Abstract
Inactivation of hemB in Staphylococcus aureus strain Newman resulted in a small-colony phenotype and was accompanied by an altered expression pattern of global regulators and control of virulence factor production. Transcription profiles followed over 15 h by Northern blot analyses revealed that transcripts of the global regulators arl, rot, sae, sarR, sarS, srr, svrA, and sigB disappeared after the exponential phase and that both agr transcripts were completely absent in the hemB mutant. Apart from a general concentration of transcriptional activity to the exponential phase, premature gene expression was observed for rot, hla, and spa. Nevertheless, reported σB-dependent transcripts, such as sarC and clfA, were produced throughout the 15-h growth period monitored. The absence of these transcripts in a hemB sigB double mutant demonstrated their dependence on σB and indicated an unexpected, permanent σB activity in the hemB mutant. Variations in the extents of the directly σB-controlled asp23, rsbVW-sigB, and sarC transcripts argue for additional factors modulating σB activity. This study provides the first extended synopsis of the transcriptional patterns of different regulators over the entire growth cycle in the widely used Newman strain.
Staphylococcus aureus is an opportunistic pathogen and one of the major causes of nosocomial and community-acquired infections. Adaptation to changing conditions and a wide spectrum of diseases ranging from superficial to serious life-threatening infections and toxicoses is conferred by a formidable arsenal of pathogenicity and virulence factors. Besides their remarkable ability to acquire multiple resistance determinants, staphylococci adopt additional strategies to evade antibiotic challenge and host defenses. The formation of small-colony variants (SCVs) and the capacity of these SCVs to persist intracellularly are regarded as survival mechanisms employed by staphylococci (55). SCVs are recovered from clinical specimens, particularly from patients with chronic, persisting, and/or relapsing infections (54, 61). Characteristically, SCVs grow slowly and form tiny, unpigmented, nonhemolytic colonies on solid medium. Clinical SCVs are often auxotrophic for hemin or menadione (1, 61), compounds involved in the synthesis of the electron carriers cytochrome and menaquinone, respectively, and therefore have an impaired electron transport. Disruption of the hemB gene, encoding the essential aminolevulinic acid dehydratase for hemin biosynthesis, leads to a stable S. aureus model of SCV, avoiding undefined genetic backgrounds and reversible phenotypes, which occur in clinical SCVs (3, 10, 31, 58, 66).
An interrupted electron transport chain causes accumulation of the reducing equivalents NAD(P)H and FADH2, a low membrane potential, and low ATP concentration (34, 53). Metabolic pathways dependent on the availability of the oxidizing equivalents NAD(P)+ and FAD are inhibited, which reduces the range of carbohydrate utilization. Several genes involved in glycolysis and fermentation are up-regulated in a COL hemB mutant, whereas tricarbonic acid cycle enzymes are down-regulated, manifesting divergent metabolic activity from that of the wild type and leading to a lower energy production (34). Consequently, the ATP-requiring biosynthesis of proteins, cell wall, or nucleotides is limited and finally cell growth stalls. However, the reduced transmembrane potential protects hemB mutants and clinical SCVs from cationic antibiotics (3), slow growth lowers the efficacy of substances targeting metabolically active cells, and the low toxin production allows them to persist in the host cell, which is a relatively protected environment (2).
Controlled and coordinate expression of virulence determinants during S. aureus infection is regulated by a multitude of global regulators (12, 48). The major accessory gene regulator locus agr represses via two divergently transcribed mRNAs (RNAII and RNAIII) in post-exponential phase cell-surface associated proteins, such as protein A (spa) or fibronectin-binding protein A (fnbA), and activates exotoxin production, e.g., α-toxin (hla). RNAII encodes a quorum-sensing two-component system, which upon increasing cell density induces the expression of the agr effector molecule RNAIII (29, 45, 50, 60). Staphylococcal accessory regulator A (sar) encodes three overlapping and differentially regulated transcripts (sarB, sarC, and sarA). While sarB and sarA are σA dependent and transcribed mainly during exponential growth, the σB-dependent sarC transcript appears later and increases towards stationary phase. However, all three transcripts encode the global regulator SarA (4, 15, 39), positively influencing fnbA and hla (13, 68) but inhibiting spa expression (20). σB, an alternative sigma factor mediating stress response, controls target genes via σB-dependent promoters, as in the case of sarC. Indirectly, σB inhibits agr and hla (7, 8, 13, 27); on the other hand, σB positively influences fnbA expression (7, 46). Additional two-component systems and transcription factors take part in a complex regulatory network, further modulating the above-described regulators and virulence determinants. As a result, in vitro, wild-type S. aureus first expresses surface-associated proteins, such as FnbA, followed by secreted protective proteins, like protein A, as well as exoenzymes, exemplified by α-toxin. Thereby, the transition from a colonizing to an invasive behavior is represented in a growth-phase-dependent manner (56). hemB mutants were found to express protein A and α-toxin only weakly (31, 66), whereas the adhesion factor FnbA and its homologue FnbB, as well as the fibrinogen-binding clumping factor A (ClfA), were up-regulated (65). The surface-associated virulence determinants FnbA and ClfA are important for colonization, escape from host defense, and persistence (44, 52, 62). Also of interest, clfA expression requires the alternative sigma factor σB (18, 46, 47), for which reason it seemed likely that this stress response regulator may be active in hemB mutants. However, the overall sar mRNA levels, including that of the σB-dependent sarC transcript, were reported to be reduced in the 8325-4 hemB mutant (65). Since no distinction was made between the three overlapping, SarA encoding, but differentially regulated sarB, sarC, and sarA transcripts originating from the sar locus, σB activity in hemB mutants remained open.
In addition to σB, none of the staphylococcal regulators have been analyzed in hemB mutants in detail. We therefore investigated in this study the expression not only of σB but also of most known global regulators and selected virulence determinants in strain Newman and its hemB mutant over an extended time period. As several of the analyzed loci have multiple, differently controlled promoters, Northern blot analyses were preferred to reverse transcription-PCR techniques to conveniently identify individual transcripts. For monitoring σB activity in the hemB mutant, a luciferase reporter construct under the control of a σB-dependent promoter was used. To determine the impact of σB absence, a hemB sigB double mutant was constructed and the expression profiles of σB-dependent transcripts were compared to those in the hemB background.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains were constructed in a Newman background (17). Strain MS17 was obtained by transducing the hemB::ermB mutation with phage 85 (5) from the strain 8325-4 derivative I10 (66) into the parent strain MB79, which is a Newman strain with an integrated reporter construct carrying the firefly luciferase gene (luc+) fused to the three σB-dependent alkaline shock protein 23 promoters (asp23P::luc+) (23). Strain MS62 was constructed by transducing the tetL-linked sigB1(Am) mutation from the 8325-4 derivative GP266 (6) into strain III33, a hemB::ermB mutant of strain Newman (31). The constructs were confirmed by PCR, Southern blot analysis, and SmaI-pulsed-field gel electrophoresis chromosomal pattern.
Bacteria were grown aerobically at 37°C. Agar plates containing Luria-Bertani broth (LB), Columbia blood agar base with sheep blood, or Mueller-Hinton (MH) broth were used for cultivation on solid media. Liquid cultures were grown in LB, and good aeration was assured by vigorously shaking flasks with an air-to-liquid ratio of at least 4. Transductants were selected on 10 μg/ml tetracycline and 2.5 μg/ml erythromycin. Hemin (1 μg/ml) was used to supplement hemB mutants where needed (66).
