Abstract
The Bacillus subtilis enzymes ExoA and Nfo (originally termed YqfS) are endonucleases that can repair apurinic/apyrimidinic (AP) sites and strand breaks in DNA. We have analyzed how the lack of ExoA and Nfo affects the resistance of growing cells and dormant spores of B. subtilis to a variety of treatments, some of which generate AP sites and DNA strand breaks. The lack of ExoA and Nfo sensitized spores (termed α−β−) lacking the majority of their DNA-protective α/β-type small, acid-soluble spore proteins (SASP) to wet heat. However, the lack of these enzymes had no effect on the wet-heat resistance of spores that retained α/β-type SASP. The lack of either ExoA or Nfo sensitized wild-type spores to dry heat, but loss of both proteins was necessary to sensitize α−β− spores to dry heat. The lack of ExoA and Nfo also sensitized α−β−, but not wild-type, spores to desiccation. In contrast, loss of ExoA and Nfo did not sensitize growing cells or wild-type or α−β− spores to hydrogen peroxide or t-butylhydroperoxide. Loss of ExoA and Nfo also did not increase the spontaneous mutation frequency of growing cells. exoA expression took place not only in growing cells, but also in the forespore compartment of the sporulating cell. These results, together with those from previous work, suggest that ExoA and Nfo are additional factors that protect B. subtilis spores from DNA damage accumulated during spore dormancy.
Dormant spores of Bacillus species are often exposed to conditions that can cause DNA damage, including high temperatures, desiccation, and oxidizing chemicals. Consequently, spores have many mechanisms to protect their DNA and ensure spore survival (21). The spore coats, the low permeability of spores to DNA-damaging chemicals, and the saturation of spore DNA with α/β-type small, acid-soluble spore proteins (SASP) account for much of the prevention of spore DNA damage (21, 30, 34, 35). The α/β-type SASP play a key role, as spores (termed α−β−) lacking the great majority of these proteins are much more sensitive than are wild-type spores to wet and dry heat, UV radiation, desiccation, and a number of genotoxic chemicals (9, 29, 34, 35). In addition, treatment of α−β−, but not wild-type, spores with wet heat, hydrogen peroxide (H2O2), and lyophilization causes DNA damage and mutagenesis (8, 9, 19, 21, 23, 30). The DNA damage generated in α−β− spores by wet heat includes apurinic/apyrimidinic (AP) sites, while H2O2 generates strand breaks but not AP sites (30, 31). Dry heating kills both α−β− and wild-type spores, and desiccation kills α−β− spores, at least in part by DNA damage, with this damage likely including AP sites (31, 32). The AP sites may be generated not only by direct depurination and depyrimidination of DNA in the dormant spore, but also by the action of DNA glycosylases during spore outgrowth. A fourth factor that is important in spore resistance to DNA-damaging treatments is DNA repair during spore outgrowth. Both spore-specific proteins and RecA-dependent processes can be important in spore resistance (21, 33).
Damage to DNA can include AP sites, as noted above, and chemical modification of AP sites can also generate 3′ blocking groups at DNA strand breaks, including phosphoglycoaldehyde, phosphate, deoxyribose-5-phosphate, and 4-hydroxy-2-pentenal. These DNA lesions are also processed by AP endonucleases to generate a 3′-OH group on the damaged DNA strand (11, 15). B. subtilis has at least two AP endonucleases, ExoA and YqfS (25, 36, 40). ExoA belongs to the Apn endonuclease family with homologs in organisms from Escherichia coli to humans. YqfS possesses 53% amino acid sequence homology with E. coli Nfo and was recently shown to be a new member of family IV of the AP endonucleases (25, 36). Consequently, based on the nomenclature for the EndoIV family member in E. coli, we have renamed YqfS as Nfo. Although ExoA and Nfo have been functionally and biochemically characterized (25, 36), only nfo regulation has been thoroughly studied (40). This gene is expressed only during sporulation in the developing forespore, and Nfo is present in the dormant spore (25).
As mentioned above, strand breaks and AP sites are two of the most common lesions generated in spore DNA by wet heat and probably by dry heat (31, 32). Since either of these lesions can inhibit DNA replication and be mutagenic, AP sites are usually eliminated from DNA. In most species, these lesions are processed by AP endonucleases that are essential components of the base excision repair (BER) pathway. Accordingly, in the present work, we have investigated whether mutations in exoA and/or nfo affect the resistance of growing cells and spores of B. subtilis to treatments that can generate AP sites and strand breaks in DNA.
MATERIALS AND METHODS
Bacterial strains and spore preparation.
