Abstract
Prior studies have described a morphologically diverse group of intestinal microorganisms associated with surgeonfish. Despite their diversity of form, 16S rRNA gene surveys and fluorescent in situ hybridizations indicate that these bacteria are low-G+C gram-positive bacteria related to Epulopiscium spp. Many of these bacteria exhibit an unusual mode of reproduction, developing multiple offspring intracellularly. Previous reports have suggested that some Epulopiscium-like symbionts produce dormant or phase-bright intracellular offspring. Close relatives of Epulopiscium, such as Metabacterium polyspora and Clostridium lentocellum, are endospore-forming bacteria, which raises the possibility that the phase-bright offspring are endospores. Structural evidence and the presence of dipicolinic acid demonstrate that phase-bright offspring of Epulopiscium-like bacteria are true endospores. In addition, endospores are formed as part of the normal daily life cycle of these bacteria. In the populations studied, mature endospores were seen only at night and the majority of cells in a given population produced one or two endospores per mother cell. Phylogenetic analyses confirmed the close relationship between the endospore-forming surgeonfish symbionts characterized here and previously described Epulopiscium spp. The broad distribution of endospore formation among the Epulopiscium phylogenetic group raises the possibility that sporulation is a characteristic of the group. We speculate that spore formation in Epulopiscium-like symbionts may be important for dispersal and may also enhance survival in the changing conditions of the fish intestinal tract.
Large microbial symbionts found in the intestinal tract of the herbivorous surgeonfish Acanthurus nigrofuscus (family Acanthuridae) caught in the Red Sea were first described in 1985 (18). The unusual morphology of these organisms, particularly with respect to their production of multiple intracellular offspring or daughter cells, warranted the description of a new genus and species, Epulopiscium fishelsoni (30). Subsequently, Clements and colleagues discovered a morphologically diverse group of Epulopiscium-like intestinal microorganisms in a variety of surgeonfish species from the South Pacific around the Great Barrier Reef (14).
Ten morphological categories of symbionts (see Table 1) were described from a variety of host fish species. These “morphotypes” were defined by several criteria: cell length and shape, the production of intracellular offspring, and whether or not binary fission was regularly observed. Large cigar-shaped cells, such as E. fishelsoni, were categorized as the A morphotype. This morphotype is found in A. nigrofuscus and several other surgeonfish species caught in the Pacific. In total, the survey described Epulopiscium-like symbionts in 21 species of surgeonfish; a number of additional species and locations have been surveyed since (W. L. Montgomery and K. D. Clements, unpublished data). An individual fish often harbors multiple morphotypes and each Epulopiscium morphotype is found in multiple host species. Surgeonfish that harbor these symbiont populations are either herbivorous or detritivorous (primarily feeding on detritus) (14). None of the plankton-feeding surgeonfish species examined harbored Epulopiscium-like symbionts, nor did other herbivorous reef fish outside the surgeonfish family.
TABLE 1.
Surgeonfish symbiont morphotypes as described by Clements et al. (14)
| Type | Cell length observed (μm) | Cell shape | No. of daughter cells | Binary fission |
|---|---|---|---|---|
| A | 70-417 | Cigar-oval | 0-2 | No |
| B | 70-230 | Oval | 0-5 | No |
| C | 40-80 | Cigar | 0-2 | No |
| D | 8-40 | Cigar | 0-2 | No |
| E | 8-120 | Elongate | 0-2 | Yes |
| F | 15-60 | Cigar | 0-7 | No |
| G | 9-60 | Cigar | — | Yes |
| H | 10-100 | Cigar | ? | ? |
| I | 20-100 | Cigar | — | Yes |
| J | 10-240 | Elongate | —a | Yes |
Daughter cells of this morphotype can be difficult to see under light microscopy alone.
Small-subunit rRNA gene sequence comparisons and fluorescent in situ hybridization (FISH) with rRNA-based probes have been used to determine the phylogeny of these unusual microorganisms (5). Sequences obtained from the largest Epulopiscium morphotypes (A and B) form a monophyletic group within the low-G+C gram-positive bacteria (Firmicutes). This surprising result demonstrated that A morphotype Epulopiscium are among the largest known bacteria, with cigar-shaped cells attaining lengths in excess of 600 μm (5, 13). The analyses demonstrated that A morphotype cells from A. nigrofuscus taken from the Red Sea and the Pacific are closely related; some clones derived from geographically disparate hosts showed >98% 16S rRNA gene sequence identity (approximately 1,440 nucleotides used in pairwise comparisons). The analysis also revealed that A morphotype 16S rRNA gene sequences obtained from an individual fish vary by as much as 9%. These phylogenetically distinct types can be distinguished from one another with clade-specific rRNA probes, and in some instances differences in cell morphology, which demonstrates that the A morphotype is comprised of genetically distinct lineages (2).
We developed Epulopiscium group probes based on the A and B morphotype-derived sequences. When used in FISH surveys of surgeonfish intestinal contents, these rRNA probes hybridize to a variety of the described symbiont morphotypes (A, B, C, E, F, and J), indicating that these symbionts comprise a morphologically diverse but phylogenetically coherent group of low-G+C gram-positive bacteria (2). As yet, no member of this group is available in laboratory culture.
A distinguishing feature of Epulopiscium spp. and many of these related surgeonfish symbionts is their ability to produce multiple offspring intracellularly (14, 18, 30). Remarkably, A morphotype populations are synchronized with respect to offspring formation and development follows a circadian cycle (30). Small internal offspring are observed near the tips of the mother cell early in the morning. During the day the offspring cells grow within the mother cell cytoplasm and are eventually released through a perforation in the mother cell envelope. The mother cell is destroyed in the process.
The intracellular offspring of Epulopiscium spp. bear a striking resemblance to the offspring of Metabacterium polyspora (28). Unlike many other endospore-forming low-G+C gram-positive bacteria, M. polyspora regularly produces multiple endospores per mother cell (10). The ability to form multiple endospores is a primary means of cellular propagation for these guinea pig symbionts (6). The close phylogenetic relationship between M. polyspora and Epulopiscium spp. and morphological similarities between intracellular offspring production in these lineages suggest that the two reproductive processes are related (1, 3).
To investigate this hypothesis, the initial stages of intracellular offspring development in the Epulopiscium B morphotype were described using immunolocalization of the cell division protein FtsZ and DNA staining (4). The analysis provided evidence for reorganization of cellular DNA, asymmetric division, and DNA packaging which appear to be shared between endospore formation in Bacillus subtilis and intracellular offspring formation in Epulopiscium spp. The study also demonstrated that, as in morphotype A populations, offspring development in B morphotype populations is synchronized (4).
