mononuclear iron (structure 2) similar to the indole in NDO-P (PDB id 1EG9). The π-π interaction between F307 in NDO-R and the nonhydroxylated ring of indole seemingly prevents indole from burying itself deeper into the cavity.
In BPDO-R, the nonhydroxylated ring of biphenyl appears to have a π-π interaction with F368 (corresponding residues are F352 and F362 in NDO-P and NDO-R, respectively) and a sulfur-π (S-π) interaction with M222 (A206 and A214 in NDO-P and NDO-R, respectively) (Fig. 6).
From Table 3 and Fig. 6, it is clear that, while most interactions of the two NDO enzymes with indole are conserved, the effect of these on substrate selectivity is not known at present. It is interesting that most of the variations result in different modes of interaction with the phenyl ring, the one farther away from the mononuclear iron. The ring that gets dihydroxylated is in a very similar position, suggesting that the possible dihydroxylation mechanism of NDO-R is similar to that of NDO-P (26).
These data clearly suggest that there are many possible similarities in substrate specificity between NDO-R and NDO-P. However, until a reasonable level of heterologous expression and a better understanding of electron transport in this system are achieved, reliable comparisons of substrate specificity backing up our observations may not be evident.
Acknowledgments
Use of the Argonne National Laboratory SBC-CAT beamlines at the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Biological and Environmental Research, under contract no. W-31-109-ENG-38. Structure 2 data were collected at beamline 17-ID in the facilities of the IMCA-CAT at the Advanced Photon Source. These facilities are supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Illinois Institute of Technology (IIT), executed through IIT's Center for Synchrotron Radiation Research and Instrumentation. S.R. and his lab are funded by U.S. Public Health Services grant GM62904. D.A.L. and C.C.R.A. acknowledge the QUESTOR Center. L.G. was funded by a graduate student fellowship from the Center for Biocatalysis and Bioprocessing (CBB), University of Iowa.
We thank Eric Brown and Daniel Ferraro for their help in analyzing the interactions and in rendering of figures. Ruslan Sanishvili at the SBC-CAT helped with data collection.
REFERENCES
- 1.Arnone, M. I., L. Birolo, S. Pascarella, M. V. Cubellis, F. Bossa, G. Sannia, and G. Marino. 1997. Stability of aspartate aminotransferase from Sulfolobus solfataricus. Protein Eng. 10:237-248. [DOI] [PubMed] [Google Scholar]
- 2.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohm, G., and R. Jaenicke. 1994. Relevance of sequence statistics for the properties of extremophilic proteins. Int. J. Pept. Protein Res. 43:97-106. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, D. R., N. D. Sharma, and C. C. Allen. 2001. Aromatic dioxygenases: molecular biocatalysis and applications. Curr. Opin. Biotechnol. 12:564-573. [DOI] [PubMed] [Google Scholar]
- 5.Carredano, E., A. Karlsson, B. Kauppi, D. Choudhury, R. E. Parales, J. V. Parales, K. Lee, D. T. Gibson, H. Eklund, and S. Ramaswamy. 2000. Substrate binding site of naphthalene 1,2-dioxygenase: functional implications of indole binding. J. Mol. Biol. 296:701-712. [DOI] [PubMed] [Google Scholar]
- 6.Cerniglia, C. E. 1992. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3:351-368. [Google Scholar]
- 7.Chen, H. M., Y. Li, T. Panda, F. U. Buehler, C. Ford, and P. J. Reilly. 1996. Effect of replacing helical glycine residues with alanines on reversible and irreversible stability and production of Aspergillus awamori glucoamylase. Protein Eng. 9:499-505. [DOI] [PubMed] [Google Scholar]
- 8.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowtan, K. D. 1993. Improvement of macromolecular electron-density maps by the simultaneous application of real and reciprocal space constraints. Acta Crystallogr. D Biol. Crystallogr. 49:148-157. [DOI] [PubMed] [Google Scholar]
- 11.Dalhus, B., M. Saarinen, U. H. Sauer, P. Eklund, K. Johansson, A. Karlsson, S. Ramaswamy, A. Bjork, B. Synstad, K. Naterstad, R. Sirevag, and H. Eklund. 2002. Structural basis for thermophilic protein stability: structures of thermophilic and mesophilic malate dehydrogenases. J. Mol. Biol. 318: 707-721. [DOI] [PubMed] [Google Scholar]
- 12.DeLano, W. L. The PyMOL Molecular Graphics System. DeLano Scientific LLC, San Carlos, CA. http://www.pymol.org.