The strains and plasmids used in this work are listed in Table 1. B. subtilis strains whose growing cells or spores were tested for resistance were derived from strain PS832. The growth medium used routinely was Luria-Bertani (LB) medium (20), although some experiments used Difco sporulation medium (DSM) (28). When appropriate, ampicillin (100 μg/ml), chloramphenicol (Cm; 5 μg/ml), neomycin (Neo; 10 μg/ml), or tetracycline (Tet; 10 μg/ml) was added to the medium. Liquid cultures were incubated at 37°C with vigorous aeration. Cultures on solid media were also grown at 37°C. Spores of all strains were prepared at 37°C on 2× SG medium (2× DSM supplemented with 0.1% glucose) agar plates without antibiotics, and spores were harvested, cleaned, and stored as described previously (22). All dormant spore preparations used in this work were free (≥98%) of growing cells, germinated spores, and cell debris as determined by phase-contrast microscopy.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL10-Gold | Tetr Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac The [F′ proAB lacIqZDM15 Tn10 (Tetr) Tn5 (Kanr) Amy] | Stratagene, Cedar Creek, TX |
| PERM337 | E. coli XL10-Gold carrying plasmid pPERM337; Neor Tetr | This study |
| PERM374 | E. coli XL10-Gold carrying plasmid pPERM374; Neor Tetr | This study |
| B. subtilis | ||
| 168 | trpC2 | Laboratory stock |
| PS356 | ΔsspA ΔsspB α−β− | 17 |
| PS832 | Wild type; trp+ revertant of strain 168 | Laboratory stock |
| PS2488c | katA Cmr | 3 |
| PS2493c | ahpF Cmr | 3 |
| PS3672c | ΔyfhQ::spc Spcr | P. Setlow (27) |
| PS3673c | ΔmutM::tet Tetr | P. Setlow (27) |
| PS3677c | ΔmutM::tet ΔyfhQ::spc Spcr Tetr | (PS3672→PS3673)a(27) |
| PERM448 | ΔsspA ΔsspB ΔexoA::tet α−β− Tetr | (pPERM374→PS356)b |
| PERM449 | ΔsspA ΔsspB Δnfo::neo α−β− Neor | (pPERM337→PS356)b |
| PERM450 | ΔsspA ΔsspB ΔexoA::tet Δnfo::neo α−β− Neor Tetr | (pPERM337→PERM448)b |
| PERM452 | ΔexoA::tet Tetr | (pPERM374→PS832)b |
| PERM453 | Δnfo::neo Neor | (pPERM337→PS832)b |
| PERM454 | ΔexoA::tet Δnfo::neo Neor Tetr | (pPERM337→PERM452)b |
| PERM504 | exoA-lacZ trpC2 Cmr | (pPERM490→168)b |
| PERM527 | exoA-lacZ sigGΔ1 trpC2 Cmr | (pPERM490→WN118)b |
| WN118 | sigGΔ1 trpC2 | P. Setlow (39) |
| Plasmids | ||
| pDG1515 | Vector containing a Tetr cassette; Ampr Tetr | R. Yasbin (12) |
| pBEST501 | Vector containing a Neor cassette; Ampr Neor | R. Yasbin (16) |
| pJF751 | Integrational lacZ fusion vector; Cmr | W. L. Nicholson (10) |
| pPERM282 | pUC18 with 1.07-kb XbaI-BamHI PCR product containing nfo; Ampr | Laboratory stock |
| pPERM337 | 1.3-kb SmaI-SmaI fragment containing a Neor cassette inserted into the NaeI site of pPERM282; Ampr Neor | This study |
| pPERM359 | pDG1515 with 469-bp XbaI-BamHI PCR product containing the 5′ region of exoA; Ampr Tetr | This study |
| pPERM374 | pPERM359 with 537-bp EcoRI-HindIII PCR product containing the 3′ region of exoA; Ampr Tetr | This study |
| pPERM489 | pCR-Blunt II-TOPO with a 752-bp EcoRI-SmaI PCR fragment encompassing 434 bp upstream and 318 bp downstream of the exoA translational start codon; Neor | This study |
| pPERM490 | Translational exoA-lacZ fusion in pJF751; Ampr Cmr | This study |
Chromosomal DNA from the strain to the left of the arrow was used to transform the strain to the right.
Plasmid DNA from the strain to the left of the arrow was used to transform the strain to the right.
The background for this strain is PS832.
Genetic and molecular biology techniques.
Preparation of competent E. coli or B. subtilis cells and their transformation with plasmid DNA were as described previously (2, 26). Chromosomal DNA from B. subtilis was purified as described by Cutting and Vander Horn (5). Small-scale preparation of plasmid DNA from E. coli cells, enzymatic manipulations, and agarose gel electrophoresis utilized standard techniques (26). Medium-scale preparation and purification of plasmid DNA used commercial ion-exchange columns according to the instructions of the supplier (QIAGEN, Inc., Valencia, CA).
Construction of plasmids to interrupt the exoA and nfo genes.
Plasmid pPERM337 containing the nfo gene interrupted by a Neor cassette was constructed as follows. nfo was PCR amplified (all primer sequences are available on request) from chromosomal DNA of Bacillus subtilis 168, and the product was cloned in the SmaI site of plasmid pUC19, giving plasmid pPERM282. A Neor cassette was recovered as a SmaI fragment from plasmid pBEST501 (16) (kindly provided by R. Yasbin, University of Nevada at Las Vegas, Las Vegas, NV) and inserted in the nfo gene's unique NaeI site in plasmid pPERM282, giving plasmid pPERM337.
Plasmid pPERM374 containing the exoA gene interrupted by a Tetr cassette was constructed as follows. The 5′ region of exoA (121 bp upstream to 348 bp downstream of the exoA translation start codon) was amplified by PCR from chromosomal DNA of B. subtilis 168 and cloned into the SmaI site of plasmid pUC19. The insert was recovered as an XbaI-BamHI fragment (sites introduced into the PCR primers), and the resultant fragment was inserted between the XbaI and BamHI sites in plasmid pDG1515 (12) (kindly provided by R. Yasbin, University of Nevada at Las Vegas, Las Vegas, NV), giving plasmid pPERM359. The 3′ region of exoA (398 bp upstream to 139 bp downstream of the exoA translation stop codon) was amplified by PCR from chromosomal DNA of B. subtilis 168 and cloned into SmaI-digested pUC19. The insert was recovered as an EcoRI-HindIII fragment (sites introduced into the PCR primers), and the fragment was inserted between the EcoRI and HindIII sites in plasmid pPERM359, giving plasmid pPERM374.