Antibiotic susceptibilities were determined with Etest strips (AB-Biodisk, Solna, Sweden), covering an exponential gradient ranging from 0.016 to 256 μg/ml, on MH agar plates with an inoculum of a 0.5 McFarland standard, corresponding to 108 cells/ml. MICs were read after 24 h of incubation and, in the case of the mutants, after 24, 48, and 72 h, during which time MICs did not change.
To sample RNA, protein, and luciferase probes, cells from overnight cultures were washed in LB at 37°C and used to inoculate prewarmed LB to an optical density at 600 nm (OD600) of 0.1, corresponding to a concentration of 107 cells/ml. After 15 h, CFU as well as phenotype were determined on blood agar and selective plates to assure that no contamination or reversion had occurred during the experiment.
Northern blot analyses.
Total RNA was isolated as described previously (14) by using a FastRNA kit and a Fastprep reciprocating shaker (Bio 101, Vista, Calif.). For Northern blots, 5 to 10 μg of total RNA per lane was separated on a 1.5% agarose-20 mM guanidine thiocyanate gel and transferred overnight onto a positively charged nylon membrane (Roche, Rotkreuz, Switzerland). The blots were hybridized with specific digoxigenin-labeled DNA probes which were produced using a PCR DIG probe synthesis kit (Roche, Rotkreuz, Switzerland). Primers used are listed in Table 1. Membranes were stripped by being boiled twice in 0.1 SSC-0.5% sodium dodecyl sulfate (SDS) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 30 min and were reprobed up to four times. Data shown were confirmed in at least two independent experiments.
TABLE 1.
Primers used for amplification of specific digoxigenin-labeled DNA probesa
| Probe | Primer | Sequence (5′-3′) and reference | Probe | Primer | Sequence (5′–3′) and reference | |
|---|---|---|---|---|---|---|
| arlS | MS43 | GATAACACAGTGAGAGTTGAACC | ||||
| MS61 | GAGTCCATTACCGCCTTGAC | |||||
| asp23 | SAasp23f | ATGACTGTAGATAACAATAAAGC (23) | ||||
| SAasp23r | TTGTAAACCTTGTCTTTCTTGG (23) | |||||
| clfA | MS33 | CGTGGCTTCAGTGCTTGTAG | ||||
| MS34 | GAGTTGTTGCCGGTGTATTAGC | |||||
| fnbA | MS16 | GGGATGGGACAAGATAAAGAAGC | ||||
| MS17 | ACGACACGTTGACCAGCATG | |||||
| hla | MS20 | AGAAAATGGCATGCACAAAAA | ||||
| MS21 | TGTAGCGAAGTCTGGTGAAAA | |||||
| isaA | MS75 | GGCATCATCATTAGCAGTGG | ||||
| MS74 | GAATCCCCAAGCACCTAAAC | |||||
| RNAII | MS14 | CGAAGACGATCCAAAAC | ||||
| MS15 | TTATCTAAATGGGCAATGAGT | |||||
| RNAIII | RNAIIIf | GTGATGGAAAATAGTTGATGAG (11) | ||||
| RNAIIIr | GTGAATTTGTTCACTGTGTCG (11) | |||||
| rot | MS39 | CAAGTTTTGGGATTGTTGGGATG (adapted from reference 59) | ||||
| MS40 | GCTCCATTCATTTGTGCCATAG | |||||
| saeR | MS45 | GACCCACTTACTGATCGTG | ||||
| MS46 | CCTAATCCCCATACAGTTGTG | |||||
| sar | SasarAf | AGGGAGGTTTTAAACATGGC (11) | ||||
| SasarAr | CTCGACTCAATAATGATTCG (11) | |||||
| sarR | sarR+f | CTTCTAATTCTGAAATCAG (57) | ||||
| sarRr | GACATTAATGATTTAGTCAAC (57) | |||||
| sarS | MS42 | CAAGCCTGAAGTCGATATGAC | ||||
| MS41 | CAGCATGGTCTTGCTGC | |||||
| sigB | IK14 | ACGCGAAGGTGGCCTAG (35) | ||||
| IK15 | ATGGTCATCTTGTTGCCCC (35) | |||||
| spa | spaf | TGAATTCGTAAACTAGGTGTAGG (57) | ||||
| spar | CGGTACCAGGCTTGTTATTGTCTTCC (57) | |||||
| srrA | MS59 | CGAAATACTTATCGTAGATGATGAGGATAG | ||||
| MS60 | CAGCAAGTACGCGATGTGC | |||||
| svrA | MS31 | CATTGCCAATGATGATAGGGAC | ||||
| MS64 | CATTGCTGCTAAAGCACAAAG | |||||
| yycFG | MS67 | GAAGGATACGATGTGTACTGTGC | ||||
| MS68 | CGTTTCGACCTCTACTCATGTTG |
f, forward, r, reverse.
Western blot analyses.
Pelleted bacteria were resuspended in 0.07 M phosphate buffer, pH 6.8, containing lysozyme, lysostaphin, and DNase (each at 0.018 mg/ml) as well as 2 mM phenylmethylsulfonyl fluoride. Protein (10 μg) from cytoplasmic fractions was loaded for each lane and separated by SDS-10% polyacrylamide gel electrophoresis. Precision Plus Protein all blue standards (Bio-Rad) were used as molecular size markers. Gels were either stained with Coomassie blue dye (R25; Réactifs IBF, Villeneuve-la-Garenne, France) or transferred onto nitrocellulose membranes (Hybond; Amersham Biosciences). Rabbit anti-Asp23 antibodies were used to detect Asp23. For σB detection, blocked membranes were preincubated with 40 μg/ml human immunoglobulin G (Calbiochem) to saturate protein A and thereby prevent cross-reactivity of antigen-purified rabbit antibodies against σB. Horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories, Inc.) was diluted 1:10,000 and detected with SuperSignal West Pico solutions (Pierce). Representative blots from three independent experiments are shown.
Luciferase assay.
Bacteria were harvested by centrifugation, and the pellet was snap-frozen in liquid nitrogen. Cells were resuspended to an OD600 of 10, corresponding to 109 cells/ml, in 0.7 M phosphate buffer, pH 6.8, containing lysozyme and lysostaphin (each at 0.036 mg/ml). After shaking for 10 min at 37°C, 10 μl supernatant of lysed cells was mixed with an equal volume of luciferase assay substrate (Promega) and luminescence was measured for 15 s after a delay of 3 s on a Turner Designs TD-20/20 luminometer (Promega). According to the manufacturer's information, the luciferase assay substrate contains excess ATP; addition of 2 mM ATP did not increase signals. Protein concentration of the supernatant was determined by the Bradford method (9), with bovine serum albumin as a standard. Representative data from three independent experiments are shown.
Determination of mRNA stability.
Bacteria grown for 5 h in LB were supplemented with 300 μg/ml rifampin, and samples were taken at 3-min intervals. Total mRNA was isolated and analyzed by Northern blotting, as described above. Band intensities were quantified densitometrically with an ImageMaster VDS-CL (Amersham Pharmacia Biotech) using ImageQuant version 5.2. Values were corrected against the background and normalized. Half-life of mRNA was determined from regression lines obtained by plotting mean values of at least two independent experiments against time on a semilogarithmic graph. The regression line was calibrated to intercept with the initial amount of transcripts, which was set to 100.