- 13.Del Vecchio, P., G. Graziano, V. Granata, G. Barone, L. Mandrich, M. Rossi, and G. Manco. 2002. Denaturing action of urea and guanidine hydrochloride towards two thermophilic esterases. Biochem. J. 367:857-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong, X., S. Fushinobu, E. Fukuda, T. Terada, S. Nakamura, K. Shimizu, H. Nojiri, T. Omori, H. Shoun, and T. Wakagi. 2005. Crystal structure of the terminal oxygenase component of cumene dioxygenase from Pseudomonas fluorescens IP01. J. Bacteriol. 187:2483-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyle, S. M., E. H. Braswell, and C. M. Teschke. 2000. SecA folds via a dimeric intermediate. Biochemistry 39:11667-11676. [DOI] [PubMed] [Google Scholar]
- 16.Finnerty, W. R. 1992. The biology and genetics of the genus Rhodococcus. Annu. Rev. Microbiol. 46:193-218. [DOI] [PubMed] [Google Scholar]
- 17.Friemann, R., M. M. Ivkovic-Jensen, D. J. Lessner, C. L. Yu, D. T. Gibson, R. E. Parales, H. Eklund, and S. Ramaswamy. 2005. Structural insight into the dioxygenation of nitroarene compounds: the crystal structure of nitrobenzene dioxygenase. J. Mol. Biol. 348:1139-1151. [DOI] [PubMed] [Google Scholar]
- 18.Furusawa, Y., V. Nagarajan, M. Tanokura, E. Masai, M. Fukuda, and T. Senda. 2004. Crystal structure of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. strain RHA1. J. Mol. Biol. 342:1041-1052. [DOI] [PubMed] [Google Scholar]
- 19.Gibson, D. T., and R. E. Parales. 2000. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 11:236-243. [DOI] [PubMed] [Google Scholar]
- 20.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305-308. [DOI] [PubMed] [Google Scholar]
- 21.Habe, H., and T. Omori. 2003. Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci. Biotechnol. Biochem. 67: 225-243. [DOI] [PubMed] [Google Scholar]
- 22.Hecht, K., A. Wrba, and R. Jaenicke. 1989. Catalytic properties of thermophilic lactate dehydrogenase and halophilic malate dehydrogenase at high temperature and low water activity. Eur. J. Biochem. 183:69-74. [DOI] [PubMed] [Google Scholar]
- 23.Hooft, R. W., C. Sander, and G. Vriend. 1996. Positioning hydrogen atoms by optimizing hydrogen-bond networks in protein structures. Proteins 26: 363-376. [DOI] [PubMed] [Google Scholar]
- 24.Irvine, V. A., L. A. Kulakov, and M. J. Larkin. 2000. The diversity of extradiol dioxygenase (edo) genes in cresol degrading rhodococci from a creosote-contaminated site that express a wide range of degradative abilities. Antonie Leeuwenhoek 78:341-352. [DOI] [PubMed] [Google Scholar]
- 25.Jones, T. A., J. Y. Zou, S. W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 26.Karlsson, A., J. V. Parales, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 2003. Crystal structure of naphthalene dioxygenase: side-on binding of dioxygen to iron. Science 299:1039-1042. [DOI] [PubMed] [Google Scholar]
- 27.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571-586. [DOI] [PubMed] [Google Scholar]
- 28.Kleywegt, G. J., and T. A. Jones. 1994. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 50:178-185. [DOI] [PubMed] [Google Scholar]
- 29.Kosa, T., T. Maruyama, and M. Otagiri. 1998. Species differences of serum albumins: II. Chemical and thermal stability. Pharm. Res. 15:449-454. [DOI] [PubMed] [Google Scholar]
- 30.Kumar, S., and R. Nussinov. 2002. Relationship between ion pair geometries and electrostatic strengths in proteins. Biophys. J. 83:1595-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar, S., and R. Nussinov. 1999. Salt bridge stability in monomeric proteins. J. Mol. Biol. 293:1241-1255. [DOI] [PubMed] [Google Scholar]
- 32.Kumar, S., C. J. Tsai, and R. Nussinov. 2000. Factors enhancing protein thermostability. Protein Eng. 13:179-191. [DOI] [PubMed] [Google Scholar]
- 33.Larkin, M. J., C. C. Allen, L. A. Kulakov, and D. A. Lipscomb. 1999. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. J. Bacteriol. 181:6200-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larkin, M. J., M. R. De, L. A. Kulakov, and I. Nagy. 1998. Applied aspects of Rhodococcus genetics. Antonie Leeuwenhoek 74:133-153. [DOI] [PubMed] [Google Scholar]