Construction of exoA and nfo mutant strains.
Plasmids pPERM337 and pPERM374 described above were used to transform B. subtilis strains PS356 (α−β−) and PS832 (wild type), generating strains PERM448 (α−β− exoA), PERM449 (α−β− nfo), PERM452 (exoA), and PERM453 (nfo). The exoA nfo strains in the PS356 and PS832 genetic backgrounds were generated by transforming strains PERM448 and PERM452 with plasmid pPERM337. The double recombination events leading to inactivation of the appropriate genes were confirmed by PCR (data not shown).
Construction of B. subtilis strains with an exoA-lacZ fusion.
Construction of a translational fusion between exoA and E. coli lacZ was carried out using the integrative plasmid pJF751 (10). A 752-bp EcoRI-SmaI fragment (encompassing 434 bp upstream to 318 bp downstream of the exoA translational start codon) from plasmid pPERM489 was inserted in pJF751 digested with EcoRI and SmaI. The resulting construct (pPERM490) containing the exoA-lacZ fusion was cloned in E. coli XL10-Gold. The correct insertion of the exoA fragment in pJF751 was confirmed by analysis of small-scale plasmid preparations digested with BglI. Plasmid pPERM490 was used to transform B. subtilis strains 168 and WN118 (sigG), selecting transformants on DSM containing Cm. Integration of the exoA-lacZ fusion at the chromosomal exoA locus was confirmed by PCR with primers from upstream of exoA and within the lacZ coding region (data not shown).
Spore treatments.
For hydrogen peroxide treatment, spores at an optical density at 600 nm (OD600) of 5 were incubated at 24°C in 5% H2O2. At various times, samples were diluted 1/100 in phosphate-buffered saline (PBS) (pH 7.4) (22), and the H2O2 was destroyed by addition of catalase (29). For t-butylhydroperoxide (tBHP) treatment, spores at an OD600 of 10 in PBS were incubated at 47°C in 730 mM tBHP, and at various times aliquots were diluted 1/100 in PBS (30). For wet-heat treatment, spores at an OD600 of 1 in water were incubated at 90°C, and at various times aliquots were diluted 1/100 in 24°C water. For dry-heat treatment, spores (0.1 to 0.2 ml) at an OD600 of 2 in water were lyophilized in glass tubes, and the dry spores were heated in an oil bath for various times. The heated tubes were cooled, and the spores were rehydrated with 1 ml sterile water. For desiccation, spores at an OD600 of 50 (0.1 to 0.2 ml) were lyophilized and rehydrated with 200 μl of sterile water, and the lyophilization/rehydration cycle was repeated. In all cases, spore survival was assessed by plating aliquots of appropriate dilutions on LB medium plates and incubating the plates for 18 to 48 h at 30°C prior to enumeration of colonies. Further incubation gave no increases in colony numbers.
Experiments measuring spore resistance to heat and desiccation were repeated twice, and values were plotted as averages of duplicate determinations ±standard deviations. Values presented in plots of spore resistance to H2O2 and tBHP are averages from two experiments, with variations between experiments of ≤15%. In all cases, killing curves were performed with two different spore preparations, and these gave essentially similar (±20%) results.
Zone of inhibition assays.
Cells were grown at 37°C in LB medium to an OD600 of 0.5, 200 μl of cells was mixed with 3 ml of soft agar, and the mixture was overlaid on an LB medium plate. An 8-mm filter disk impregnated with 10 μl of 3% H2O2 or 330 mM tBHP was placed in the centers of the plates, the plates were incubated overnight at 30°C, and the diameter of the zone of inhibition of growth around each disk was measured.
Cell growth and enzyme assays.
B. subtilis strains carrying the exoA-lacZ fusion were grown and sporulated in liquid DSM containing Cm, and 1.5-ml samples were harvested by centrifugation during growth and sporulation. The cell pellets were washed with 1 ml of cold 0.1 M Tris-HCl (pH 7.5), frozen, and stored at −20°C. β-Galactosidase specific activity (in Miller units) was determined with o-nitrophenyl-β-d-galactoside as the substrate (18, 20, 22).
Analysis of spontaneous mutation frequencies.
Spontaneous mutation frequencies to rifampin resistance of growing cells were determined as follows. Strains were grown for 12 h at 37°C in PB (antibiotic 3; Difco) medium supplemented with appropriate antibiotics. Mutation frequencies were determined by plating aliquots on six LB plates containing 5 μg/ml rifampin, and rifampin-resistant (Rifr) colonies were counted after 1 day of incubation at 37°C. The experiment was repeated at least two times.
RESULTS
Properties of growing cells of exoA and nfo strains.
The exoA, nfo, and exoA nfo strains grew at the same rate as the parental strain (either PS356 or PS832) at 37°C in LB medium (data not shown). Determination of the frequency of spontaneous mutation to rifampin resistance further revealed that neither exoA nor nfo strains exhibited mutator phenotypes (Table 2). Even the spontaneous mutation frequency of an exoA nfo strain did not differ significantly from the value for the isogenic wild-type strain (Table 2). In contrast, under the same experimental conditions, a strain lacking the DNA glycosylases MutM and YfhQ (a MutY homolog) had a mutation frequency ∼100-fold higher than that of the wild-type strain (Table 2).
TABLE 2.
