Abstract
Salmonella enterica modulates resistance to antimicrobial peptides in part via covalent modifications of the lipopolysaccharide (LPS). The two-component systems PhoP/PhoQ and PmrA/PmrB are activated during infection and regulate several genes involved in LPS modifications by responding to signals such as pH, iron, magnesium, and antimicrobial peptides. A recombination-based in vivo expression technology approach was adopted to analyze the spatial-temporal patterns of in vivo expression of genes of the PhoP and PmrA regulons and to identify the in vivo signals modulating their transcription. In vitro, we showed PhoP- and/or PmrA-dependent induction of pmrH (LPS aminoarabinose modification operon) by acidic pH, low levels of magnesium, or high levels of Fe(III). Upregulation in cultured J774A.1 macrophages was shown for pmrH, pagP (LPS palmitate addition), and ssaB (pathogenicity island II secretion) but not for prgH (pathogenicity island I secretion). Increased levels of pmrH, phoP, and prgH transcription but not ssaB were observed in bacteria isolated from the lumen of the distal ileum. Bacteria isolated from spleens of orally inoculated mice showed no further induction of prgH but had the highest expression of pmrH, pagP, and ssaB. In vivo induction of pmrH was fully dependent on pmrA and phoP, and buffering stomach acidity, iron chelation, or low-iron diets did not affect the expression of pmrH in the intestinal lumen. The observation of pmrH and pagP expression in the intestine refutes the paradigm of PhoP/PhoQ and PmrA/PmrB in vivo expression as solely intracellularly induced and supports previous data demonstrating peroral virulence attenuation of pmrH mutants.
Salmonella enterica serovar Typhimurium is a facultative intracellular enteric pathogen that causes gastroenteritis in humans and an enteric fever-like disease in mice. A major mechanism of virulence is the ability to actively counteract host cationic antimicrobial peptides (CAMP), an important component of the nonoxidative killing mechanisms of the innate immune system (40, 41). CAMPs are found in mucosal secretions, skin surfaces, and granules of professional phagocytes (5, 18). In response to these defenses, S. enterica serovar Typhimurium modifies the lipopolysaccharide (LPS) and other components of the outer membrane. In particular, the PhoP/PhoQ two-component system modulates a large regulon (reviewed in reference 20) that includes genes involved in resistance to α-helical CAMPs, epithelial cell invasion, bile resistance, and cation transport (4, 10, 21, 22, 34-36, 47, 54). The genes encoding PmrA/PmrB two-component system are among the loci regulated by PhoP/PhoQ, as they are indirectly activated via the small protein PmrD (23, 27). PmrA/PmrB is required for resistance to CAMPs, including polymyxin B, by regulating the addition of aminoarabinose to lipid A via pmrE and the pmrHFIJKLM operon (hereafter referred to as the pmrH operon) (24, 59). Other LPS modifications induced by the activation of PmrA/PmrB are the addition of phosphoethanolamine to lipid A via PmrC (29) and the addition of phosphoethanolamine to the LPS core via CptA (52).
The signals that regulate PhoP/PhoQ and PmrA/PmrB are diverse. In vitro, millimolar concentrations of Mg2+ (and some other divalent cations) activate the phosphatase activity of PhoQ (9, 19, 57), therefore downregulating PhoP-activated genes, including pmrAB and its regulon. In addition, high concentrations of iron(III) (in the range of hundreds of micromolars) activate PmrB in a PhoP/PhoQ-independent manner (58). Iron sensing by PmrB is presumed to be important for survival outside the host in soil and waters (9). The relevance of sensing elevated concentration of iron during infection is unclear, because the bacteria experience such high levels of bioavailable iron only transiently in the stomach and proximal duodenum but not in the distal ileum, the preferred site of infection (44, 45). Acidic pH and/or sublethal concentrations of certain CAMPs (polymyxin B, protegrin-1, and the α-helical peptide C18G) have also been shown to activate the pmrA and phoP regulons (3).