Previous studies suggested that some Epulopiscium and related surgeonfish symbionts produce dormant or phase-bright offspring (14, 18). Here, we build on those observations and report the characterization of endospores in some Epulopiscium-like morphotypes and describe the diurnal life cycle of these particular lineages. Our study focuses on morphotypes C and J obtained from the surgeonfish Naso lituratus. This host is found around tropic coral reefs throughout the Pacific Ocean and harbors four distinct morphotypes; C, E, H, and J (14) (see Table 1 for morphotype descriptions).
Endospore-specific staining and dipicolinic acid assays indicate that the phase-bright offspring of the Epulopiscium-like symbionts are true endospores. In addition, ultrastructural analyses of these phase-bright offspring reveal features similar to endospores of Bacillus and Clostridium spp. Finally, we document the development of endospores in these populations and the discovery that mature endospores are only seen at night. We propose that some natural populations of Epulopiscium-like symbionts produce endospores as part of their daily life cycle. Phylogenetic analyses indicate that the ability to produce endospores is widespread in the Epulopiscium phylogenetic group. Combined with previous studies, our results support the concept of a symbiotic interaction coordinating fish behavior and the life cycles of their intestinal bacteria to enhance dispersal of these symbionts to other host fish.
MATERIALS AND METHODS
Sample collection.
Intestinal tract samples were obtained from the surgeonfish Naso lituratus collected with spear or hand net at reefs around Hilo, Hawaii. Captured live fish were held in aquaria containing seawater at ambient temperature. At night, aquarium-held fish were shielded from exposure to light. Fish were sacrificed with a sharp blow to the head and pithing. Intestinal contents were removed and fixed in 80% ethanol, 4% formaldehyde or suspended in 20% sterile glycerol. Ethanol-fixed samples and glycerol suspensions were stored at −20°C. Formaldehyde-fixed samples were stored at 4°C until processed.
Preparation of Bacillus subtilis endospores.
B. subtilis strain PY79 inoculated into Luria-Bertani medium was incubated at 37°C for 7 days. Phase-contrast microscopy revealed the presence of mature endospores. Cells and spores were collected by centrifugation at 2,260 × g for 15 min. The pellet was washed with phosphate buffered saline (PBS) containing, per liter, 8.0 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4 · 7H2O, and 0.24 g KH2PO4, pH 7.4, and resuspended in 5 ml PBS. The suspension was then sonicated for 1 min to dislodge endospores from mother cells and cellular debris. The cell debris and endospores were collected by centrifugation, washed once with PBS and resuspended in 1.0 ml PBS. The suspension was layered on top of 10 ml of Percoll (Amersham Biosciences Corp.) diluted 9:1 in 1.5 M NaCl. A buoyant density gradient was created with centrifugation at 10,000 × g for 15 min. The bottom 3 ml of the gradient containing the endospores was collected and washed with 5 volumes PBS.
Microscopic analyses of endospores.
Surgeonfish intestinal contents, guinea pig fecal samples or B. subtilis cultures with endospores were washed once in PBS prior to testing. Three endospore tests, the popping test, Schaeffer-Fulton (42) staining, and Dorner (16) staining methods were performed as previously described (31) with minor modifications. These three tests have all been used to identify endospores in a variety of endospore-forming species. Specifically, the popping test has been used to distinguish endospores from vegetative cells in B. subtilis, Bacillus megaterium, and Metabacterium polyspora (3, 40). For the popping test, samples were incubated in 200 μl of 1 M HCl for 10 min at 60°C. The pH of the solution was adjusted with the addition of 200 μl 1 M NaOH and 10 μl 1 M Tris, pH 8.0. After each procedure, cells were mounted in glycerol on a microscope slide and viewed using phase-contrast (for the popping test) or bright-field (for both endospore staining procedures) microscopy.
DNA staining.
Fixed surgeonfish intestinal contents were washed with and resuspended in TE (10 mM Tris, 1 mM EDTA; pH 8). Cells were stained by adding 4′,6′-diamidino-2-phenylindole (DAPI) to a concentration of 2 μg/ml and incubating for 15 min. The suspension was washed once with TE and mounted in glycerol on a microscope slide. For DNase pretreatment, cells were washed in sterile water and suspended in 50 μl of 10 mM Tris, pH 8.0; 10 mM MgCl2; and DNase, 1 mg/ml, and incubated for 20 min at 37°C. Cells were washed with 10 mM Tris, pH 8.0, prior to staining with DAPI.
Bright-field, phase-contrast, Nomarski differential interference contrast (DIC) and fluorescence microscopy were performed using an Olympus BX61 epifluorescent microscope, with 20× UPlanFl (N.A. 0.50), 40× UPlanFl (N.A. 0.75), 60× PlanApo (N.A. 1.4), and 100× UPlanApo (N.A. 1.35) objectives. The microscope is equipped with filter cubes for viewing fluorescein and DAPI fluorescence. Images were acquired using a Cooke SensiCam with a Sony Interline chip and Slidebook software (Intelligent Imaging Inc.). Cell and endospore measurements were determined with the software (calibrated with a stage micrometer).
Transmission electron microscopy.
Surgeonfish intestinal contents that had been stored as frozen glycerol stocks were used for electron microscopy studies. A sample (collected at 00:00, midnight) was thawed, washed in 100 mM cacodylate buffer (pH 7.0) and immediately suspended in 100 mM cacodylate buffer containing 4.0% paraformaldehyde, 4.0% gluteraldehyde, and 2 mM CaCl2. The sample was fixed overnight at 4°C then washed 3 times with 100 mM cacodylate buffer. The sample was then fixed overnight with 1.0% osmium tetroxide in cacodylate buffer. Endospores were embedded in Spurr's resin as previously described (21). Resin blocks were thin sectioned and stained with uranyl acetate and lead citrate at the Cornell Integrated Microscopy Center and imaged in a Morgagni transmission electron microscope (Philips) operated at 80 kV.
Dipicolinic acid assay.
Dipicolinic acid (DPA) extractions were performed as described (34) with modifications. The extraction procedure was performed on a 600-μl aliquot of surgeonfish intestinal contents that had been stored frozen in glycerol. One sample collected at 00:00 (midnight) contained approximately 106 phase-bright offspring in a variety of Epulopiscium-like symbionts; another sample tested had been collected at 13:00 and contained comparable numbers of Epulopiscium-like cells but no phase-bright offspring. Extracts from 0.3 g guinea pig feces containing mature endospores of M. polyspora (1.1 × 107 endospores) and 1 ml of B. subtilis culture (4.1 × 108 endospores) as well as vegetative B. subtilis and guinea pig fecal samples without M. polyspora were analyzed.