Spontaneous-mutation frequencies of wild-type, exoA, nfo, and exoA nfo strainsa
| Strain (genotype) | Frequency of Rifr mutants (10−9)b
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| PS832 (wild type) | 1.2 ± 0.30 | 1.3 ± 0.13 |
| PERM 452 (exoA) | 1.6 ± 0.32 | 1.24 ± 0.32 |
| PERM 453 (nfo) | 1.1 ± 0.05 | 1.2 ± 0.16 |
| PERM 454 (exoA nfo) | 1.15 ± 0.25 | 1.92 ± 0.70 |
| PS3677 (mutM yfhQ) | 322 ± 26 | 284 ± 16 |
The mutation frequencies in growing cells were measured as described in Materials and Methods.
Average of six different selection plates ± standard deviations.
To investigate the roles played by ExoA and Nfo in oxidative stress resistance during vegetative growth, the sensitivity of cells of exoA, nfo, and exoA nfo strains to H2O2 and tBHP was assessed using zone of inhibition assays. There were slight decreases in the H2O2 and tBHP resistances of strains lacking ExoA and/or Nfo, but these differences were not great (Table 3). In contrast, growing cells lacking their major catalase (KatA) or alkylhydroperoxide reductase (AhpF) were much more sensitive to H2O2 and tBHP, respectively, than were exoA, nfo, and exoA nfo strains (Table 3).
TABLE 3.
Effects of exoA, nfo, katA, and ahpF mutations on growing cell resistance to tBHP and H2O2 as determined by zone of inhibition assaysa
| Strain (genotype) | Diameter of zone of inhibition (mm)b
|
|
|---|---|---|
| tBHP | H2O2 | |
| PS832 (wtc) | 31.2 ± 0.8 | 16.2 ± 0.8 |
| PERM 452 (exoA) | 31.8 ± 0.8 | 17.2 ± 0.8 |
| PERM 453 (nfo) | 33.0 ± 1.6 | 18.0 ± 0.7 |
| PERM 454 (exoA nfo) | 38.2 ± 1.4 | 20.6 ± 1.1 |
| PS433 (ahpF) | 58.6 ± 1.2 | ND |
| PS2488 (katA) | NDd | 34.6 ± 1.4 |
Cells were grown and growth inhibition was tested as described in Material and Methods.
Values represent the average of five different experiments ± standard deviations.
wt, wild type.
ND, not determined.
Role of ExoA and Nfo in B. subtilis spore resistance.
Although loss of ExoA and/or Nfo had little effect on the resistance of growing cells to several oxidizing agents, this was not surprising for Nfo, as this enzyme is present only in dormant spores (25). A number of treatments, including heat and oxidizing agents, kill spores of some strains by DNA damage, in some cases by generating AP sites (21, 30, 31, 32). Consequently, it seemed worthwhile to investigate whether the lack of ExoA and Nfo sensitizes α−β− or wild-type spores to treatments that damage spore DNA.
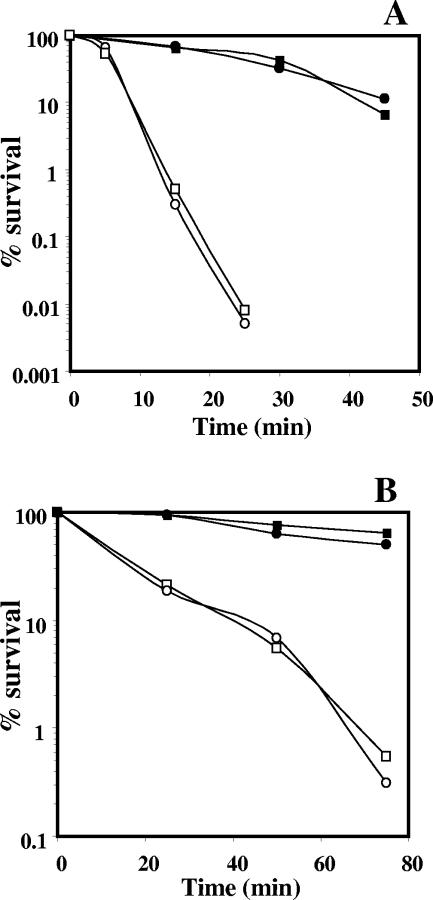
The mechanism(s) whereby oxidizing agents kill wild-type B. subtilis spores is unknown, but it does not involve DNA damage (19, 29, 30). Consequently, it was not surprising that the resistance of exoA nfo spores to H2O2 or tBHP was the same as that of wild-type spores (Fig. 1A). α−β− spores were considerably more sensitive to H2O2 and tBHP than were wild-type spores (Fig. 1A and B), as was found previously (29, 30). However, the lack of ExoA and Nfo had no effect on the H2O2 or tBHP resistance of α−β− spores (Fig. 1B), even though both of these agents kill α−β− spores by DNA damage (29, 30).
FIG. 1.
Resistance of wild-type, exoA nfo, α−β−, and α−β− exoA nfo spores to H2O2 (A) and tBHP (B). Spores of various strains were incubated with H2O2 or tBHP, and viability was determined as described in Materials and Methods. ▪, PS832 (wild type); •, PERM454 (exoA nfo); □, PS356 (α−β−); ○, PERM450 (α−β− exoA nfo).