Most of the information on signals regulating PhoP/PhoQ and PmrA/PmrB is based on in vitro experiments, and little is known about the actual signals perceived in vivo. Several lines of evidence suggest that PhoP- and PmrA-regulated genes are activated in the phagosome of professional phagocytes in response to its acidification (1) and that this represents the major site of expression of both regulons. However, recent experiments showing that null mutations in pmrH attenuate S. enterica serovar Typhimurium when inoculated perorally but not when injected intraperitoneally question this model (24). This result implies that pmrH-dependent aminoarabinose modifications of LPS may be important for overcoming innate immune responses in the gastrointestinal tract much earlier than the establishment of an intracellular lifestyle by the bacterium and colonization of the reticuloendothelial system. Moreover, loss of the ability to add aminoarabinose to LPS conferred a more marked virulence defect compared to that of a pmrA mutant, implying that other regulatory factors and/or signaling pathways may be involved in activating pmrH.
In this study, we adapted resolvase-in vivo expression technology (RIVET) to S. enterica serovar Typhimurium to facilitate the analysis of in vivo-expressed genes. RIVET was originally developed as a genetic screening method for in vivo-induced genes (7). In the RIVET system, a DNA library or the promoter of the gene of interest is cloned into a promoterless tnpR gene, which encodes the resolvase of Tn1000 and catalyzes recombination between tandem res sites (7). A reporter consisting of a tetracycline resistance gene flanked by res1 sites is located in a single copy in the chromosome (7, 46). When a sufficient amount of TnpR is produced, it catalyzes recombination between the res1 sites flanking the gene encoding tetracycline resistance. This recombination results in heritable loss of the gene encoding tetracycline resistance and thus sensitivity to tetracycline. RIVET is also a sensitive method for studying the spatial and temporal expression of genes in vivo, as shown by studies in Vibrio cholerae (31), but applications to other pathogens have been limited (6, 42, 56). In this work, we demonstrate the use of RIVET in S. enterica serovar Typhimurium for analyzing the spatial-temporal patterns of in vivo expression of selected PmrA- and PhoP-regulated genes, concluding that the PhoP and PmrA regulons are expressed not only inside macrophages but also in the intestine of BALB/c mice. We also show that activation of these two regulons in the lumen occurs upon perception of signals other than acidic pH or iron.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains used in this study are shown in Table 1. All S. enterica serovar Typhimurium strains are derivatives of the virulent isolate ATCC 14028. For inoculum preparation and molecular genetics experiments, Escherichia coli and salmonellae were grown in Luria-Bertani (LB) broth with the addition of the appropriate antibiotics when required (12.5 μg ml−1 tetracycline [Tc]; 50 μg ml−1 kanamycin [Km]; 100 μg ml−1 ampicillin [Ap]; 25 μg ml−1 chloramphenicol [Cm]). For in vitro expression studies of the phoP and pmrA regulons, we employed N-minimal medium (NMM), pH 5.6 or pH 7.7, containing 0.1% Casamino Acids, 38 mM glycerol, 10 μM or 10 mM MgCl2 (58). FeSO4 in acidic solution stocks was added at 100 μM when required. For low-pH shock experiments, the inoculum was grown overnight in LB-morpholinepropanesulfonic acid (40 mM MOPS, pH 7.7) with Tc, resuspended to an optical density at 600 nm of ∼0.1 in low-pH NMM (pH 5.5) for 15, 30, and 60 min, and then spun down and grown in NMM (pH 7.7) overnight.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Escherichia coli | ||
| SM10λpir | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu (Kanr) λ::pir | |
| DH5α | F−supE44 lacU169 (φ80lacZΔM15) hasR17 recA1 endA1 gyrA96 (Nalr) thi-1 relA1 | |
| DH5αλpir | F−lacU169 (φ80lacZΔM15) recA1 endA1 hsdR17 supE44 thi-1 gyrA96 (Nalr) relA1 λ::pir | |
| HB101 | F−thi-1 hsd20 (r−B m−B) supE44 recA13 ara-14 leuB6 proA2 lacY1 rpsL20 (Smr) xyl-5 mtl-1 | |
| Salmonella enterica serovar Typhimurium | ||
| 14028 | Wild type | ATCC |
| JS246 | 14028 yjeP8103::res1-tetAR-res1 | This study |
| JSG2270 | 14028 pmrA::kan | This study |
| JSG1744 | 14028 invA::cat | 16 |
| JSG206 | phoP::Tn10d-cam (CS015) | 35 |