Samples were washed with 10 mM Tris (pH 8.0), suspended in 0.5 ml of extraction buffer (40 mM acetic acid, 30% methanol; pH 6.5) in a 2.0-liter microcentrifuge tube and placed in a boiling water bath for 15 min to release the DPA. The samples were then centrifuged 5 min at 14,000 × g. The supernatant was collected and adjusted to pH 2 with 1.0 M HCl. The resulting solution was extracted once with 1.0 ml ethyl acetate and then reextracted with 0.5 ml ethyl acetate. The ethyl acetate fractions were pooled and evaporated to dryness under a stream of nitrogen gas. The resulting pellet was suspended in 10 μl ethyl acetate and treated with 10 μl bis(trimethylsilyl)trifluoroacetamide (BSTFA) at room temperature.
A 0.2 μl aliquot of the sample was analyzed with gas chromatography/mass spectroscopy (GC/MS) as described previously (9). Purified DPA (Acros Organics, New Jersey) was used as a standard. Since the retention time and mass spectral fragmentation pattern for DPA is not in the library of compounds included with the instrument software, the mass to charge ratio (m/e) of predicted products were calculated. These calculations were based on the assumption that both carboxyl groups on DPA would be trimethylsilyated by reaction with BSTFA, yielding a parent compound with an m/e of 311. In addition, DPA derivatives were confirmed by comparison of chromatographic retention time and MS fragmentation patterns of the DPA standard.
Phylogenetic analyses of morphotypes.
Genomic DNA was extracted from a 0.35 ml aliquot of ethanol-fixed intestinal contents. Cells were washed twice with PBS and suspended in 250 μl of lysis buffer (50 mM Tris, 5.0 mM EDTA; pH 8.0) containing 1.0 mg/ml proteinase K (Merck) and incubated for 30 min at 37°C followed by 30 min at 50°C. An aliquot of 25 μl of 10% sodium dodecyl sulfate (SDS) was added and the suspension was treated with 4 cycles of freeze-thaw, by transfer between a dry ice-ethanol bath and a 70°C heating block. The lysate was extracted twice with 2 volumes phenol-chloroform-isoamyl alcohol (25:24:1) (Amresco) and nucleic acids were precipitated from the aqueous phase. DNA was dissolved in 10 mM Tris, pH 8.0 and stored at −20°C.
The 16S rRNA genes were PCR amplified from a 1:100 dilution of the extracted DNA using oligonucleotide primers designed to conserved regions of the gene, specifically 27F with 1492R (Table 2). PCR products were cloned into a plasmid vector using the TOPO TA cloning kit (Invitrogen) according to the protocol provided by the manufacturer. Clones were screened using PCR amplification with the primer set 27F and 1423R (Epulo probe). Unique clones were selected based on restriction fragment length polymorphisms of digests of PCR-amplified inserts using HaeIII, HhaI, TaqαI, and MseI (New England Biolabs). Plasmid DNA from selected clones was isolated using the RPM kit (Q-BIOgene). The nucleotide sequence of the cloned DNA was determined. All DNA sequencing reactions employed Big Dye Terminator chemistry with AmpliTaq-FS DNA polymerase (Applied Biosystems) and were performed at the Cornell University BioResource Center. Sequences were determined using an Applied Biosystems Automated 3730 DNA Analyzer.
TABLE 2.
Oligonucleotide PCR primers and FISH probes
| Primer name | Oligonucleotide sequencea (5′-3′) | Reference | Morphotypes hybridized |
|---|---|---|---|
| 27F | AGAGTTTGATCCTGGCTCAG | 29 | |
| 1492R | GGTTACCTTGTTACGACTT | 29 | |
| 1423R Epulo probe | *TTGCGGTTAGGTCACTGACGTTGGGCCCCCT | 5 | All |
| C1 probe (198R) | *TTTGCCGTAGCCACCATGC | This study | C1 |
| C2 probe (198R) | *CTTTGCTATACACACCATGC | This study | C2 |
| J1 probe (198R)b | *ATGCGATAAAATTAGTGTATGC | This study | J1 |
| J2 probe (827R) | *ATCCTTAAATCCCGACACCT | This study | J2 |
| J group probe (1260R) | *CGTCACCGCTATGCTT | This study | J1 and J2 |
| 515F probe | *GTGCCAGCMGCCGCGG | 15 | None |
| Euk probe | *GGGCATCACGGACCTG | 15 | None |
5′ 6-carboxyfluorescein-modified primers are marked with an asterisk.
Weak hybridization signal was observed.
DNA sequences were aligned using ClustalX (11) and analyzed using the PHYLIP (Phylogenetic Inference Package version 3.6; J. Felsenstein, University of Washington [http://evolution.genetics.washington.edu/phylip.html]). 16S rRNA gene alignments were visually scanned for variable regions suitable for targets of clade-specific oligonucleotide probes. Probes complementary to rRNA sequences were designed based on Tm, the absence of significant intermolecular complementarity and the position of the probe in the predicted rRNA secondary structure (7, 20). The rRNA probes were compared with DNA sequences in GenBank using BLAST (8). Oligonucleotide probes were synthesized as 5′-phosphoramidite conjugates of fluorescein by Integrated DNA Technologies. Fluorescent in situ hybridizations were performed as previously described (15) with minor modifications.
Nucleotide sequence accession numbers.
The nearly complete 16S rRNA gene sequences were deposited in GenBank (accession numbers AY844964 to AY844991).
RESULTS
Daily life cycle of Epulopiscium-like symbionts of N. lituratus.
Periodically throughout the day and into the night, fish were sacrificed, intestinal contents were collected and Epulopiscium-like cells from these samples were observed. During the day, the C and J morphotypes were motile and some possessed pole-associated internal bodies (offspring or daughter cells). The Epulopiscium-like cells were uniformly phase dark or phase bright and internal offspring were difficult to see (Fig. 1A). However, dramatic differences were observed in the Epulopiscium-like populations collected from 00:00 (midnight) to 04:00; C and J morphotype cells in these samples were not motile and most Epulopiscium-like cells contained distinct, phase-bright intracellular offspring (Fig. 1B). In samples collected at 06:45, few phase-bright offspring were seen.
FIG. 1.
Micrographs of Epulopiscium-like symbionts of N. lituratus. Collection times are provided in the upper left of each set of images. (A and B) Phase-contrast (PC) images of surgeonfish intestinal contents. In these panels, representatives of the C and J morphotypes are indicated with arrows. Note the prominent phase-bright intracellular offspring seen in the midnight (00:00) sample only. (C to H) Higher magnification images of individual type C (C and D) or J (E to H) cells. The type of image is indicated above each column: DAPI, fluorescence; DIC, Nomarski differential interference contrast. For some, PC images are also provided. PC images were taken with short exposure times to prevent saturation of pixels and to preserve cell structure. (C) Bright DNA staining within the cytoplasm of two large, phase-dark forespores. (D) DAPI cannot penetrate the phase-bright spores but instead associates with the outer most layers of these offspring. (E to H) Developmental progression of a type J endospore. Each panel focuses on the tip of a type J cell. Drawings are provided in the DIC panels for E and G to illustrate the position of the forespore within a mother cell. (E) Bright DAPI staining within the cytoplasm of a forespore. (F) The forespore DNA takes on a rope-like configuration (upper cell). A mature endospore (lower cell in this series) shows smooth, peripheral staining with DAPI. (G) PC and DIC show an offspring with a slightly rough or irregular surface appearance. DAPI can enter the cytoplasm of this forespore and stain the DNA. (H) A mature endospore is phase-bright and has a smooth outer surface in DIC and PC optics. DAPI cannot enter the core of the mature spore but instead staining of the cortex is observed. Scale bars represent 20 μm. Scale bar in A also applies to B. The scale in D applies to C also. The bar in H applies to panels E to H.