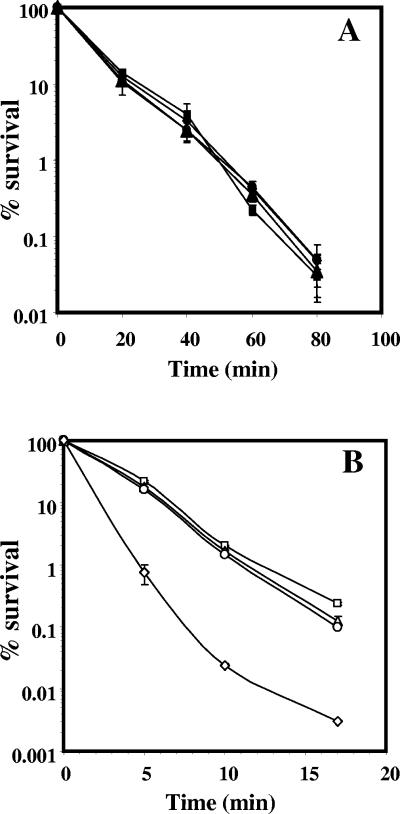
Wet-heat treatment of α−β−, but not wild-type, spores generates strand breaks and AP sites in DNA, lesions that are likely processed by ExoA and Nfo (8, 25, 31, 36). As predicted from these results, the lack of both ExoA and Nfo had no effect on the wet-heat resistance of wild-type spores (Fig. 2A), in which wet heat does not cause DNA damage (8). However, the lack of both enzymes significantly reduced the wet-heat resistance of α−β− spores, although lack of either enzyme alone had almost no effect (Fig. 2B).
FIG. 2.
Effects of exoA and nfo mutations on the wet-heat resistance of (A) wild-type and (B) α−β− spores. Spores of wild-type and α−β− strains were incubated at 90°C, and viability was determined as described in Materials and Methods. ▪, PS832 (wild type); ▴, PERM452 (exoA); •, PERM453 (nfo); ⧫, PERM454 (exoA nfo); □, PS356 (α−β−); ▵, PERM448 (α−β− exoA); ○, PERM449 (α−β− nfo); ◊, PERM450 (α−β− exoA nfo). The error bars are the standard deviations as described in Materials and Methods.
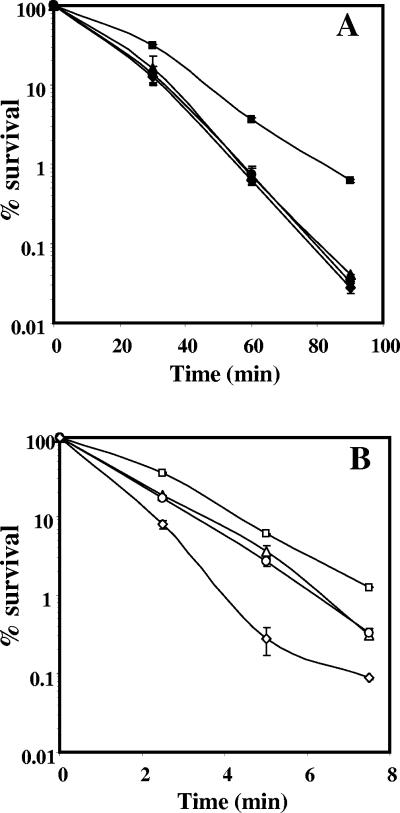
Dry-heat treatment of α−β− and wild-type spores also generates DNA strand breaks and probably AP sites (32). In a wild-type background, the lack of ExoA or Nfo sensitized spores to dry heat (Fig. 3A). However, spores of the exoA nfo strain were as sensitive to dry heat as spores lacking either ExoA or Nfo (Fig. 3A). The lack of ExoA or Nfo also sensitized α−β− spores to dry heat, and the lack of both enzymes rendered α−β− spores even more dry heat sensitive (Fig. 3B).
FIG. 3.
Effects of exoA and nfo mutations on the dry-heat resistance of (A) wild-type and (B) α−β− spores. Spores of wild-type and α−β− strains were lyophilized, the dry spores were heated for various times at (A) 120°C or (B) 90°C, and their viability was determined as described in Materials and Methods. ▪, PS832 (wild type); ▴, PERM452 (exoA); •, PERM453 (nfo); ⧫, PERM454 (exoA nfo); □, PS356 (α−β−); ▵, PERM448 (α−β− exoA); ○, PERM449 (α−β− nfo); ⋄, PERM450 (α−β− exoA nfo). The error bars are the standard deviations as described in Materials and Methods.
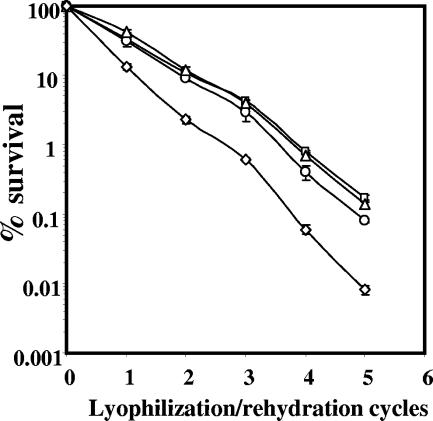
Previous work has shown that wild-type spores are not killed by repeated cycles of desiccation/rehydration and accumulate no DNA damage (9). Consequently, it was not surprising that the loss of ExoA and Nfo did not result in killing of wild-type spores through at least 12 desiccation/rehydration cycles (data not shown). However, loss of both ExoA and Nfo slightly sensitized α−β− spores to desiccation/rehydration, although loss of either ExoA or Nfo did not (Fig. 4).
FIG. 4.
Effects of exoA and nfo mutations on the desiccation resistance of α−β− spores. Spores of α−β− strains were lyophilized and rehydrated five times, and spore viability was determined after each cycle as described in Materials and Methods. □, PS356 (α−β−); ▵, PERM448 (α−β− exoA); ○, PERM449 (α−β− nfo); and ⋄, PERM450 (α−β− exoA nfo). The error bars are the standard deviations as described in Materials and Methods.