| JSG2427 | JS246 Φ(pmrH′-tnpRwt-lacZY+)540 pmrH+ | This study |
| JSG2428 | JS246 Φ(pmrH′-tnpRmut135-lacZY+)566 pmrH+ | This study |
| JSG2429 | JS246 Φ(pmrH′-tnpRmut168-lacZY+)557 pmrH+ | This study |
| JSG2436 | JS246 pmrA::kan Φ(pmrH′-tnpRmut135-lacZY+) pmrH+ | This study |
| JSG2437 | JS246 phoP::Tn10d-Cm Φ(pmrH′-tnpRmut135-lacZY+) pmrH+ | This study |
| JSG2502 | JS246 Φ(pagP′-tnpRmut135-lacZY+)608-6 pagP+ | This study |
| JSG2504 | JS246 Φ(prgH′-tnpRmut135-lacZY+)610-9 prgH+ | This study |
| JSG2524 | JS246 Φ(ssaB′-tnpRmut135-lacZY+) ssaB+ | This study |
| JSG2540 | JS246 invA::cat Φ(pmrH′-tnpRmut135-lacZY+)566 pmrH+ | This study |
| JSG2579 | JS246 Φ(fepA′-tnpRmut135-lacZ+)2535 fepA+ | This study |
| Plasmids | ||
| pJS225 | pSC101 ori Apr 5-kb region around purA | 32 |
| pGH436 | ColE1 ori Aprres1-Tet-res1 | 51 |
| pCE61 | ColE1 ori res1-tetAR-res1 | This study |
| pCE63 | pJS225 yjeP8103::res1-tetAR-res1 | This study |
| pCE70 | oriR6K FRT-tnpRwt-lacZY aph (Kanr) | This study |
| pCE72 | oriR6K FRT-tnpRmut168-lacZY aph (Kanr) | This study |
| pCE74 | oriR6K FRT-tnpRmut135-lacZY aph (Kanr) | This study |
| pCE71 | oriR6K FRT-tnpRwt-lacZY aph (Kanr) | This study |
| pCE73 | oriR6K FRT-tnpRmut168-lacZY aph (Kanr) | This study |
| pCE75 | oriR6K FRT-tnpRmut135-lacZY aph (Kanr) | This study |
| pIVET5 | oriR6K mobRP4 promoterless tnpR+-lacZY+ (Apr) | 8 |
| pIVET5n | pIVET5 derivative | 38 |
| pGOA1193 | pIVET5n promoterless tnpR+-lacZY+ (Apr) | 38 |
| pGOA1194 | pIVET5n promoterless tnpRmut135-lacZY+ (Apr) | 38 |
| pGOA1195 | pIVET5n promoterless tnpRmut168-lacZY+ (Apr) | 38 |
| pRK2013::Tn7 | ColE1 mob+traRK2 ΔrepRK2repE kan::Tn7 (Tpr Smr Spr) | 14 |
| pJSG2412 | pGOA1193 with a 270-bp BamHI fragment carrying PpmrH | This study |
| pJSG2413 | pGOA1194 with a 270-bp BamHI fragment carrying PpmrH | This study |
| pJSG2414 | pGOA1195 with a 270-bp BamHI fragment carrying PpmrH | This study |
| pJSG2483 | pGOA1194 with a 378-bp BamHI fragment carrying PpagP | This study |
| pJSG2484 | pGOA1194 with a 321-bp BamHI fragment carrying PprgH | This study |
| pJSG2485 | pGOA1194 with a 406-bp BamHI fragment carrying PssaB | This study |
| pJSG2535 | pGOA1194 with a 391-bp BamHI fragment carrying PfepA | This study |
DNA cloning and construction of reporter fusion strains.
General molecular cloning procedures were adopted for the construction and verification of plasmids (2). Marked mutations were moved into the various S. enterica serovar Typhimurium genetic backgrounds via transduction with the phage P22 HT105 int-102 (26, 43). The construction of the novel res1-tetAR-res1 cassette carrying the Tn10 antibiotic resistance genes to produce strain JS246 is described in the Results section. Gene fusions were constructed by cloning BamHI-digested PCR fragments carrying the promoter regions under investigation into the R6K ori suicide vectors pGOA1193 (pIVET5n tnpRwt), pGOA1194 (pIVET5n tnpRmut135), and pGOA1195 (pIVET5n tnpRmut168) (38). These vectors are pIVET5 (8) derivatives that carry a promoterless tnpR-lacZ operon with the various ribosomal binding site mutants described by Lee et al. (31). Table S2 in the supplemental materials shows the primers used for each promoter (pmrH, pagP, prgH, ssaB, and fepA). The promoter fusions were exchanged into the Salmonella chromosome by Campbell-type recombination, creating partial merodiploids that maintain a wild-type copy of the gene. PCR, Southern blot, and DNA sequence analyses were used to confirm that the correct fusions were located at the appropriate genomic site. As an alternative to the pIVET5n derivative plasmids, we also constructed suicide tnpR reporter plasmid derivatives of those described in reference 15 for the construction of targeted single-copy lac fusions using the FLP-FRT (FLP recognition target)-mediated site-specific recombinase. These plasmids contain an FRT site in front of a tnpR+-lacZY+ cassette. For each tnpR allele, we constructed two plasmids that differ only in the orientation of the FRT site: FRT orientation A was pCE70(tnpRwt), pCE72(tnpRmut168), and pCE74(tnpRmut135); the FRT orientation B was pCE71(tnpRwt), pCE73(tnpRmut168), and pCE75(tnpRmut135). These plasmids were tested in vitro with an iron-regulated promoter as a proof of concept (see Table S1 in supplemental materials) but were not used for the in vivo experiments described in this work.