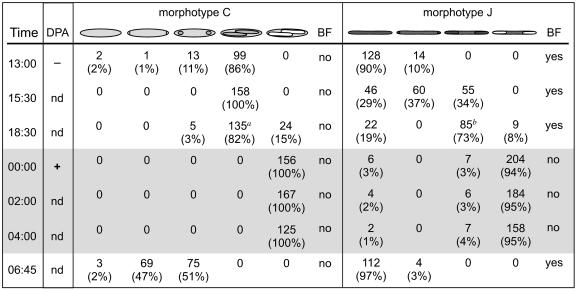
Fixed samples of surgeonfish intestinal contents were further evaluated using DAPI staining and phase-contrast microscopy. A tally of developmental stages for the C and J morphotypes collected at intervals starting at 13:00 and ending at 06:45 are presented in Fig. 2. We found that offspring development in the C and J morphotypes from N. lituratus follows a circadian cycle and populations within a fish are synchronized with respect to development. But unlike previously described cycles, these cells produce phase-bright offspring at night. C morphotype populations are more developmentally synchronized than J morphotype populations. Nearly all C morphotype cells produce phase-bright offspring at night while approximately 95% of the J morphotype cells produce phase-bright offspring.
FIG. 2.
Developmental stages seen in C and J morphotype cells collected periodically throughout the day and night. Collection times are shown on the left, followed by DPA assay results, a plus sign indicates DPA was present in the sample, minus is for no DPA, “nd” not determined. Across the top of the chart are drawings representing developmental stages; forespores are shown in dark gray, mature endospores are shown in white. For binary fission (BF) data, “no” indicates binary fission was not observed in the cells surveyed; “yes” indicates binary fission was observed in some cells. Numbers of cells and percentages of total cells counted are provided separately for each morphotype. a indicates the only time point where either rough, phase-bright forespores (in 29 cells) or rope-like DNA in forespores (in 23 cells) was observed for the C morphotype; b indicates the only time where either rough, phase-bright forespores (in 10 cells) or rope-like DNA in forespores (in 28 cells) was observed in J morphotype cells. The shaded part of the chart indicates samples taken from fish held in the dark.
In samples collected from 13:00 to 15:30 viewed with phase-contrast microscopy alone, intracellular offspring were often difficult to see. Cells with intracellular offspring had bright DAPI staining within the cytoplasm of their offspring and less-bright DNA staining associated with the mother cell cytoplasm (Fig. 1C, for example). These observations are similar to the patterns of DNA staining in large Epulopiscium morphotypes A and B (4, 30, 38).
Samples collected at 18:30 contained Epulopiscium-like populations that appeared to be in transition, with some cells containing phase-bright offspring. A portion of each population had mother cells with offspring that exhibited DNA staining throughout the cytoplasm (Fig. 1E), as seen in the 15:30 samples. But some cells possessed an unusual form of peripherally located DNA within the offspring (Fig. 1F, upper cell). A similar form of DNA has been described in forespores of Metabacterium polyspora (6). This rope-like conformation is only seen in large forespores and may be attributed to the association of forespore DNA with DNA-binding proteins, such as small acid soluble proteins (44, 45).
The 18:30 sample contained two forms of phase-bright offspring. The surfaces of some phase-bright offspring (approximately 18% of the C morphotype, 9% of the J morphotype) had a rough appearance when viewed using Nomarski DIC or phase-contrast microscopy (Fig. 1G). DAPI staining of these rough, phase-bright cells revealed coiled, peripherally located structures within these offspring. In contrast, all other phase-bright offspring had a very smooth surface and exhibited DAPI staining localized to the outer surface of the offspring cell (Fig. 1F, lower cell). This type of offspring and pattern of staining was prevalent in samples taken from midnight (00:00) to 04:00 (Fig. 1D and H).
DAPI and other DNA dyes have an apparent affinity for the cortex of mature dormant endospores (43). Pretreatment with DNase eliminates the DAPI staining of DNA in phase-dark offspring. Coiled DNA was no longer seen in phase-dark offspring however phase-bright cells retained their DAPI-stained coiled DNA. The peripheral DAPI staining of smooth phase-bright offspring was still observed (data not shown). This result indicates that the coiled structures within phase-dark and rough, phase-bright offspring cells are probably comprised of DNA. The DNA in rough, phase-bright cells is still accessible to small molecules (like DAPI) but protected from larger molecules such as DNase. Full refractility develops over time during the later stages of endospore development, coincident with cortex development and coat formation (51).
We believe that the rough, phase-bright offspring of the Epulopiscium-like symbionts in the 18:30 sample are forespores that have started to become phase bright but have not yet fully developed resistance to chemical fixatives such as ethanol and therefore the DNA in these ethanol-fixed cells can be readily stained with DAPI. Samples collected later at night (from midnight to 04:00) contained some vegetative cells but the majority of Epulopiscium-like cells contained phase-bright offspring with only the smooth, surface DAPI staining of mature endospores.
Characterization of the phase-bright offspring.
Samples of Epulopiscium-like cells with phase-bright offspring were tested with the endospore-specific Schaeffer-Fulton and Dorner staining methods (31). Mature endospores of gram-positive bacteria resist staining (22). Both the Schaeffer-Fulton and Dorner methods heat the cells in the presence of stain which allows malachite green and carbolfuchsin, respectively, to enter endospores. Destaining is performed at room temperature. At this lower temperature stain is trapped within mature endospores but not vegetative cells. The vegetative cells are then counterstained. The choice of counterstains in both procedures allows one to readily differentiate endospores and vegetative cells.
Using these procedures the phase-bright offspring stained bright green with malachite green and red with carbolfuchsin (data not shown). These results are indicative of endospores. In addition, fixed samples containing either phase-bright offspring or phase-gray offspring were heated in the presence of a strong acid. This treatment weakens the endospore cortex (31). As a result cytoplasm with DNA is extruded from the spore core, forming a characteristic mushroom-shaped structure while having little effect on the morphology of vegetative cells (39-41). Acid treatment caused cytoplasm extrusion of the phase-bright offspring of all of the morphotypes (data not shown), but had no apparent affect on mother cells or phase-gray offspring.