Analysis of exoA expression during growth and sporulation.
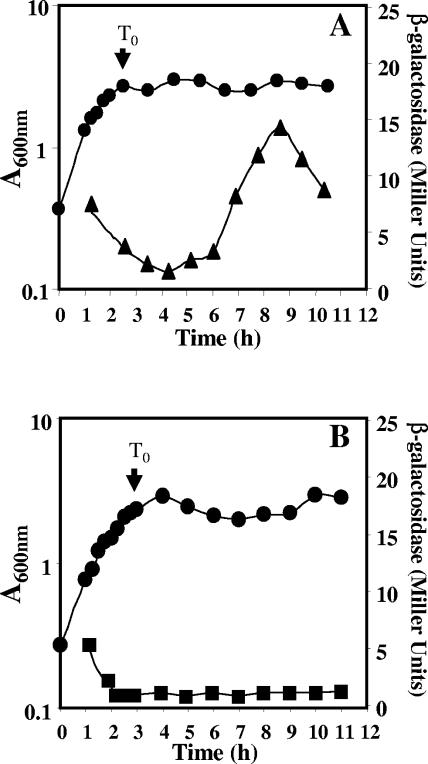
The results described above suggest that both ExoA and Nfo repair at least some types of DNA damage during spore outgrowth, and further, that both enzymes are likely present in dormant spores. nfo expression is known to be specific to the forespore compartment of the sporulating cell, and Nfo is in dormant spores (25, 40). exoA is also known to be transcribed during vegetative growth (36), but exoA expression during sporulation has not been studied. Consequently, an exoA-lacZ fusion was inserted at the exoA locus, and β-galactosidase levels were determined during growth and sporulation. The exoA reporter gene showed two peaks of expression, one during vegetative growth and another at approximately hour 9 of sporulation (Fig. 5A). The second peak of expression was followed by a decrease in the specific activity, suggesting that the β-galactosidase had been sequestered in the forespore, as this cell becomes resistant to lysis by lysozyme (18). Northern blot experiments also detected exoA mRNA, not only during vegetative growth, but also late in sporulation (data not shown). exoA-lacZ expression in sporulation was almost completely abolished by mutation of the gene (sigG) that encodes σG, the RNA polymerase sigma factor that directs the expression of most forespore-specific genes in B. subtilis, including nfo (14, 40) (Fig. 5B).
FIG. 5.
Levels of β-galactosidase from an exoA-lacZ translational fusion during growth and sporulation of (A) wild-type and (B) sigG strains. B. subtilis strains (A) PERM504 (exoA-lacZ wild type) and (B) PERM527 (exoA-lacZ sigG) were grown and sporulated in liquid DSM. Cell samples were collected at different times and treated with lysozyme, and the extracts were assayed for β-galactosidase as described in Materials and Methods. Shown are β-galactosidase specific activities in (▴) wild-type and (▪) sigG strains and (•) A600.
DISCUSSION
Intracellular reactive oxygen species and exogenous agents, such as heat, UV, and γ-radiation and oxidizing chemicals, damage DNA, leading directly or indirectly to the formation of AP sites and strand breaks (24, 37, 38). In most organisms, these DNA lesions are repaired by AP endonucleases, important components of the BER pathway (1, 7). B. subtilis possesses at least two AP endonucleases, ExoA and Nfo (originally termed YqfS). In E. coli, the loss of either Xth (the homolog of B. subtilis ExoA) or Nfo has little phenotypic effect (4). However, the loss of both proteins sensitizes E. coli to methyl methanesulfonate, H2O2, and tBHP and increases mutagenesis by methyl methanesulfonate (4). In contrast, with growing B. subtilis cells, loss of ExoA and Nfo neither decreased resistance to H2O2 or tBHP nor increased the spontaneous-mutation frequency. The dramatic difference in the relative susceptibilities to oxidizing agents of wild-type and xth nfo E. coli and wild-type and exoA nfo B. subtilis may be due to the fact that (i) in B. subtilis, nfo expression is confined to the forespore (25, 40) and (ii) a mutation in exoA alone does not sensitize growing B. subtilis cells to oxidizing and alkylating agents (36). Therefore, either B. subtilis has AP endonucleases in addition to ExoA and Nfo, or other DNA repair systems are sufficient to compensate for the lack of ExoA and Nfo in growing cells.
The results in this work indicate that expression of exoA takes place not only in vegetative cells, as shown previously (36), but also during sporulation. After the level of β-galactosidase from exoA-lacZ peaked in sporulation, there was a rapid decline. This finding, and the lack of sporulation-associated exoA-lacZ expression in a sigG strain, indicates that the sporulation-specific exoA transcription is in the forespore. However, it remains to be determined whether ExoA itself is present in the dormant spore. Interestingly, between nucleotides 76 and 107 upstream of the translational start codon of exoA, there are sequences with high similarity to those in promoters of genes of the σG regulon (13). Except for a 1-base insertion, a putative −10 region (CATACgTA; consensus in capitals) perfectly matches the consensus for σG-dependent promoters, while a putative −35 region (CGCAcG) contains five of the six residues conserved in these promoters (13). These putative −10 and −35 sequences are also separated by 17 bp, typical of σG-dependent promoters (13). Experiments are in progress to determine whether these sequences do indeed function as the promoter for exoA expression during sporulation.