In vitro and in vivo RIVET transcription assays.
To prepare the inoculum, RIVET reporter strains were grown in LB broth with selection for Ap and Tc (to maintain the selection for the reporter fusion and the target res1-tetAR-res1 cassette) for variable amounts of time at 37°C. For in vitro expression experiments, the inoculum was washed in saline solution and inoculated at 1:5,000 into NMM (pH 5.6 or pH 7.7). In all cases, to determine the level of transcription of the RIVET fusions, about 200 CFU were plated on LB agar buffered with MOPS-Tris (25 mM Tris-HCl, pH 7.7) containing Ap. After growth overnight at 37°C, colonies were then replica plated or patched onto LB-MOPS agar containing Tc and the extent of induction of the transcriptional fusion to tnpR was measured as the percentage of tetracycline-sensitive (Tets) CFU (Tets CFU divided by the total CFU). For each determination we replicated 50 to 100 colonies. Standard errors of individual counts were estimated by the formula (fq/x)[1/2], where f and q are the percentages of Tets and Tetr, respectively, in the total sample of patched Ampr colonies (x). Counts from replicates were averaged, with n equal to the number of mice employed. A χ2 test was used for statistical analysis of proportions.
β-Galactosidase activity determination.
β-Galactosidase activities in soluble cell extracts were determined by using o-nitrophenyl-β-d-galactopyranoside (Sigma) as the substrate and were expressed as Miller units, as previously described (33).
Animal studies.
Female BALB/c mice (weighing 16 to 18 g) were kept on a normal rodent diet (Harlan-Teklad cat. 2014) or an iron-deficient diet (Harlan-Teklad cat. TD80396) as required by the specific experiment, and food and water were withdrawn from 4 to 24 h before inoculation. Mice were inoculated either perorally (by oral gavage with a 22-gauge feeding needle) or intraperitoneally. Dilutions of the stationary-phase cultures were plated to determine the number of bacteria present in the inoculum. For the in vivo temporal gene fusion induction experiments, 108 CFU were perorally (or 103 CFU intraperitoneally) inoculated into female BALB/c mice as described below. At various time points after inoculation, the small intestine distal lumenal contents, Peyer's patches, mesenteric lymph nodes, spleen, and livers were removed and homogenized, and the percentage of Tets CFU was determined as described above. When neutralization of the stomach pH was required, mice were orally fed with 100 μl 10% sodium bicarbonate 60 min before, at the time of Salmonella inoculation, and 30 min afterward. In experiments with iron chelators, mice were administered desferrioxamine mesylate (DFO) orally by either oral gavage or as an addition in the drinking water. Infected mice were sacrificed at various time points postinoculation. The lumenal content or the Peyer's patches, mesenteric lymph nodes, spleen, and liver were homogenized in phosphate-buffered saline (PBS), serially diluted, and plated onto the appropriate ampicillin-containing agar plates as described above.
Animal cell cultures and invasion assay.
J774A.1 macrophage cell lines were obtained from the American Type Culture Collection (ATCC). Cells were grown to a monolayer at 37°C, 5% CO2 in Dulbecco's modified Eagle medium (DMEM) with 10% heat-inactivated fetal bovine serum. Cells were then infected at an multiplicity of infection of 10 in 24-well plates. Upon infection, cells were spun down and incubated at 4°C for 30 min and then incubated at 37°C for 1 h. Extracellular bacteria were killed with 100 μg ml−1 gentamicin for 2 h and washed, and the cells were incubated for an additional 16 h at 37°C with 10 μg ml−1 gentamicin. Cells were lysed with 1% Triton X-100.