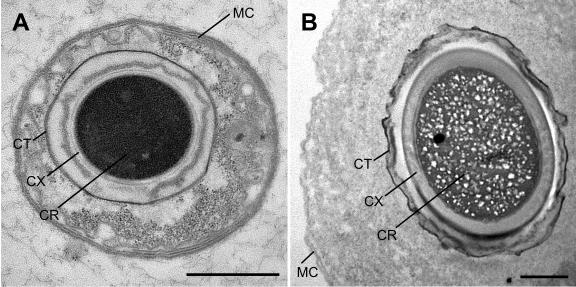
Transmission electron microscopy of thin sections of samples collected at 00:00 (midnight) was used to examine the ultrastructure of phase-bright offspring from Epulopiscium-like symbionts (Fig. 3). Each of the characteristic layers and structures associated with mature endospores of low-G+C gram-positive bacteria (41) was found in most of the intracellular offspring examined. Most small intracellular offspring (with core cross sectional diameter of approximately 1 μm or less) possessed electron-dense cores (Fig. 3A), however, the cores of many larger offspring (>1 μm in cross-section) appeared less dense and contained irregular structures (Fig. 3B). The nature of these internal structures is not known.
FIG. 3.
Ultrastructure of intracellular offspring of Epulopiscium-like surgeonfish symbionts. (A) Thin section of an Epulopiscium-like cell harboring an offspring with an electron-dense core. (B) A wider cell containing an endospore with electron-dense spore coat and a mottled spore core. Features common to all cells are provisionally labeled: MC = mother cell envelope, CT = outer layer of the coat(s), CX = cortex, and CR = core. Scale bars represent 1.0 μm.
Dipicolinic acid is a small molecule found in high concentrations in dormant endospores (36, 37) and occasionally it is produced by some fungi (25, 49). GC/MS analysis was used to test for the presence of DPA. Extracts from surgeonfish intestinal samples collected at midnight yielded a large peak with a retention time of 5.64 min (Fig. 4A, upper panel). This retention time corresponded to that of the major analyte peak obtained from GC analysis of pure DPA (Fig. 4A, lower panel). A peak with an identical retention time was the major analyte seen in extracts of B. subtilis and M. polyspora endospores (data not shown). It should be noted that the B. subtilis endospores used as a control in this assay were isolated from LB-grown cultures and LB is not commonly used for sporulation of B. subtilis. Although B. subtilis PY79 will produce endospores when grown for extended periods in LB, the frequency of endospore formation is lower compared to sporulation in Difco sporulation medium (33), and development is not synchronized within the culture. No peak at 5.64 min was seen in extracts from samples without phase-bright endospores (data not shown).
FIG. 4.
Comparison of GC/MS analyses of dipicolinic acid extracted from surgeonfish intestinal contents with spores (upper) and purified DPA (lower; inversed). (A) Gas chromatograph and (B) mass spectrum analysis of GC peak of 5.64-min retention time. The y axes are in arbitrary units (106).
Since DPA was not included in the GC/MS software library of compounds we calculated the m/e of the trimethylsilyated DPA to be 311, assuming both carboxyl groups of DPA would be silyated upon reaction with BSTFA. Accordingly we observed a minor product peak at m/e = 311, corresponding to this parent compound (Fig. 4B, upper panel). The major product peak, m/e = 296, corresponds to the parent compound minus one methyl group. The peak of m/e = 73 corresponds to the trimethylsilyl ion, which is characteristic of trimethylsilylated compounds (9). The peak of m/e = 147 represents a fragmentation product that we could not ascertain. This fragmentation pattern was comparable to fragmentation patterns of the DPA standard (Fig. 5B, lower panel).
FIG. 5.
Phylogenetic analyses of C and J morphotypes from N. lituratus. (A) Phylogenetic tree constructed from 16S rRNA gene sequence comparisons demonstrating the relationship of endospore-forming C1, C2, J1, and J2 clades to previously characterized Epulopiscium A1, A2, and B. Bootstrap values, based on 100 replicates, are provided at nodes. GenBank accession numbers follow names. Sequences obtained from samples from two separate fish (N.l. 1 and N.l. 2) were used in the phylogenetic analysis. The scale represents 0.02 nucleotide changes per position. (B) Identification of morphotypes using clade-specific oligonucleotide probes. Pairs of micrographs are shown with phase-contrast images on the left and epifluorescent images of the same cells to the right. Note the regions in the cells without fluorescence where the oligonucleotide probes were unable to enter the endospore. Cell type designations that correspond to the four major clades shown in A are provided in the upper left of each phase-contrast image. Scale bars represent 20 μm.
Phylogenetic diversity of Epulopiscium-like populations in Hawaiian surgeonfish.
A variety of Epulopiscium-like morphotypes was consistently found in N. lituratus. Qualitatively, this fish species harbors a similar variety of morphotypes as reported for N. lituratus collected around the Great Barrier Reef (14). However, overall community composition, that is the relative ratios of morphotypes, varied from one fish to another.
A 16S rRNA gene-based phylogenetic analysis was performed to characterize the predominant morphotypes found in N. lituratus and to determine their relatedness to previously described Epulopiscium. Clone libraries generated from DNA extracted from intestinal contents of two individual fish were used in the analyses. Several distinct lineages were identified (Fig. 5A). To match phylogenetic groupings with morphological types, oligonucleotide probes, representing each major clade, were designed and used in hybridizations with surgeonfish intestinal contents. Although clone libraries were derived from vegetative cells, hybridizations were performed on both vegetative cells and samples of cells with phase-bright offspring.
For cells with phase-bright offspring, hybridization was seen within the mother cell cytoplasm but not within the offspring themselves (Fig. 5B). All morphotypes hybridize with the Epulo group probe 1423R and no fluorescence above background was viewed with negative control probes (rRNA-like 515F and Euk probes, Table 2). Each clade-specific probe hybridizes to a distinct subset of morphotypes, which allows us to recognize subtypes within the C and J morphotypes. Clones belonging to the C1 group were by far the most abundant in our clone libraries even though they were not the most abundant cells in these communities. This may be due to the large amount of DNA found within this cell type compared with the other morphotypes examined here, or due to differential extraction of DNA from the various morphotypes. A description of the C1, C2, J1, and J2 populations is provided in Table 3. Although we were unable to assign a phylogenetic position to all of the morphotypes found in the fish we found that the relative proportions of the characterized C and J populations vary from one fish to another (Table 4). It is worth noting that the newly characterized C and J clades are more closely aligned with the A2 and B morphotypes than the A1 clade is to the A2 or B clades. Therefore, the diversity of sequences derived from this analysis falls within the previously described Epulopiscium group.