Neither ExoA nor Nfo protected B. subtilis spores with or without α/β-type SASP from H2O2 and tBHP. This result reinforces previous conclusions that oxidizing agents do not kill wild-type spores by DNA damage (19, 30). While hydrogen peroxide treatment does kill α−β− spores by DNA damage, this treatment does not generate AP sites, although it does result in the formation of DNA strand breaks (30, 31). In agreement with these observations, a recent study revealed differences in the spectra of mutations to nalidixic acid resistance induced by H2O2 or wet-heat treatment of α−β− spores, suggesting that these treatments generate different types of DNA damage (6). Since a recA mutation sensitizes α−β− spores to H2O2, RecA-dependent processes may be most important in repairing H2O2-induced DNA lesions, and RecA is in dormant spores (33). Alkyl hydroperoxide reductase, superoxide dismutase (SodA), and catalase (KatX) are also present in dormant spores, but these enzymes play no role in protecting spores against H2O2 and tBHP (3).
Previous work has detected at most a very low level of AP sites and strand breaks in DNA from wet-heat-killed wild-type spores (31). Thus, in otherwise wild-type spores, the lack of ExoA and Nfo was not expected to have any effect on wet-heat resistance, and it did not. Spores were sensitized to wet heat by lack of ExoA and Nfo only when the spores also lacked α/β-type SASP. These results indicate that ExoA and Nfo are dispensable in wet-heat-treated wild-type spores due to protection against wet-heat-induced DNA damage by α/β-type-SASP. However in α−β− spores, wet heat damages DNA, and ExoA and Nfo are important in repair of such damage during spore outgrowth.
Dry heat kills α−β−, as well as wild-type, spores by DNA damage, including strand breaks and probably AP sites (32). Thus, dry-heated spores with or without α/β-type SASP would be expected to require AP endonuclease(s) to repair damaged DNA. Although α−β− spores lacking either ExoA or Nfo showed only a modest increase in their susceptibility to dry heat, loss of both proteins rendered spores significantly more sensitive to this treatment. In spores containing α/β-type SASP, the lack of ExoA or Nfo sensitized the spores to dry heat, and the dry-heat sensitivity was similar for exoA nfo spores. Thus, these results strongly suggest that ExoA and Nfo are important in protecting spores against dry-heat-induced DNA damage.
The lack of effect of ExoA and/or Nfo on wild-type spore resistance to desiccation was expected, since this treatment does not kill wild-type spores, presumably due to DNA protection by α/β-type SASP (9). However, α−β− spores are killed by desiccation through DNA damage, including strand breaks (9). Thus, it was not surprising that α−β− spore resistance to desiccation was decreased by loss of ExoA and Nfo.
The increased sensitivity of exoA and/or nfo spores to treatments that damage spore DNA through generation of AP sites and strand breaks, the transcription of exoA and nfo (40) in the forespore compartment, and the fact that ExoA and Nfo have structural and enzymatic properties required to repair AP sites and 3′ blocking groups in DNA (25, 36) suggest that these proteins are additional factors that contribute to spore resistance by repairing DNA damage in spore outgrowth.
Thus, in addition to SplB, RecA, and the UVR system, ExoA and Nfo are also part of the arsenal of DNA repair proteins that increase the potential for survival of B. subtilis spores. This is the first evidence that the BER pathway is important in the repair of damage accumulated by B. subtilis spores during dormancy.
Acknowledgments
This work was supported by grant 43644 from the Consejo Nacional de Ciencia y Tecnología (CONACYT) of México to M. Pedraza-Reyes. J. M. Salas-Pacheco was supported by doctoral fellowships from CONACYT and CONCYTEG. B. Setlow and P. Setlow were supported by grants from the Army Research Office and the National Institutes of Health (GM19698).
We thank J. A. Rojas and V. Calderón for excellent technical assistance.
REFERENCES
- 1.Barzilay, G., and L. D. Hickson. 1995. Structure and function of apurinic/apyrimidinic endonucleases. Bioessays 17:713-719. [DOI] [PubMed] [Google Scholar]
- 2.Boylan, R. J., N. H. Mendelson, D. Brooks, and F. E. Young. 1972. Regulation of the bacterial cell wall: analysis of a mutant of Bacillus subtilis defective in the biosynthesis of teichoic acid. J. Bacteriol. 173:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casillas-Martinez, L., and P. Setlow. 1997. Alkyl hydroperoxide reductase, catalase, MrgA, and superoxide dismutase are not involved in resistance of Bacillus subtilis spores to heat or oxidizing agents. J. Bacteriol. 179:7420-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunningham, R. P., S. M. Saporito, S. G. Spitzer, and B. Weiss. 1986. Endonuclease IV (nfo) mutant of Escherichia coli. J. Bacteriol. 168:1120-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutting, S. M., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Sussex, England.
- 6.del Huesca-Espitia, L., C., C. Caley, I. Bagyan, and P. Setlow. 2002. Base-change mutations induced by various treatments of Bacillus subtilis spores with and without DNA protective small, acid-soluble spore proteins. Mut. Res. 503:77-84. [DOI] [PubMed] [Google Scholar]
- 7.Demple, B., and L. Harrison. 1994. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 63:915-948. [DOI] [PubMed] [Google Scholar]
- 8.Fairhead, H., B. Setlow, and P. Setlow. 1993. Prevention of DNA damage in spores and in vitro by small acid-soluble spore proteins from Bacillus species, J. Bacteriol. 175:1367-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fairhead, H., B. Setlow, W. M. Waites, and P. Setlow. 1994. Small, acid-soluble proteins bound to the DNA protect Bacillus subtilis spores from being killed by freeze-drying. Appl. Environ. Microbiol. 60:2647-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari, E., S. M. H. Haward, and J. A. Hoch. 1985. Effect of sporulation mutations on subtilisin expression, assayed using a subtilisin-β-galactosidase gene fusion, p. 180-184. In J. A. Hoch and P. Setlow (ed.), Molecular biology of microbial differentiation. American Society for Microbiology, Washington, D.C.