Gentamicin protection assay.
Lumenal samples were collected from infected mice at 48 h postinoculation, exposed to 100 μg ml−1 gentamicin for 60 min, washed, serially diluted, and plated. The percentages of gentamicin-sensitive cells were calculated by comparison to untreated samples.
Determination of lumenal iron.
Lumen content was extracted with 1 M MgSO4 to analyze the bioavailable iron fraction as described by Simpson et al. (45). Iron(III) levels in the intestinal lumen chyme were analyzed using the Ferrozine protocol after reduction in sodium ascorbate. Concentrations are expressed in micromoles Fe per kilogram of intestine content (chyme or fecal matter).
RESULTS
Adaptation of RIVET to Salmonella enterica: construction of a functional res1-tetAR-res1 reporter strain.
While attempting to adapt the RIVET system for use in S. enterica serovar Typhimurium, we discovered that the original res1-Tetr-res1 reporter construct (7), which utilizes the tetR gene from pBR322, supplies only minimal resistance to tetracycline when located in a single copy in the chromosome. Such low levels of tetracycline resistance made this construct impractical for use in S. enterica serovar Typhimurium. The problem was corrected by amplifying and cloning of the tetRA genes from Tn10 between the res1 sites of pGH436 to generate pCE61. The res1-tetRA-res1 fragment was cloned from pCE61 into pJS225, creating pCE63. Allelic exchange was used to create strain JS246, a derivative of S. enterica serovar Typhimurium ATCC 14028 in which the res1-tetRA-res1 reporter was located in a single copy in the chromosome, at a location nearby the purA gene (yjeP8103::res1-tetRA-res1) previously shown to play no role in virulence (50). This cassette can easily be moved between different Salmonella serovars using P22-mediated transduction. Additionally, the use of a tetAR resolution reporter could allow the design of RIVET screening schemes by positive selection in fusaric acid-containing Bochner medium. In the experiments described below, the resolution of the reporter gene is caused by the expression of tnpR from cointegrated pIVETn derivative plasmids (Fig. S1, panel A, in the supplemental material). Alternatively, the plasmid pCE70-75 can be utilized to construct chromosomal promoter fusions to tnpR+- lacZY+ (Table 1; see also Table S1 and Fig. S1, panel B) using the lambda red recombinase and FLP-FRT technologies as previously described (13, 15).
Expression of Φ(pmrH-tnpR) in vitro.
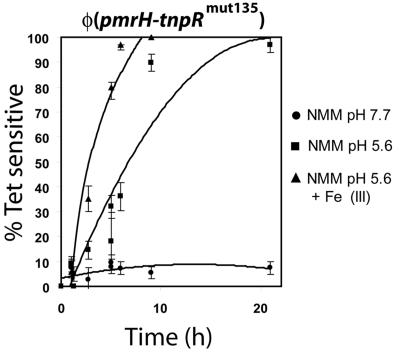
The pmrH operon, which encodes enzymes necessary for modifying LPS with aminoarabinose, is directly regulated by PmrA. The pmrCAB operon is autoregulated and upregulated upon growth in acidic conditions, in medium low in Mg2+, and within phagocytic cells. To analyze the expression of the pmrH operon, we constructed a pmrH-tnpR+-lacZY+ operon fusion in the suicide plasmids pIVET5n tnpR, pIVET5n tnpRmut135, and pIVET5n tnpRmut168 and integrated the constructs in JS246. These three vectors have different ribosomal binding sites that allow various levels of translation of the TnpR resolvase. The expression of the promoter fusion was initially verified in vitro under conditions known to repress or activate the PhoP/PhoQ and PmrA/PmrB regulons. Upon overnight growth, all fusions showed similar transcription patterns of induction when the level of the β-galactosidase reporters were measured (Fig. S2 in supplemental material), but when the level of resolution of the tetRA cassette was evaluated, only the pmrH-tnpRmut135 fusion in strain JSG2428 gave a transcription pattern of induction-repression that correlated with lacZY reporter expression. In particular, with this fusion the resolution ratios increased from 12% (x = 25, where x is the number of Ampr colonies counted) to 80% (x = 50) upon induction in NMM pH 5.6 compared to NMM pH 7.7. Fusions to tnpRwt showed percentages of resolution from 90 to 100% (x = 233) independent of the growth conditions, while tnpRmut168 fusions showed no resolution (x = 172) (Fig. S2). When this system is adapted to other promoters, similar experiments will be necessary to properly match the appropriate ribosomal binding site mutant to get measurable levels of resolution. The pmrH-tnpRmut135 fusion in strain JSG2428 was then assayed in a repressing medium and two different inducing media in time-course experiments (Fig. 1). Under repressing conditions (basic pH and a high concentration of Mg2+), the resolution of the reporter was low, at around 6% (x = 95), even after 24 h of growth, roughly corresponding to ∼25 generations. When grown in acidic media with micromolar concentrations of Mg2+, the resolution of the reporter increased progressively to a maximum resolution of 95% after 24 h. In a similar inducing medium, the addition of micromolar concentrations of FeSO4 led to increased levels of expression of the pmrH-tnpRmut135 fusion, with resolution levels nearing 100% after less than 10 h (Fig. 1). These patterns of in vitro regulation measured using reporter resolution are consistent with data published in the literature for PmrA/PmrB-regulated genes using classical reporter fusions and show the usefulness of this system for monitoring gene expression.