RESULTS
Phenotypes and growth characteristics of the hemB mutants.
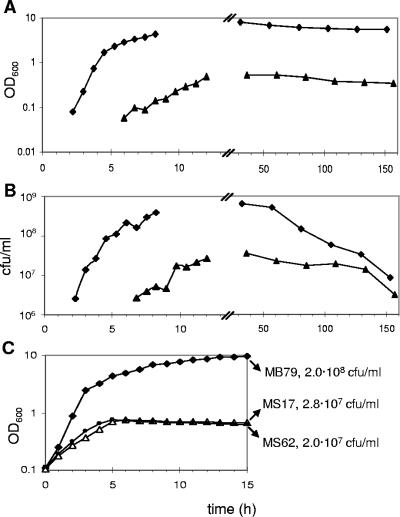
To evaluate the importance of σB in hemB mutants, strain MS17, carrying a σB-dependent luciferase reporter fusion (23), and the reporterless hemB sigB double mutant MS62 were constructed. Both mutants formed tiny colonies on MH plates, whereas on sheep blood agar, cells could scavenge enough hemin to form wild-type-sized, hemolytic colonies. In LB, they reached a maximal OD600 of 0.8, while their parent, MB79, grew up to an OD600 of 10 (Fig. 1A, data not shown for MS62). Viability was not reduced in the hemB mutants, as judged from CFU determined over several days, which corresponded to the respective OD600 values (Fig. 1B and C). Both mutants were four and eight times more resistant to the aminoglycosides gentamicin and amikacin, respectively. However, they were slightly more susceptible to oxacillin and teicoplanin by factors of 2 and 4, respectively, but not to vancomycin (Table 2). By supplementing the growth medium with hemin, all phenotypes reported here and further could be restored to a wild-type pattern in the mutants, indicating that inactivation of hemB was solely responsible for the characteristics of the mutants and that no unexpected mutations or rearrangements had occurred during the construction of the strains.
FIG. 1.
Growth and viability of MB79 (wild type), MS17 (hemB), and MS62 (hemB sigB). (A) Growth curves and (B) CFU of cultures, inoculated with overnight cells and an initial OD600 of 0.01, were monitored over 6 days. (C) Growth curves from cultures with an inoculum 10 times higher (OD600 of 0.1) than that for panels A and B, obtained by using washed overnight cells. CFU/ml were determined after 15 h. MB79, diamonds; MS17, triangles; MS62, circles.
TABLE 2.
Antibiotic susceptibilities of strains MB79 (wild type), MS17 (hemB), and MS62 (hemB sigB)
| Antibiotic | MIC (μg/ml) of:
|
||
|---|---|---|---|
| MB79 | MS17 | MS62 | |
| Amikacin | 4 | 32 | 32 |
| Gentamicin | 1 | 4 | 4 |
| Oxacillin | 0.38 | 0.19 | 0.19 |
| Teicoplanin | 3 | 0.75 | 0.75 |
| Vancomycin | 2 | 2 | 2 |
In cultures with an initial OD600 of 0.01, the lag phase of hemB mutants was more than 4 h longer than that of the parent strain (Fig. 1A). Cultures were therefore generally started with an inoculum of OD600 of 0.1, which synchronized the beginning of exponential growth of wild-type bacteria and hemB mutants (Fig. 1C). Under these conditions, the mutants entered the stationary growth phase 2 h later than the parent strain. Carryover of extracellular signal molecules, which might mask growth-dependent processes, was avoided by washing cells before inoculation. Growth was followed over 15 h to include late-stationary-phase data.
Transcription of two-component systems is restricted to the exponential phase in the hemB mutant.
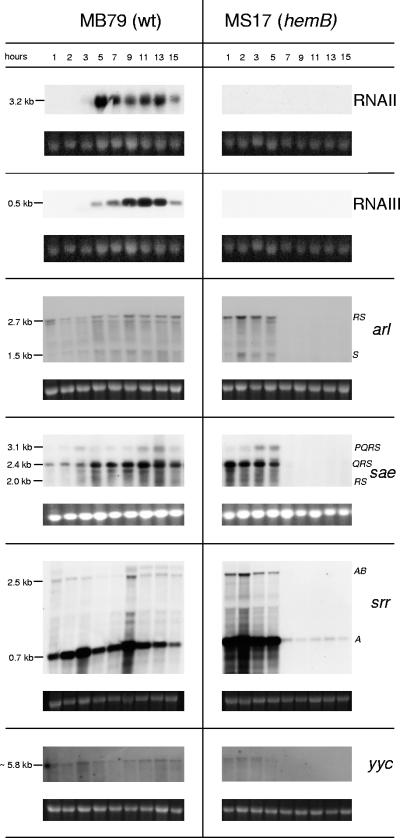
Expression patterns regarding agr, arl, sae, and srr were in agreement with previous studies of the wild type, MB79 (Fig. 2) (19, 24, 49, 63, 70). The 2.7-kb arlRS transcript, coding for an autolysis-related sensor transducer involved in biofilm formation, was seen throughout growth and increased slightly in the late stationary phase. Of the three sae transcripts, saePQRS (3.1 kb), saeQRS (2.4 kb), and saeRS (2 kb), whose products influence several virulence factors presumably in response to environmental stimuli, saeQRS was the most prominent and peaked in the late post-exponential phase. The weaker 2.5-kb srrAB (synonym, srhSR) and the generally stronger 0.7-kb srrA mRNAs, reported to be produced until post-exponential phase and encoding a system reacting to environmental oxygen changes, showed here peaks at 3 and 9 h and were expressed throughout the 15 h monitored. The expression profile of yycFG (synonym, vicRK), coding for an essential two-component system affecting cell permeability as well as resistance against macrolide-lincosamide-streptogramin B antibiotics (16, 41), was similar to that of arl. A transcript of approximately 5.8 kb was observed, suggesting that yycFG was cotranscribed with at least two of the yet uncharacterized downstream genes.
FIG. 2.
Northern blot analyses of the two-component systems agr (RNAII and RNAIII), arl, sae, srr, and yyc in MB79 (wild type [wt]) and MS17 (hemB). Ethidium bromide-stained 16S rRNA is shown as an indication of RNA loading.
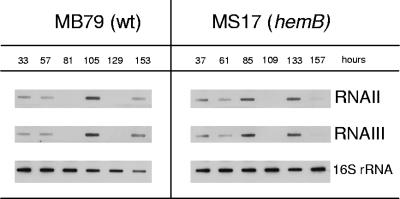
Transcripts of arl, sae, srr, and yyc disappeared in the hemB mutant after 5 h, except for a very faint srrA band detected throughout growth. In this short period of expression, arl, sae, and srr transcription levels were up-regulated in the hemB mutant compared to levels in MB79, while yyc was expressed at similar levels (Fig. 2). In the Newman hemB mutant MS17, unlike as reported for a hemB mutant in a strain 8325-4 background (65), not only was the agr transcript RNAIII (0.5 kb) absent, but RNAII (3.2 kb) was missing as well (Fig. 2). Supplementation of the growth medium with hemin restored the agr profile in MS17 to the wild-type pattern, ruling out an unrelated deletion or mutation affecting agr (data not shown). Interestingly, preliminary long-term monitoring revealed both agr transcripts to be expressed after 30 h in the hemB mutant MS17 (Fig. 3). As in the parent MB79, agr transcription fluctuated in MS17, stopping after 3 or 4 days, respectively, to resume at day 5 or 6, respectively. A second transcriptional gap was observed for both strains after an additional day, reminiscent of the oscillating agr expression levels observed with S. aureus biofilms (69), and here might be caused by growth of a new subpopulation living on dead cells and debris once nutrients had been depleted from the medium.