TABLE 3.
Measurements of cell types identified with FISH
| Cell type (no. of cells) | Range, minimum-maximum, μm (mean ± SD)
|
Binary fission | ||||
|---|---|---|---|---|---|---|
| Cell length | Cell width | Spore length | Spore width | No. of offspring | ||
| C1 (88) | 42-131 (72.9 ± 8.2) | 8-20 (12.4 ± 1.4) | 19-70 (38.4 ± 5.0) | 3-11 (5.8 ± 0.9) | 0-2 | No |
| C2 (71) | 39-81 (57.8 ± 8.2) | 3.5-12 (6.2 ± 1.3) | 15-43 (24.7 ± 5.5) | 2-5 (3.3 ± 0.7) | 0-2 | No |
| J1 (99) | 53-341 (133.1 ± 46.2) | 2-5.5 (3.5 ± 0.5) | 10-60 (31.1 ± 6.5) | 1.5-5 (2.3 ± 0.4) | 0-2 | Yes |
| J2 (83) | 44-362 (129.6 ± 49.7) | 1.5-3 (2.0 ± 0.4) | 13-55 (33.7 ± 10.9) | 0.5-2 (1.0 ± 0.4) | 0-2 | Yes |
TABLE 4.
Relative numbers of Epulopiscium-like symbiont cell types identified in fish
| Fish no. (no. of cells) | No. of cell type (% of total)
|
||||
|---|---|---|---|---|---|
| J1 | J2 | C1 | C2 | Other | |
| 30 (566) | 53 (9) | 164 (29) | 83 (15) | 69 (12) | 197 (35) |
| 2 (477) | 62 (13) | 171 (36) | 0 | 0 | 244 (51) |
| 25 (400) | 38 (9) | 59 (15) | 0 | 26 (7) | 277 (69) |
| 24 (594) | 100 (17) | 74 (12) | 70 (12) | 55 (9) | 295 (50) |
| 16 (589) | 33 (6) | 86 (15) | 1 (0.2) | 34 (6) | 435 (74) |
DISCUSSION
Our attention became focused on possible endospore formation in Epulopiscium-like symbionts when night collections of intestinal contents from surgeonfish in Hawaii were viewed with phase-contrast microscopy; strikingly, almost the entire Epulopiscium-like community taken from any of these fish harbored phase-bright offspring. Phase-bright offspring in Epulopiscium-like populations had been reported previously (14) but not to the extent that was observed in these night collections.
All tests of the phase-bright offspring of symbionts from N. lituratus reveal characteristics common to endospores. The positive results of endospore-specific staining and the presence of DPA support the conclusion that the nocturnal, phase-bright offspring of Epulopiscium-like symbionts are true endospores.
Transmission electron micrographs of thin sections of the Epulopiscium-like cells collected at midnight showed structures comparable to the coat, cortex and core of a mature endospore (19, 41). Smaller intracellular offspring (<1 μm in cross-sectional diameter) contained an electron-dense core, however most large offspring possessed cores with a mottled, lighter appearance. The electron-dense granular appearance of a spore core can be indicative of an immature or germinated spore, while mature endospore cores often have a poor affinity for stains and are more electron lucent (27, 51). The nature of the white granular structures within the core is difficult to determine. These structures may represent storage products or crystals of DPA. Some large forespores in these samples contain structures that stain bright yellow with DAPI, which may be indicative of polyphosphates (26), although further tests are needed to confirm this.
Another striking feature of the developing endospores of Epulopiscium-like cells is the conversion of forespore DNA into a rope-like structure within the internal periphery of the cell. A similar conformation of genomic DNA is present in large forespores of M. polyspora late in development (6). We speculate that these structures are similar to the highly condensed DNA observed in stage III B. subtilis forespores (35, 47).
As yet, no Epulopiscium-like intestinal symbiont can be maintained in laboratory culture, which limits our ability to test for resistance traits in these endospores. However, these cells resist the penetration of dyes and appear to have a mineralized core that contains substantial quantities of DPA, which is consistent with at least some of the qualities of other endospores (33, 46). Our attempts to germinate endospores from these Epulopiscium-like surgeonfish symbionts under anaerobic laboratory culture conditions were unsuccessful (data not shown). This is not too surprising since we have not yet determined the nutritional requirements for Epulopiscium spp. or germinant signals for these endospores.
Endospores from Epulopiscium-like symbionts are the largest described thus far. Large C1 morphotype endospores (70 by 11 μm) are greater than 4,000 times the volume of spores of B. subtilis (2 by 1 μm). Endospore formation and maturation in the C morphotype resemble spore formation in many low-G+C gram-positive bacteria but may include stages specialized for developing full resistance traits in enormous cells. Despite these differences, such large endospores may provide an exceptional model for studies of protein localization and endospore maturation.
The 16S rRNA gene-based phylogenetic analysis revealed that N. lituratus is host to at least four distinct lineages of symbionts that are closely related to previously described Epulopiscium spp. The C1, C2, J1, and J2 clades differ from one another, with identity of 94.6 to 97.9% over 1,430 nucleotides compared, which raises the possibility that these subtypes represent different “species” (50) that are discernible from the previously identified lineages. These four newly identified clades are allied with the previously described A2 and B lineages. The previously characterized A1 lineage forms the deepest divergence in the Epulopiscium clade, followed by the divergence of the J morphotypes. This configuration separates the A1 clade from the A2 clade, and places the A2 and B clades among the endospore-forming symbionts.
The Epulopiscium A and B morphotypes are described as live-offspring-bearing, or viviparous. The phylogenetic data presented here suggest that endospore formation is widespread within the Epulopiscium group and raises the possibility that other Epulopiscium morphotypes produce endospores as a part of their daily life cycle or that they possess a cryptic sporulation program. Furthermore, preliminary genome sequence data from Epulopiscium type B has identified homologues of late sporulation genes, suggesting that this large morphotype has the capacity to produce mature endospores. Another possibility is that mechanisms exist that allow these cells to alternate between viviparity and endospore formation. Such alternative reproductive strategies have been described in other endospore-forming Firmicutes such as the segmented filamentous bacteria of rodents (1). Appropriate samples for assessing these possibilities in other Epulopiscium morphotypes have yet to be examined.