FIG. 1.
Expression of pmrH in vitro. S. enterica serovar Typhimurium JSG2428 carrying a cointegrated pmrH-tnpRmut135 fusion was grown in LB (pH 7.7) in the presence of Ap and Tc and then was diluted 1:100 in NMM (pH 7.7) with 10 mM MgCl2, NMM (pH 5.6)- 10 μM MgCl2, or NMM (pH 5.6)-10 μM MgCl2 with 100 μM FeSO4. At various time points, aliquots were diluted and plated on LB-Ap, and 100 colonies were patched to LB-Tc from each sample. The proportion of Tets colonies was calculated, and the background resolution in the original inoculum was subtracted.
Expression of Φ(pmrH-tnpR) in BALB/c mice and mouse macrophage cell lines.
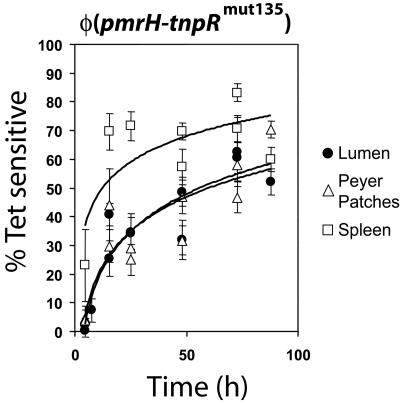
The level of induction of pmrH-tnpRmut135 was measured in bacterial populations recovered from orally and intraperitoneally inoculated female BALB/c mice and compared to in vitro repressing conditions (NMM [pH 7.7], 10 mM MgCl2). Bacteria were grown overnight in buffered LB (pH 7.7) selecting for the reporter Tetr marker, washed in PBS, and inoculated into mice. The average level of background tetRA resolution in the inoculum was 1 to 2%. Bacteria were recovered from the distal ileum lumenal space, ileal Peyer's patches, mesenteric lymph nodes, spleen, and liver. At the 4-h time point, the level of resolution of the Tetr reporter was generally low (from 0 to ∼10%) at all anatomical locations analyzed, with the exception of a few small-size samples (<20 CFU recovered) from spleens that showed around 25% ± 12% resolution (n = 2; n, number of mice) (Fig. 2). The expression of the pmrH operon increased in a hyperbolic fashion over time at all sites until reaching maximum resolution at 72 h postinoculation. Such a maximum resolution time point is likely promoter dependent. While lumenal samples showed net levels of resolution usually ranging from 45 to 55% (n = 4) at 48 h to ∼60% (n = 4) at >72 h, bacteria recovered from spleen (Fig. 2) and liver (data not shown) showed higher expression of pmrH, with many samples reaching levels of >80%.
FIG. 2.
Expression of pmrH during infection of BALB/c mice. S. enterica serovar Typhimurium JSG2428 carrying a cointegrated pmrH-tnpRmut135 fusion was grown in LB (pH 7.7) in the presence of Ap and Tc. Female BALB/c mice were infected with 108 CFU each by oral gavage. At various time points, groups of two mice were sacrificed, and selected organs were dissected and homogenized. The proportion of Tets colonies was determined as described in Materials and Methods.