FIG. 3.
agr expression during prolonged cultivation. MB79 (wild type [wt]) and MS17 (hemB) were monitored over 7 days. RNA samples were taken every 24 h, prepared as described in Materials and Methods, and transferred onto a nylon membrane by slot blotting. Bacteria were grown as described in the legend for Fig. 1A and B.
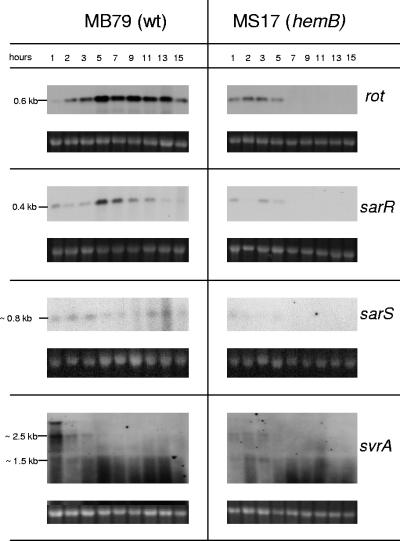
Expression of agr requires svrA, encoding a putative membrane-associated protein affecting virulence (21), which recently has been identified to be a multidrug export protein (therefore renamed mepA) belonging to the MATE family of efflux pumps (42). The transcript(s) of yet unknown size, reported to be expressed in post-exponential phase (21), were here however detected only during early growth in the MB79 wild type. A faint band of approximately 2.5 kb suggested that svrA (1.35 kb) is cotranscribed with two flanking genes, encoding a MarR-like transcriptional regulator (mepR) and a hypothetical protein of yet-unknown function (mepB) (Fig. 4). A shorter, approximately 1.5-kb transcript might be present, paralleling the larger transcript. In MS17, the transcripts were almost undetectable.
FIG. 4.
Northern blot analyses of the SarA homologues rot, sarR, and sarS as well as the virulence regulator svrA transcripts in MB79 (wild type [wt]) and MS17 (hemB). Ethidium bromide-stained 16S rRNA is shown as an indication of RNA loading. The observed band at the height of approximately 2.8 kb in svrA blots might be caused by interference of bulk 23S rRNA.
Reduced transcription of SarA homologues in the hemB mutant.
The SarA family of transcriptional regulators constitutes a further set of major regulatory elements. They include Rot, SarA, SarR, SarS, SarT, and SarU (12). sarT and sarU, not being expressed in the Newman background according to microarray data (M. Bischoff, unpublished data), were not analyzed here.
In MB79, both rot (0.6 kb) and sarR (0.4 kb) were transcribed maximally in the post-exponential phase, only to decline towards the stationary phase (Fig. 4), as reported previously (38, 67). The sarS transcript (0.8 kb) showed an antiparallel behavior, being present in low amounts in early exponential phase (64) and then again towards stationary phase. In contrast to these wild-type patterns, the hemB mutant MS17 displayed reduced transcription levels of rot, sarR, and sarS and only during exponential phase (Fig. 4).
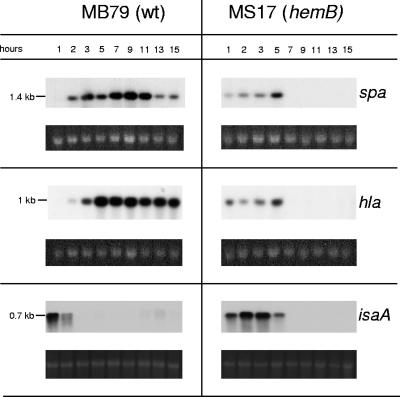
Virulence determinants spa, hla, and isaA are transcribed only in exponential phase in the hemB mutant.
Besides complex mutual interactions, the above-presented regulators, together with the below-discussed SarA and σB, affect virulence gene expression directly or indirectly. From the transcriptional data presented here, it was possible neither to predict the amount of active regulator nor to attribute net effects to a single regulator. The different resulting transcription patterns are exemplified for the virulence factors spa, hla, and isaA.
In MB79, spa was maximally transcribed in post-exponential phase; hla transcription started shortly after that of spa but reached its maximum level earlier and was thereafter rather constant (Fig. 5). Overall transcription levels of both spa and hla were reduced in the hemB mutant MS17, in agreement with previous findings (66); yet, surprisingly we observed that transcription started earlier than in the parent (Fig. 5), a phenomenon also seen for rot (Fig. 4).
FIG. 5.
Northern blot analyses of the virulence determinants spa, hla, and isaA in MB79 (wild type [wt]) and MS17 (hemB). Ethidium bromide-stained 16S rRNA is shown as an indication of RNA loading.
Transcription of the immunodominant antigen A (isaA), whose product had previously been reported to be up-regulated and present in stationary growth phase in a COL hemB mutant (34), was prolonged and increased in MS17 compared to transcription in MB79 (Fig. 5). However, transcription stopped upon entry into stationary phase, as in the case of spa and hla.
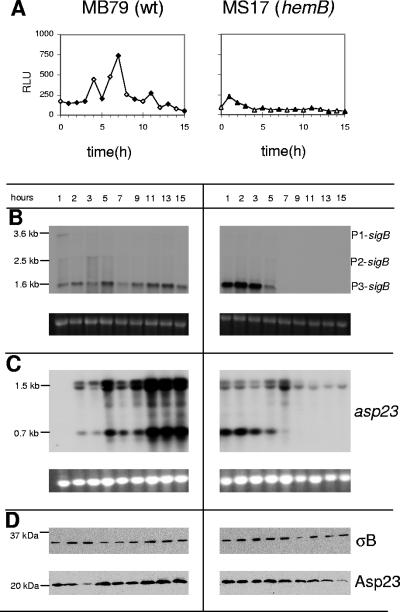
σB expression and activity in the hemB mutant.
σB mediates various stress responses and also plays a role in antibiotic resistance as well as biofilm formation (summarized in reference 7). Transcription of the sigB operon is presumably initiated by three promoters, producing a 3.6-kb transcript containing sas067-sa1873-rsbUVW-sigB (P1-sigB), an rsbUV-sigB- covering 2.5-kb transcript (P2-sigB), and a third, 1.6-kb, stress-inducible, autoregulated transcript covering rsbVW-sigB (P3-sigB) that is σB dependent (22, 23). In strain MB79, the P1- and P2-driven mRNAs were expressed weakly and only during early growth, while the 1.6-kb P3-sigB transcript displayed a more complex pattern: it peaked in late exponential phase and in early post-exponential phase and increased again towards stationary phase (Fig. 6B). In contrast, in the hemB mutant MS17, the P1- and P2-driven transcripts were almost undetectable and the 1.6-kb P3-sigB transcript, initially stronger than in the parent, disappeared after the exponential phase.