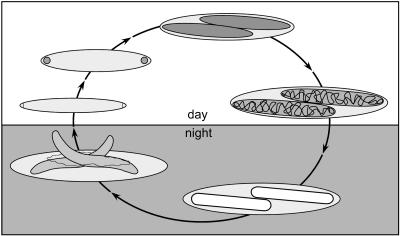
Based on observations of samples collected throughout the day and into the night we found that endospore development in Epulopiscium-like symbiont populations within N. lituratus follows a predictable diurnal cycle. In the C morphotype, small intracellular offspring (forespores) form early and grow throughout the day, while the host surgeonfish is active (Fig. 6). In the late afternoon (our 18:30 collection, for example), the DNA within each forespore takes on a rope-like configuration, which may indicate the formation of DNA-protein complexes in preparation for dormancy (23, 32, 45). Shortly thereafter, the forespores mature into phase-bright endospores. By midnight, almost all type C cells and most J's contain mature endospores. By this stage, we believe, the mother cell begins to deteriorate. Ribosomes are still present in the mother cell, but the cells are no longer motile and apparently never regain motility. The endospores are retained in the mother cell throughout the night. Germination begins early in the morning. By the time the sun rises (just before our 06:45 collections), endospores are no longer abundant.
FIG. 6.
Life cycle of the endospore-forming C morphotype. Starting around sunrise, the cell divides at both cell poles. Intracellular offspring form by engulfment of these polar cells. Throughout the day offspring enlarge within the mother cell. Late in the afternoon, prior to sunset, the DNA within these offspring (forespores) takes on a rope-like configuration at the periphery of the forespore cytoplasm. The offspring mature into phase-bright endospores and remain dormant throughout the night. Prior to sunrise, the endospores begin to germinate and are released from the mother cell to repeat the cycle.
It is apparent that a symbiotic relationship exists between Epulopiscium-like populations and their surgeonfish host. These are large, thick-bodied fish, so no light reaches the intestinal microbial community. Therefore, intercellular signaling within the community and/or signals from the host must entrain cell cycle events. A host-symbiont interaction as described here would coordinate symbiont biomass increase, i.e., the rapid growth of mother cells and their intracellular offspring, with times when the host is actively feeding. The lysis of mother cells may also provide the host with nutrients at the end of their nightly fast.
Endospore formation is a developmental process exhibited by some members of the low-G+C gram-positive bacteria. In the model organism for studying endospore formation, B. subtilis, an endospore is produced in response to nutrient stress, allowing the vegetative cell to form a highly resistant, dormant cell (17, 24, 48). Morphological and molecular features exclusive to endospores provide resistance to environmental stresses to ensure survival during periods of dormancy. Signals from the environment induce germination of the endospore, which leads to hydration and outgrowth of a metabolically active vegetative cell. While endospore formation is brought on by nutrient deprivation in many sporeforming bacterial species, some low-G+C gram-positive bacteria, such as M. polyspora, use endospore formation for propagation and for survival outside of its host, the guinea pig (1). In M. polyspora, endospore formation is part of the normal life cycle of the bacterium. Germination and sporulation are coordinated with passage of M. polyspora through the gastrointestinal tract of its natural host (6). Binary fission occurs during a brief stage in the life cycle of M. polyspora and the ability to form multiple endospores per mother cell is a significant form of reproduction.
Sporulation would assist dispersal of surgeonfish intestinal symbionts too. Previous studies have shown that juveniles of some surgeonfish exhibit coprophagy toward feces of adults and other juvenile surgeonfish, demonstrating a means of obtaining complex intestinal microbiota (12). Even brief exposure to seawater could be detrimental to vegetative cells of strictly anaerobic intestinal symbionts. Like M. polyspora, the ability of Epulopiscium-like cells to develop endospores provides protection from oxygen and injury during passage through the upper gastrointestinal tract of the host.
While we regularly observed evidence of binary fission in J morphotype cells, we never saw C morphotype cells undergo binary fission. It appears that C morphotype cells rely exclusively on the daily formation of two endospores for propagation. It is remarkable that these cells live in a nutrient rich intestinal environment but spend a large portion of their life, perhaps as much as 12 h a day, in a dormant state. This dormancy corresponds to periods when the fish is less active and perhaps when nutrients would be less available to the symbionts. Although spore formation is an energetically expensive process, either some aspects of this life style allow for this expense or, alternatively, dormancy provides an advantage to the C morphotype. It is possible that some of this apparent expense to the symbiont, that is, casting off the mother cell remains early in the morning, may provide the host some nutritional benefit, thus reinforcing the symbiotic association.
Acknowledgments
We thank Bill Ghiorse for his enthusiastic support and guidance with TEM studies and Anatol Eberhard for his guidance and use of the GS/MS. We also thank Kendall Clements and Ralph Slepecky for helpful comments, Anita Aluisio for thin sectioning, and Mark Albins for assistance in the field. We are particularly grateful for the support and hospitality of our hosts in Hawaii: Karla McDermid of the Marine Sciences Department, University of Hawaii, Hilo, and Margaret McFall-Ngai and Edward Ruby of the Kewalo Marine Laboratory of the Pacific Biomedical Research Center.
This project was supported by National Science Foundation awards MCB 0075738 (to W.L.M.) and MCB 0237025 (to E.R.A.).
REFERENCES
- 1.Angert, E. R. 2005. Alternatives to binary fission in bacteria. Nat. Rev. Microbiol. 3:214-224. [DOI] [PubMed] [Google Scholar]
- 2.Angert, E. R. 1995. Molecular phylogenetic analyses of Epulopiscium-like symbionts. Ph.D. thesis. Indiana University, Bloomington, Ind.
- 3.Angert, E. R., A. E. Brooks, and N. R. Pace. 1996. Phylogenetic analysis of Metabacterium polyspora: clues to the evolutionary origin of daughter cell production in Epulopiscium species, the largest bacteria. J. Bacteriol. 178:1451-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angert, E. R., and K. D. Clements. 2004. Initiation of intracellular offspring in Epulopiscium. Mol. Microbiol. 51:827-835. [DOI] [PubMed] [Google Scholar]
- 5.Angert, E. R., K. D. Clements, and N. R. Pace. 1993. The largest bacterium. Nature 362:239-241. [DOI] [PubMed] [Google Scholar]
- 6.Angert, E. R., and R. M. Losick. 1998. Propagation by sporulation in the guinea pig symbiont Metabacterium polyspora. Proc. Natl. Acad. Sci. USA 95:10218-10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behrens, S., C. Ruhland, J. Inacio, H. Huber, A. Fonseca, I. Spencer- Martins, B. M. Fuchs, and R. Amann. 2003. In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes. Appl. Environ. Microbiol. 69:1748-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2004. GenBank: update. Nucleic Acids Res. Database Issue 32:D23-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brencic, A., A. Eberhard, and S. C. Winans. 2004. Signal quenching, detoxification and mineralization of vir gene-inducing phenolics by the VirH2 protein of Agrobacterium tumefaciens. Mol. Microbiol. 51:1103-1115. [DOI] [PubMed] [Google Scholar]
- 10.Chatton, E., and C. Pérard. 1913. Schizophytes du caecum du cobaye. II Metabacterium polyspora n. g., n. s. C. R. Hebd. Soc. Biol. (Paris) 74: 1232-1234. [Google Scholar]
- 11.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clements, K. D. 1997. Fermentation and gastrointestinal microorganisms of fishes, p. 156-198. In R. I. Mackie and B. A. White (ed.), Gastrointestinal microbiology: gastrointestinal ecosystems and fermentations, vol. 1. Chapman & Hall, New York, N.Y. [Google Scholar]
- 13.Clements, K. D., and S. Bullivant. 1991. An unusual symbiont from the gut of surgeonfishes may be the largest known prokaryote. J. Bacteriol. 173:5359-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clements, K. D., D. C. Sutton, and J. H. Choat. 1989. Occurrence and characteristics of unusual protistan symbionts from surgeonfishes Acanthuridae of the Great Barrier Reef, Australia. Mar. Biol. 102:403-412. [Google Scholar]
- 15.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 16.Dorner, W. 1922. Ein neues verfahren für isolierte sporenfärbung. Landwirts. Jahrb. Schweiz. 36:595-597. [Google Scholar]
- 17.Errington, J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117-126. [DOI] [PubMed] [Google Scholar]
- 18.Fishelson, L., W. L. Montgomery, and A. A. Myrberg. 1985. A unique symbiosis in the gut of a tropical herbivorous surgeonfish (Acanthuridae: Teleostei) from the Red Sea. Science 229:49-51. [DOI] [PubMed] [Google Scholar]
- 19.Fitz-James, P., and E. Young. 1969. Morphology of sporulation, p. 39-72. In G. W. Gould and A. Hurst (ed.), The bacterial spore. Academic Press, London, England.