- 11.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. American Society for Microbiology, Washington, D.C.
- 12.Guérout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 13.Haldenwang, W. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilbert, D. W., and P. J. Piggot. 2004. Compartmentalization of gene expression during Bacillus subtilis sporulation. Microbiol. Mol. Biol. Rev. 68:234-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosfield, D. J., Y. Guan, B. J. Haas, R. P. Cunningham, and J. A. Tainer. 1999. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell 98:397-408. [DOI] [PubMed] [Google Scholar]
- 16.Itaya, M., K. Kondo, and T. Tanaka. 1989. A neomycin resistance gene cassette selectable in a single copy state in Bacillus subtilis. Nucleic Acids Res. 17:4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mason, J. M., and P. Setlow. 1987. Essential role of small, acid-soluble spore proteins in resistance of Bacillus subtilis spores to ultraviolet light. J. Bacteriol. 167:174-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mason, J. M., R. H. Hackett, and P. Setlow. 1988. Regulation of expression of genes coding for small acid-soluble spore proteins of Bacillus subtilis spores: studies using lacZ gene fusions. J. Bacteriol. 170:239-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melly, E., A. E. Cowan, and P. Setlow. 2002. Studies on the mechanism of killing of Bacillus subtilis spores by hydrogen peroxide. J. Appl. Microbiol. 93:316-325. [DOI] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Nicholson, W. L., N. Munakata, G. Horneck, H. G. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination, and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.). Molecular biological methods for Bacillus. John Wiley and Sons, Sussex, England.
- 23.Popham, D. L., S. Sengupta, and P. Setlow. 1995. Heat, hydrogen peroxide, and UV resistance of Bacillus subtilis spores with increased core water content and with or without major DNA-binding proteins. Appl. Environ. Microbiol. 61:3633-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riley, P. A. 1994. Free radicals in biology: oxidative stress and the effects of ionizing radiation. Int. J. Radiat. Biol. 65:27-37. [DOI] [PubMed] [Google Scholar]
- 25.Salas-Pacheco, J. M., N. Urtiz-Estrada, G. Martínez-Cadena, R. E. Yasbin, and M. Pedraza-Reyes. 2003. YqfS from Bacillus subtilis is a spore protein and a new functional member of the type IV apurinic/apyrimidinic-endonuclease family. J. Bacteriol. 185:5380-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Sasaki, M., Y. Yonemura, and Y. Kurusu. 2000. Genetic analysis of Bacillus subtilis mutator genes. J. Gen. Appl. Microbiol. 46:183-187. [DOI] [PubMed] [Google Scholar]
- 28.Schaeffer, P., J. Millet, and J. P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Setlow, B., C. Setlow, and P. Setlow. 1997. Killing bacterial spores by organic hydroperoxides. J. Ind. Microbiol. 18:384-388. [Google Scholar]
- 30.Setlow, B., and P. Setlow. 1993. Binding of small, acid-soluble spore proteins to DNA plays a significant role in the resistance of Bacillus subtilis spores to hydrogen peroxide. Appl. Environ. Microbiol. 59:3418-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Setlow, B., and P. Setlow. 1994. Heat inactivation of Bacillus subtilis spores lacking small, acid-soluble spore proteins is accompanied by generation of abasic sites in spore DNA. J. Bacteriol. 176:2111-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Setlow, B., and P. Setlow. 1995. Small, acid-soluble proteins bound to DNA protect Bacillus subtilis spores from killing by dry heat. Appl. Environ. Microbiol. 61:2787-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Setlow, B., and P. Setlow. 1996. Role of DNA repair in Bacillus subtilis spore resistance. J. Bacteriol. 178:3486-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Setlow, P. 1988. Small acid-soluble spore proteins of Bacillus species: structure, synthesis, genetics, function and degradation. Annu. Rev. Microbiol. 42:319-338. [DOI] [PubMed] [Google Scholar]
- 35.Setlow, P. 1995. Mechanisms for the prevention of damage to the DNA in spores of Bacillus species. Annu. Rev. Microbiol. 49:29-54. [DOI] [PubMed] [Google Scholar]
- 36.Shida, T., T. Ogawa, N. Ogasawara, and J. Sekiguchi. 1999. Characterization of Bacillus subtilis ExoA protein: a multifunctional DNA-repair enzyme similar to the Escherichia coli exonuclease III. Biosci. Biotechnol. Biochem. 63:1528-1534. [DOI] [PubMed] [Google Scholar]
- 37.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 38.Storz, G., and M. Zheng. 2000. Oxidative stress, p. 47-59. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASMy Press, Washington, D.C.
- 39.Sun, D., P. Stragier, and P. Setlow. 1989. Identification of a new sigma factor which allows RNA polymerase to transcribe the sspE gene and other forespore specific genes during sporulation of Bacillus subtilis. Genes Dev. 3: 141-149. [DOI] [PubMed] [Google Scholar]
- 40.Urtiz-Estrada, N., J. M. Salas-Pacheco, R. E. Yasbin, and M. Pedraza-Reyes. 2003. Forespore-specific expression of Bacillus subtilis yqfS which encodes type IV apurinic/apyrimidinic endonuclease, a component of the base excision repair pathway. J. Bacteriol. 185:340-348. [DOI] [PMC free article] [PubMed] [Google Scholar]