FIG. 6.
σB expression in the Newman background. (A) σB activities in MB79 (wild type [wt]) and MS17 (hemB). σB activities were measured by the σB-dependent asp23P::luc+ reporter gene fusion. Luciferase activity is given as RLU per μg protein of cleared lysate. Filled symbols indicate time points of sampling for Northern and Western blot analyses. (B and C) Northern blot analyses of (B) sigB and of (C) asp23. The sizes of relevant bands are given on the left. Ethidium bromide-stained 16S rRNA is shown as an indication of RNA loading. The observed double band at the site of the 1.5-kb asp23 mRNA might be caused by interference of bulk 16S rRNA. (D) Western blot analyses of σB (∼35 kDa) and Asp23 (∼23 kDa). Cytoplasmic protein fractions were separated by SDS-10% polyacrylamide gel electrophoresis, transferred onto a nitrocellulose membrane, and analyzed with antigen-purified anti-σB antibodies (upper panels) or anti-Asp23 antibodies (lower panels). The molecular sizes of the Precision Plus Protein all blue standard (Bio-Rad) markers are indicated on the left.
Despite the difference in transcriptional activities, Western blot analyses of cytoplasmic protein fractions of the MB79 wild type and the hemB mutant MS17 revealed that the σB protein was present at similar, constant levels in both strains throughout growth (Fig. 6D). Presence of the σB protein, however, is not necessarily indicative of its activity, due to posttranslational control by RsbU, RsbV, and RsbW (23, 43, 51). A reporter construct consisting of the σB-dependent promoters of the alkaline shock protein 23 (asp23) fused to the firefly luciferase gene (luc+) was therefore used to analyze the actual σB activity. The luciferase activity profile followed the peaks of the 1.6-kb P3-sigB transcript in MB79 with an approximately-1-h delay (Fig. 6A). In the hemB mutant MS17, σB activity could be detected only during the first 2 h of growth, where relative light unit (RLU) values were higher in the hemB mutant than in MB79. Thereafter, RLUs dropped quickly below the values measured in the parent (Fig. 6A). Western blot analysis showed that Asp23, which was here under the same control as luciferase, decreased in the mutant over the time course as well, while in MB79, Asp23 seemed to increase from the end of the exponential phase towards stationary phase (Fig. 6D).
The resident asp23 gene is preceded by two σB consensus sequence promoters producing 0.7- and 1.5-kb transcripts with identical 3′ ends (23). In the parent strain MB79, both asp23 transcripts paralleled the expression profile of the 1.6-kb P3-sigB mRNA, peaking at 5 h and reaching maximal levels during the last 3 h (Fig. 6B and C). In the hemB mutant, however, only the 0.7-kb asp23 mRNA paralleled the hemB mutant-specific 1.6-kb P3-sigB transcription profile, with transcription stopping after 5 h of growth. Surprisingly, the upper asp23 transcript was present at low levels throughout the 15 h. Contrary to what the 1.6-kb P3-sigB mRNA and luciferase measurements had suggested, these data indicated a possible σB activity in the mutant. We therefore compared transcription levels of other known σB-controlled genes with the expression levels in the hemB mutant MS17 and the hemB sigB mutant MS62.
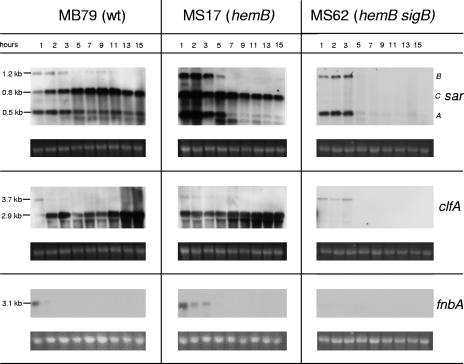
Transcription of the σB-influenced genes sarC, clfA, and fnbA requires σB in the hemB background.
For MB79, we observed characteristic sar profiles (4). sarA (0.5 kb) and a weak sarB (1.2-kb) transcript were mainly present in exponential phase, while the σB-dependent sarC (0.8-kb) transcript appeared in mid-exponential phase, reaching a maximum intensity in post-exponential phase (Fig. 7). Interestingly, in the hemB mutant MS17, all three sar transcripts were maximally expressed in the beginning and were strongly increased compared to in MB79. While sarA and sarB were restricted to the exponential phase, sarC was transcribed throughout growth, although slightly decreasing towards stationary phase. We confirmed the sarC transcript to be σB dependent in the hemB background as well, since it was abolished in the hemB sigB double mutant MS62 (Fig. 7).
FIG. 7.
Northern blot analyses of the σB-influenced genes sar, clfA, and fnbA in MB79 (wild type [wt]), MS17 (hemB), and MS62 (hemB sigB). Ethidium bromide-stained 16S rRNA is shown as an indication of RNA loading.
The clfA gene is preceded by σA- and σB-dependent promoters, which initiate transcripts of 3.7 kb and 2.9 kb, respectively (18, 46). The 3.7-kb σA-dependent transcript was seen in all three strains during exponential growth phase, irrespective of the changes triggered by hemB or sigB inactivation (Fig. 7). The σB-dependent 2.9-kb clfA mRNA roughly followed the 1.6-kb P3-sigB transcription profile in MB79, with peaks at 3 h and 13 to 15 h. In the hemB mutant MS17, however, the 2.9-kb clfA transcript displayed a transcriptional pattern quite different than that of the 1.6-kb P3-sigB mRNA. After a strong initial signal, transcription diminished slightly and then increased again towards stationary phase, comparable to what was observed for the parent strain. The σB dependence of the 2.9-kb clfA mRNA was confirmed by its absence in the hemB sigB double mutant MS62.
Although not preceded by an apparent σB consensus promoter sequence, the fnbA gene is positively influenced by σB (7, 18, 46). While the recently reported σA-dependent 4.5-kb fnbA transcript (18) was not observed in the Newman background, a 3.1-kb fnbA transcript was detected during the first hours after inoculation in MB79 and MS17 but was completely missing in the hemB sigB mutant MS62 (Fig. 7). In the hemB mutant MS17, fnbA transcription was slightly prolonged compared to in the parent strain MB79.
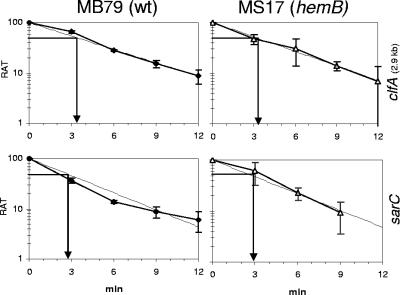
The presence of σB-dependent asp23, 2.9-kb clfA, fnbA, and sarC transcripts in the hemB mutant MS17 and their respective absences in the hemB sigB mutant MS62 (data not shown for asp23) proved that σB was required and active at all time points in MS17. Nevertheless, in this strain, striking differences in the extents of stimulation of the directly σB-regulated genes were observed. Variations in mRNA half-lives of these σB-dependent transcripts were ruled out. There was no significant difference in mRNA stability between parent and mutant or between sarC and 2.9-kb clfA (Fig. 8). The approximate half-life of transcripts was 3 min, similar to values determined for the asp23 transcripts (Bischoff, unpublished). Therefore, variations in regulation of directly σB-controlled promoters might be attributed to the involvement of additional factors modulating σB activity.