- 35.Lessner, D. J., G. R. Johnson, R. E. Parales, J. C. Spain, and D. T. Gibson. 2002. Molecular characterization and substrate specificity of nitrobenzene dioxygenase from Comamonas sp. strain JS765. Appl. Environ. Microbiol. 68:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levitt, M., and M. F. Perutz. 1988. Aromatic rings act as hydrogen bond acceptors. J. Mol. Biol. 201:751-754. [DOI] [PubMed] [Google Scholar]
- 37.Malik, Z. A., C. C. Allen, L. Gakhar, D. A. Lipscomb, M. J. Larkin, and S. Ramaswamy. 2002. Crystallization and preliminary X-ray diffraction analysis of naphthalene dioxygenase from Rhodococcus sp. strain NCIMB 12038. Acta Crystallogr. D Biol. Crystallogr. 58:2173-2174. [DOI] [PubMed] [Google Scholar]
- 38.Martins, B. M., T. Svetlitchnaia, and H. Dobbek. 2005. 2-Oxoquinoline 8-monooxygenase oxygenase component: active site modulation by Rieske-[2Fe-2S] center oxidation/reduction. Structure (Cambridge) 13:817-824. [DOI] [PubMed] [Google Scholar]
- 39.Mason, J. R., and R. Cammack. 1992. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu. Rev. Microbiol. 46:277-305. [DOI] [PubMed] [Google Scholar]
- 40.Morris, G. M., D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew, and A. J. Olson. 1998. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19:1639-1662. [Google Scholar]
- 41.Moser, R., and U. Stahl. 2001. Insights into the genetic diversity of initial dioxygenases from PAH-degrading bacteria. Appl. Environ. Microbiol. 55:609-618. [DOI] [PubMed] [Google Scholar]
- 42.Mueller, J. G., P. J. Chapman, and P. H. Pritchard. 1989. Creosote contaminated sites—their potential for bioremediation. Environ. Sci. Technol. 23:1197-1201. [Google Scholar]
- 43.Murshudov, G. N. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53: 240-255. [DOI] [PubMed] [Google Scholar]
- 44.Musafia, B., V. Buchner, and D. Arad. 1995. Complex salt bridges in proteins: statistical analysis of structure and function. J. Mol. Biol. 254:761-770. [DOI] [PubMed] [Google Scholar]
- 45.Navaza, J. 1994. AMORE—an automated package for molecular replacement. Acta Crystallogr. A 50:157-163. [Google Scholar]
- 46.Parales, R. E., K. Lee, S. M. Resnick, H. Jiang, D. J. Lessner, and D. T. Gibson. 2000. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J. Bacteriol. 182: 1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parales, R. E., J. V. Parales, and D. T. Gibson. 1999. Aspartate 205 in the catalytic domain of naphthalene dioxygenase is essential for activity. J. Bacteriol. 181:1831-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parales, R. E., S. M. Resnick, C. L. Yu, D. R. Boyd, N. D. Sharma, and D. T. Gibson. 2000. Regioselectivity and enantioselectivity of naphthalene dioxygenase during arene cis-dihydroxylation: control by phenylalanine 352 in the alpha subunit. J. Bacteriol. 182:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramaswamy, S. 2001. Naphthalene 1,2-dioxygenase, p. 613-621. In A. Messerschmidt, R. Huber, K. Wieghardt, and T. Poulos (ed.), Handbook of metalloproteins. Wiley, Chichester, United Kingdom.
- 50.Read, R. J. 1986. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A 42:140-149. [Google Scholar]
- 51.Tripos. 2004. SYBYL 6.9. Tripos Inc., St. Louis, Mo.
- 52.Vriend, G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 8:52-56, 29. [DOI] [PubMed] [Google Scholar]
- 53.Warhurst, A. M., and C. A. Fewson. 1994. Biotransformations catalyzed by the genus Rhodococcus. Crit. Rev. Biotechnol. 14:29-73. [DOI] [PubMed] [Google Scholar]
- 54.Wolfe, M. D., and J. D. Lipscomb. 2003. Hydrogen peroxide-coupled cis-diol formation catalyzed by naphthalene 1,2-dioxygenase. J. Biol. Chem. 278: 829-835. [DOI] [PubMed] [Google Scholar]
- 55.Wolfe, M. D., J. Parales, K. Lee, D. T. Gibson, and J. D. Lipscomb. 1999. Substrate binding to the mononuclear ferrous ion of the Rieske dioxygenase naphthalene 1,2-dioxygenase from Pseudomonas sp. NCIB 9816-4. J. Inorg. Biochem. 74:339. [Google Scholar]
- 56.Wolfe, M. D., J. V. Parales, D. T. Gibson, and J. D. Lipscomb. 2001. Single turnover chemistry and regulation of O2 activation by the oxygenase component of naphthalene 1,2-dioxygenase. J. Biol. Chem. 276:1945-1953. [DOI] [PubMed] [Google Scholar]
- 57.Xiao, L., and B. Honig. 1999. Electrostatic contributions to the stability of hyperthermophilic proteins. J. Mol. Biol. 289:1435-1444. [DOI] [PubMed] [Google Scholar]