- 20.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glauert, A. M. 1975. Fixation, dehydration and embedding of biological specimens. In A. M. Glauert (ed.), Practical methods in electron microscopy, vol. 3. Elsevier, Amsterdam, The Netherlands.
- 22.Gould, G. W. 1969. Germination, p. 397-444. In G. W. Gould and H. A. (ed.), The bacterial spore. Academic Press, London, England.
- 23.Hecker, M., and U. Volker. 1998. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigmaB regulon. Mol. Microbiol. 29:1129-1136. [DOI] [PubMed] [Google Scholar]
- 24.Hilbert, D. W., and P. J. Piggot. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68:234-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalle, G. P., and S. S. Adkar. 1986. Fermentative production of dipicolinic acid by Penicillium citreoviride from sugarcane molasses. J. Ferment. Technol. 64:493-497. [Google Scholar]
- 26.Kawarasaki, M., H. Tanaka, T. Kanagawa, and K. Nakamura. 1999. In situ identification of polyphosphate-accumulating bacteria in activated sludge by dual staining with rRNA-targeted oligonucleotide probes and 4′,6-diamidino-2-phenylindol (DAPI) at a polyphosphate-probing concentration. Water Res. 33:257-265. [Google Scholar]
- 27.Kay, D., and S. C. Warren. 1968. Sporulation in Bacillus subtilis morphological changes. Biochem. J. 109:819-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunstyr, I., R. Schiel, F. J. Kaup, G. Uhr, and H. Kirchhoff. 1988. Giant gram-negative noncultivable endospore-forming bacteria in rodent intestines. Naturwissenschaften 75:525-527. [DOI] [PubMed] [Google Scholar]
- 29.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, N.Y.
- 30.Montgomery, W. L., and P. E. Pollak. 1988. Epulopiscium fishelsoni n. g., n. s., a protist of uncertain taxonomic affinities from the gut of an herbivorous reef fish. J. Protozool. 35:565-569. [Google Scholar]
- 31.Murray, R. G. E., R. N. Doetsch, and C. F. Robinow. 1994. Determinative and cytological light microscopy, p. 21-41. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 32.Nair, S., and S. E. Finkel. 2004. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 186:4192-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons Ltd., Chichester, England.
- 35.Pogliano, K., E. Harry, and R. Losick. 1995. Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol. Microbiol. 18:459-470. [DOI] [PubMed] [Google Scholar]
- 36.Powell, J. F., and R. E. Strange. 1956. Biochemical changes occurring during sporulation in Bacillus species. Biochem. J. 63:661-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powell, J. F., and R. E. Strange. 1953. Biochemical changes occurring during the germination of bacterial spores. Biochem. J. 54:205-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinow, C., and E. R. Angert. 1998. Nucleoids and coated vesicles of “Epulopiscium” spp. Arch. Microbiol. 170:227-235. [DOI] [PubMed] [Google Scholar]
- 39.Robinow, C. F. 1960. Morphology of bacterial spores, their development and germination, p. 207-248. In I. C. Gunsalus and R. Y. Stanier (ed.), The bacteria, vol. 1. Academic Press, New York, N.Y. [Google Scholar]
- 40.Robinow, C. F. 1951. Observations on the structure of Bacillus spores. J. Gen. Microbiol. 5:439-457. [DOI] [PubMed] [Google Scholar]
- 41.Robinow, C. F. 1953. Spore structure as revealed by thin sections. J. Bacteriol. 66:300-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaeffer, A. B., and M. D. Fulton. 1933. A simplified method for staining endospores. Science 77:194. [DOI] [PubMed] [Google Scholar]
- 43.Setlow, B., C. A. Loshon, P. C. Genest, A. E. Cowan, C. Setlow, and P. Setlow. 2002. Mechanisms of killing spores of Bacillus subtilis by acid, alkali and ethanol. J. Appl. Microbiol. 92:362-375. [DOI] [PubMed] [Google Scholar]
- 44.Setlow, B., D. Sun, and P. Setlow. 1992. Interaction between DNA and alpha/beta-type small, acid-soluble spore proteins: a new class of DNA-binding protein. J. Bacteriol. 174:2312-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Setlow, P. 1995. Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu. Rev. Microbiol. 49:29-54. [DOI] [PubMed] [Google Scholar]
- 46.Setlow, P. 2000. Resistance of bacterial spores, p. 217-230. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 47.Sharp, M. D., and K. Pogliano. 2002. The dynamic architecture of the Bacillus cell, p. 15-20. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives from genes to cells. ASM Press, Washington D.C.
- 48.Stragier, P., and R. Losick. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297-341. [DOI] [PubMed] [Google Scholar]
- 49.Tanenbaum, S. W., and K. Kaneko. 1964. Biosynthesis of dipicolinic acid and of lysine in Penicillium citreo-viride. Biochemistry 14:1314-1322. [DOI] [PubMed] [Google Scholar]
- 50.Ward, D. M. 1998. A natural species concept for prokaryotes. Curr. Opin. Microbiol. 1:271-277. [DOI] [PubMed] [Google Scholar]
- 51.Young, I. E., and P. C. Fitz-James. 1962. Chemical and morphological studies of bacterial spore formation IV. The development of spore refractility. J. Cell Biol. 12:115-133. [DOI] [PMC free article] [PubMed] [Google Scholar]