FIG. 8.
Stability of 2.9-kb clfA and sarC mRNAs in MB79 (wild type [wt]) and MS17 (hemB). The relative amounts of transcript (RAT) are expressed as percentages of the quantities at the time of rifampin addition. Regression lines (dashed lines) are based on mean values from at least two independent experiments. mRNA half-lives were read at a RAT of 50.
DISCUSSION
Differences in genetic backgrounds as well as minor changes in growth conditions can have major impacts on the profiles of regulators. Comparison of wild-type and growth-deficient strains, like hemB mutants, is in addition complicated by a growth phase disparity. By starting cultures with a relatively high inoculum of washed cells from overnight cultures, lag phases were synchronized (Fig. 1C), having no influence on the growth phase-dependent patterns of gene expression, as judged from agr and sar profiles (Fig. 2 and 7). Cells were sampled frequently enough to obtain representative profiles for both strains and to allow comparison of the temporal gene expression profiles from the respective growth phases. Hence, this study provides for the first time a useful overview of wild-type and hemB mutant transcription profiles of the most important regulators.
We analyzed the temporal patterns of several global regulators for 15 h and found that their transcriptional profiles did not reflect any particular growth phase of the parent strain or the absence or predominance of one single regulatory element. Surprisingly, an astonishing overall reduction of transcription happened upon exit from exponential phase, which applied to the virulence factors hla and spa as well. Deviations were found for all analyzed global regulators and can be classified into three groups: (i) no transcription, as observed for agr; (ii) transcripts that were only present until cells entered post-exponential phase (during this time they were either up-regulated [arl, sae, sarA, sarB, sigB, and srr], down-regulated [rot, sarR, sarS, and svrA], or expressed as in the parent strain [yyc]), and (iii) transcription throughout the observed time course, as seen for sarC.
The complete absence during 15 h of both the agr effector molecule RNAIII (29, 50) and RNAII, encoding a quorum-sensing two-component system (37), is supposed to have an influence on the expression of several genes in the hemB mutant. It could explain the lack of sae transcription after the exponential phase in the hemB mutant, since sae transcription requires agr in post-exponential phase (24, 49). Although agr is the only known inhibitor of fnbA, it was not (yet) expressed in either strain when fnbA transcription decreased (Fig. 2 and 7). In addition, in an RNAIII-defective 8325-4 background, up-regulation of fnbA transcription in the corresponding hemB mutant was still observable (65). These findings support the idea that some additional repressor must exist (68).
sar transcription had been shown to be reduced in an 8325-4 hemB mutant, suggesting that the observed increased fnbA levels were not connected to sar but that other regulators were likely responsible for the observed phenomenon (65). By using Northern blot techniques and by monitoring the entire growth cycle, we obtained a more detailed image. We were able to show that in the hemB mutant, sarC was the only regulator transcript present throughout growth and that, in consequence, its exclusive and required activator σB was present and active as well. The absence of σB in the hemB sigB double mutant dominated over residual sar expression, as fnbA transcription ceased in that strain as well.
Since findings of reduced sar, but increased fnbA and clfA, transcription in an 8325-4 hemB mutant stem from just one time point of late log phase (65), comparison with our results is difficult. By evaluating mRNA levels in MB79 (2 and 3 h) and MS17 (3 and 5 h) during late exponential phase, we found increased fnbA transcription in the Newman hemB background. However, overall sar mRNA levels were increased, while clfA seemed to be lower in late log phase (Fig. 7). Apart from sampling differences, the diverse genetic backgrounds might have had an influence on relative expression levels as well, as seen for agr.
While transcription of most of the analyzed genes was concentrated to the exponential phase in the hemB mutant, some of the gene products were presumably present and active until stationary phase, as was found for σB. Results for fnbA and isaA transcription presented here combined with data reported for FnbA and IsaA strongly suggest that these two proteins persist as well (34, 65). Speculating that the energy-restricted hemB mutant does not express needless genes, the increased transcription levels of arl, sae, sar, and srr in exponential phase suggest that these regulators are involved in the control of its transcriptome. A subject of future work is to determine the presence and activity of the aforementioned regulators in vitro as well as in vivo. For this purpose, reporter systems have to be used with care, as seen for luciferase, which otherwise reliably detects fluctuating gene expression. Apparently, in the hemB mutant, conditions are such that the rather unstable luciferase (half-life of 2 h) (28) is hardly translated, rapidly degraded, or inactivated. Availability of ATP within the hemB mutant could be ruled out as the reason for reduced luciferase activity, as luciferase measurements were performed with cell extracts in excess of ATP. Data indicate that the used reporter system poorly reflects σB activity in wild-type stationary-phase bacteria as well. Whether some similarity between their physiological state and that encountered with the hemB mutant exists remains to be investigated.
Interestingly, both spa and hla transcription started earlier in the mutant (Fig. 4). This was also observed for rot, asp23, and 2.9-kb clfA transcription (Fig. 4, 6, and 7), arguing against the idea that SCVs are cells stalled in early growth phase, as suggested by the absence of agr and exoprotein production and the prolonged transcription of genes typically expressed in early growth phase, like fnbA and isaA (Fig. 2, 5, and 7). Whether the almost-uniform stop of transcriptional activity upon exit of the exponential phase is a coincidence or correlates with a yet uncharacterized transition process remains open. A link to reduced ATP levels seems difficult to make, since the reduced ATP levels are rather constant in a COL hemB mutant (34). Various metabolic enzymes have been reported to be transcribed after the exponential phase (34), suggesting that transcription focuses on selected and required genes.
Both the premature transcription and the drastic reduction of transcription observed in the hemB mutant, for σB-dependent and -independent loci, indicate that global alterations of gene expression do happen. Thus, the extraordinary cellular state caused by the interrupted electron transport in hemB mutants is linked to altered activity of global regulators and expression of virulence factors.
Long-term follow-up of σB-dependent transcription revealed that σB was active, with fluctuations, until stationary phase in the parent strain, possibly in response to general changes in growth conditions. With the exception of the sarC transcript having its maximal level in post-exponential phase and thereafter declining (Fig. 7), σB-dependent mRNA patterns were similar and increased towards stationary phase. This variation in expression of known σB-dependent genes was much more pronounced in the Newman hemB mutant. While sarC and the 2.9-kb clfA and 1.5-kb asp23 transcripts were detectable throughout growth, this was not the case for the 1.6-kb P3-sigB and 0.7-kb asp23 transcripts.
A recent study of clinical thymidine-auxotrophic SCVs concluded from pigmentation and sarC and asp23 transcriptional patterns that σB has a lower activity in most of these mutants than in their parents (33). While generally reduced agr and hla levels were found as well, spa levels were frequently higher. The diversity of mRNA levels and expression patterns found for the wild-type strains indicates that these clinical isolates have varying genetic backgrounds. The fact that supplementation of the SCVs with thymidine did not always fully restore the wild-type phenotype furthermore suggests that additional unknown alterations, possibly affecting σB regulation, had occurred before or during SCV formation (33). Comparison with our data is further complicated by the fact that the analyzed thymidine-dependent SCVs displayed two classes of colony morphology, the genetic reason for which is yet unknown. Lack of pigment formation is not a reliable indicator of the absence of σB activity, since many more factors can influence this trait, e.g., mutation of the synthesizing enzymes or regulators or a defective electron transport (6, 32, 36, 40). However, as judged from the presence of the σB-requiring sarC or asp23 mRNA levels, apparently reduced σB activity could be observed for several of the analyzed SCVs. The partial inconsistency of these two σB-activity indicators further hints at the presence of promoter-specific σB modulators and makes it difficult to conclude the relative amounts of σB activity from selected mRNA levels.
Based on these observations, we postulate the presence of factors modulating σB activity in the recognition of its promoter consensus sequences under certain circumstances. Such σB modulators may also explain diverging results concerning σB activities in different genetic backgrounds and under various experimental conditions.
Yet, sigB was definitely not an essential locus for the in vitro SCV phenotype, as its inactivation did not affect colony morphology, growth, or antibiotic resistance. Nevertheless, σB is strictly required for high-level expression of at least two important virulence factors in vitro, FnbA and ClfA. In vivo studies with a guinea pig model of device-related infection of wild-type Newman showed that alternative regulatory pathways seem to be active in vivo, as shown for coa, which despite its σB-dependent consensus promoter sequence (7) needs Sae in vivo (25). However, full ClfA expression still requires σB, the only clfA regulator identified so far (25). The general importance of σB for virulence is a continuing field of investigation. In several animal models, no permanent difference between the wild type and sigB mutants was observed (18, 27, 47). Analogous to the σB-independent adherence observed in vitro (18), stemming from a basal σB-independent expression of fnbA and clfA, and redundant, σB-independent, adhering proteins like FnbB and ClfB (7), σB-mediated adherence might not be required for all types of infection. Nonetheless, in a mouse model of septic arthritis, lack of σB resulted in attenuated infection (30). In that model, sigB mutants displayed a reduced ability to survive in the bloodstream and to persist in kidneys and joints, leading to a reduced mortality.
It is reasonable that the expression of FnbA, the more effective adhesion and invasion factor of the two redundant FnBPs FnbA and FnbB (26), together with ClfA, which has been shown to protect bacteria from phagocytosis by macrophages (52), might significantly contribute to persistence in the host, a characteristic feature of SCVs. Whether in vivo FnbA, ClfA, and σB indeed play a crucial role in the establishment of SCV infections remains to be determined.
Acknowledgments
This study was supported by the Swiss National Foundation grant NF31-063552 and EU LSH CT 2003-503335 (BBW 03.0098).
REFERENCES
- 1.Acar, J. F., F. W. Goldstein, and P. Lagrange. 1978. Human infections caused by thiamine- or menadione-requiring Staphylococcus aureus. J. Clin. Microbiol. 8:142-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balwit, J. M., P. van Langevelde, J. M. Vann, and R. A. Proctor. 1994. Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J. Infect. Dis. 170:1033-1037. [DOI] [PubMed] [Google Scholar]
- 3.Baumert, N., C. von Eiff, F. Schaaff, G. Peters, R. A. Proctor, and H. G. Sahl. 2002. Physiology and antibiotic susceptibility of Staphylococcus aureus small colony variants. Microb. Drug Resist. 8:253-260. [DOI] [PubMed] [Google Scholar]
- 4.Bayer, M. G., J. H. Heinrichs, and A. L. Cheung. 1996. The molecular architecture of the sar locus in Staphylococcus aureus. J. Bacteriol. 178:4563-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger-Bächi, B. 1983. Increase in transduction efficiency of Tn551 mediated by the methicillin resistance marker. J. Bacteriol. 154:533-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bischoff, M., and B. Berger-Bächi. 2001. Teicoplanin stress-selected mutations increasing σB activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bächi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bischoff, M., J. M. Entenza, and P. Giachino. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Brouillette, E., A. Martinez, B. J. Boyll, N. E. Allen, and F. Malouin. 2004. Persistence of a Staphylococcus aureus small-colony variant under antibiotic pressure in vivo. FEMS Immunol. Med. Microbiol. 41:35-41. [DOI] [PubMed] [Google Scholar]
- 11.Chan, P. F., and S. J. Foster. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180:6232-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y. Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1-9. [DOI] [PubMed] [Google Scholar]
- 13.Cheung, A. L., Y.-T. Chien, and A. S. Bayer. 1999. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect. Immun. 67:1331-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung, A. L., K. J. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from Gram-positive bacteria and mycobacteria. Anal. Biochem. 222:511-514. [DOI] [PubMed] [Google Scholar]
- 15.Deora, R., T. Tseng, and T. K. Misra. 1997. Alternative transcription factor σSB of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J. Bacteriol. 179:6355-6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubrac, S., and T. Msadek. 2004. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J. Bacteriol. 186:1175-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 18.Entenza, J.-M., P. Moreillon, M. M. Senn, J. Kormanec, P. M. Dunman, B. Berger-Bächi, S. Projan, and M. Bischoff. 2005. Role of σB in the expression of Staphylococcus aureus cell wall adhesins ClfA and FnbA and contribution to infectivity in a rat model of experimental endocarditis. Infect. Immun. 73:990-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 20.Gao, J., and G. C. Stewart. 2004. Regulatory elements of the Staphylococcus aureus protein A (spa) promoter. J. Bacteriol. 186:3738-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garvis, S., J.-M. Mei, J. Ruiz-Albert, and D. W. Holden. 2002. Staphylococcus aureus svrA: a gene required for virulence and expression of the agr locus. Microbiology 148:3235-3243. [DOI] [PubMed] [Google Scholar]
- 22.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of sigmaB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261:558-566. [DOI] [PubMed] [Google Scholar]
- 23.Giachino, P., S. Engelmann, and M. Bischoff. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giraudo, A. T., C. Mansilla, A. Chan, C. Raspanti, and R. Nagel. 2003. Studies on the expression of regulatory locus sae in Staphylococcus aureus. Curr. Microbiol. 46:246-250. [DOI] [PubMed] [Google Scholar]
- 25.Goerke, C., U. Fluckiger, A. Steinhuber, V. Bisanzio, M. Ulrich, M. Bischoff, J. M. Patti, and C. Wolz. 2005. Role of Staphylococcus aureus global regulators sae and σB in virulence gene expression during device-related infection. Infect. Immun. 73:3415-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grundmeier, M., M. Hussain, P. Becker, C. Heilmann, G. Peters, and B. Sinha. 2004. Truncation of fibronectin-binding proteins in Staphylococcus aureus strain Newman leads to deficient adherence and host cell invasion due to loss of the cell wall anchor function. Infect. Immun. 72:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ignowski, J. M., and D. V. Schaffer. 2004. Kinetic analysis and modeling of firefly luciferase as a quantitative reporter gene in live mammalian cells. Biotechnol. Bioeng. 86:827-834. [DOI] [PubMed] [Google Scholar]
- 29.Janzon, L., and S. Arvidson. 1990. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 9:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jonsson, I.-M., S. Arvidson, S. Foster, and A. Tarkowski. 2004. Sigma factor B and RsbU are required for virulence in Staphylococcus aureus-induced arthritis and sepsis. Infect. Immun. 72:6106-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonsson, I. M., C. von Eiff, R. A. Proctor, G. Peters, C. Ryden, and A. Tarkowski. 2003. Virulence of a hemB mutant displaying the phenotype of a Staphylococcus aureus small colony variant in a murine model of septic arthritis. Microb. Pathog. 34:73-79. [DOI] [PubMed] [Google Scholar]
- 32.Joyce, G. H., and D. C. White. 1971. Effect of benzo(a)pyrene and piperonyl butoxide on formation of respiratory system, phospholipids, and carotenoids of Staphylococcus aureus. J. Bacteriol. 106:403-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kahl, B. C., G. Belling, P. Becker, I. Chatterjee, K. Wardecki, K. Hilgert, A. L. Cheung, G. Peters, and M. Herrmann. 2005. Thymidine-dependent Staphylococcus aureus small-colony variants are associated with extensive alterations in regulator and virulence gene expression profiles. Infect. Immun. 73:4119-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohler, C., C. von Eiff, G. Peters, R. A. Proctor, M. Hecker, and S. Engelmann. 2003. Physiological characterization of a heme-deficient mutant of Staphylococcus aureus by a proteomic approach. J. Bacteriol. 185:6928-6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kullik, I., and P. Giachino. 1997. The alternative sigma factor sigma B in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch. Microbiol. 167:151-159. [DOI] [PubMed] [Google Scholar]
- 36.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lina, G., S. Jarraud, G. Ji, T. Greenland, A. Pedraza, J. Etienne, R. P. Novick, and F. Vandenesch. 1998. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 28:655-662. [DOI] [PubMed] [Google Scholar]
- 38.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manna, A. C., M. G. Bayer, and A. L. Cheung. 1998. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J. Bacteriol. 180:3828-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall, J. H., and G. J. Wilmoth. 1981. Proposed pathway of triterpenoid carotenoid biosynthesis in Staphylococcus aureus: evidence from a study of mutants. J. Bacteriol. 147:914-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin, P. K., T. Li, D. Sun, D. P. Biek, and M. B. Schmid. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 181:3666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McAleese, F., P. Petersen, A. Ruzin, P. M. Dunman, E. Murphy, S. J. Projan, and P. A. Bradford. 2005. A novel MATE family efflux pump contributes to the reduced susceptibility of laboratory-derived Staphylococcus aureus mutants to tigecycline. Antimicrob. Agents Chemother. 49:1865-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyazaki, E., J.-M. Chen, C. Ko, and W. R. Bishai. 1999. The Staphylococcus aureus rsbW (orf159) gene encodes an anti-sigma factor of SigB. J. Bacteriol. 181:2846-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreillon, P., J. M. Entenza, P. Francioli, D. McDevitt, T. J. Foster, P. Francois, and P. Vaudaux. 1995. Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect. Immun. 63:4738-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morfeldt, E., D. Taylor, A. von Gabain, and S. Arvidson. 1995. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 14:4569-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nair, S. P., M. Bischoff, M. M. Senn, and B. Berger-Bächi. 2003. The σB regulon influences internalization of Staphylococcus aureus by osteoblasts. Infect. Immun. 71:4167-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicholas, R. O., T. Li, D. McDevitt, A. Marra, S. Sucoloski, P. L. Demarsh, and D. R. Gentry. 1999. Isolation and characterization of a sigB deletion mutant of Staphylococcus aureus. Infect. Immun. 67:3667-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 49.Novick, R. P., and D. Jiang. 2003. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 149:2709-2717. [DOI] [PubMed] [Google Scholar]
- 50.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palma, M., and A. L. Cheung. 2001. Sigma B activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect. Immun. 69:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palmqvist, N., J. M. Patti, A. Tarkowski, and E. Josefsson. 2004. Expression of staphylococcal clumping factor A impedes macrophage phagocytosis. Microbes Infect. 6:188-195. [DOI] [PubMed] [Google Scholar]
- 53.Proctor, R. A., J. M. Balwit, and O. Vesga. 1994. Variant subpopulations of Staphylococcus aureus as cause of persistent and recurrent infections. Infect. Agents Dis. 3:302-312. [PubMed] [Google Scholar]
- 54.Proctor, R. A., D. M. Bates, and P. J. McNamara. 2001. Electron transport-deficient Staphylococcus aureus small-colony variants as emerging pathogens, p. 95-110. In W. Scheld, W. Craig, and J. Hughes (ed.), Emerging infections, vol. 5. ASM Press, Washington, D.C. [Google Scholar]
- 55.Proctor, R. A., and G. Peters. 1998. Small colony variants in staphylococcal infections: diagnostic and therapeutic implications. Clin. Infect. Dis. 27:419-422. [DOI] [PubMed] [Google Scholar]
- 56.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-82. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 57.Rossi, J., M. Bischoff, A. Wada, and B. Berger-Bächi. 2003. MsrR, a putative cell envelope-associated element involved in Staphylococcus aureus sarA attenuation. Antimicrob. Agents Chemother. 47:2558-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sadowska, B., A. Bonar, C. von Eiff, R. A. Proctor, M. Chmiela, W. Rudnicka, and B. Rozalska. 2002. Characteristics of Staphylococcus aureus, isolated from airways of cystic fibrosis patients, and their small colony variants. FEMS Immunol. Med. Microbiol. 32:191-197. [DOI] [PubMed] [Google Scholar]
- 59.Saïd-Salim, B., P. M. Dunman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saravia-Otten, P., H. Müller, and S. Arvidson. 1997. Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J. Bacteriol. 179:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seifert, H., C. von Eiff, and G. Fatkenheuer. 1999. Fatal case due to methicillin-resistant Staphylococcus aureus small colony variants in an AIDS patient. Emerg. Infect. Dis. 5:450-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sinha, B., P. P. Francois, O. Nusse, M. Foti, O. M. Hartford, P. Vaudaux, T. J. Foster, D. P. Lew, M. Herrmann, and K. H. Krause. 1999. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin α5β1. Cell. Microbiol. 1:101-117. [DOI] [PubMed] [Google Scholar]
- 63.Steinhuber, A., C. Goerke, M. G. Bayer, G. Doring, and C. Wolz. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 185:6278-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398-409. [DOI] [PubMed] [Google Scholar]
- 65.Vaudaux, P., P. Francois, C. Bisognano, W. L. Kelley, D. P. Lew, J. Schrenzel, R. A. Proctor, P. J. McNamara, G. Peters, and C. von Eiff. 2002. Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small colony variant phenotypes. Infect. Immun. 70:5428-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.von Eiff, C., C. Heilmann, R. A. Proctor, C. Woltz, G. Peters, and F. Götz. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 179:4706-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weinrick, B., P. M. Dunman, F. McAleese, E. Murphy, S. J. Projan, Y. Fang, and R. P. Novick. 2004. Effect of mild acid on gene expression in Staphylococcus aureus. J. Bacteriol. 186:8407-8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolz, C., P. Pohlmann-Dietze, A. Steinhuber, Y. T. Chien, A. Manna, W. van Wamel, and A. Cheung. 2000. Agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol. Microbiol. 36:230-243. [DOI] [PubMed] [Google Scholar]
- 69.Yarwood, J. M., D. J. Bartels, E. M. Volper, and E. P. Greenberg. 2004. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186:1838-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]