These levels of reporter resolution are comparable to the expression of pmrH in infected murine macrophage cell lines after 16 h of infection (Fig. 3). The high expression in cells recovered from reticuloendothelial system organs is consistent with previously published data indicating upregulation of PhoP- and PmrA-regulated genes in the vacuoles of eukaryotic cells. However, we were surprised to observe expression of this operon in the ileal lumen, as it is an environment characterized by a high pH, relatively high concentrations of electrolytes and detergents, and low levels of bioavailable iron(III) due to the neutral-to-alkaline pH (45). To better characterize the population of bacteria in the lumen and rule out that bacteria recovered there were growing inside transmigrating neutrophils or other cells, we performed a gentamicin protection assay on lumenal contents recovered 48 h postinoculation. We found that at least 98.2% of bacteria were gentamicin sensitive, indicative of a prevalent extracellular localization of S. enterica serovar Typhimurium at that site.
FIG. 3.
Expression of tnpR fusions within murine macrophage cell lines. Salmonella strains carrying a cointegrated tnpRmut135 fusion to various genes (pmrH, pagP, prgH, and ssaB) were grown in LB (pH 7.7) in the presence of Ap and Tc to prepare the inoculum for cell culture infections. The mouse macrophage cell line J774A.1 was infected at an multiplicity of infection of 10 and, following the invasion period, was exposed to gentamicin. After 16 h, macrophage cells were lysed with 1% Triton X-100 and aliquots were diluted and plated on LB-Ap. Approximately 100 colonies were patched to LB-Tc from each sample. As a medium control, bacteria were grown in DMEM for 16 h. The proportion of Tets colonies was calculated, and the background resolution in the original inoculum was subtracted.
Lumenal bacterial populations at later time points during the infection process may potentially be a mix of colonization from the original lumenal inoculum and reinfection through the bile duct with bacteria from the liver, which would artificially increase the measured resolution frequencies. To evaluate the level of reinfection of the ileal lumen, the reporter bacteria were introduced by intraperitoneal injection and necropsies were performed at 24 h, 48 h, and 66 h. Spleen and lumen samples were removed as described above and evaluated for the level of transcription of the pmrH-tnpRmut135 reporter. High levels of expression ranging from 54% (x = 51; n = 2) to 63% (x = 191; n = 2) were observed in the spleen at all time points (spleen infection was absent or modest, <102 bacteria per organ, before the 24-h time point). In the lumen, no bacteria or modest populations (∼102 bacteria per sample) were recovered at <24 h, and populations of ca. 2 × 102 to 4 × 102 CFU per mouse ileum were observed at 48 h and 66 h (in contrast to average populations of ca. 1 × 104 to 1 × 105 bacteria normally recovered from the ileum of perorally inoculated mice at 48 to 72 h). At those time points, the level of pmrH expression was comparable to what was observed in bacteria from spleen samples (44 to 70%). As an additional control, we transduced the invA::cat mutation into JSG2428 to construct JSG2540. This Salmonella mutant is expected to be unable to efficiently invade and replicate in enterocytes due to a structural defect in Salmonella pathogenicity island 1 (SPI-1) type III secretion (17). As a consequence, this mutation should severely impair the ability of JSG2540 to escape the lumen and reach systemic sites of infection, except for the physiological levels of antigen sampling by M cells (12) and systemic infection via CD18+ dendritic cells (55). Using mice perorally inoculated with the invA mutant, we found that expression of pmrH in the lumen occurred at levels comparable to those of the parent strain JSG2428 pmrH-tnpRmut135 (58.5% ± 4.9%; n = 2). Collectively these results imply that recycling can potentially occur but at quite a low level, probably not sufficient to affect the overall apparent lumenal expression of pmrH.
In vivo expression of Φ(pmrH-tnpR) is fully dependent on PhoP and PmrA.
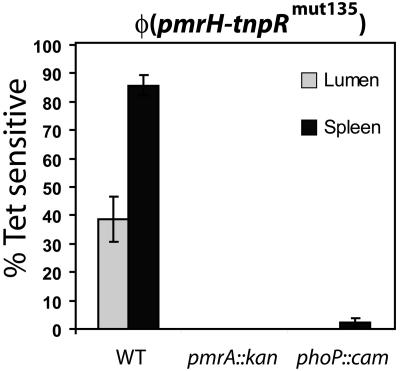
A greater oral virulence defect was observed for the pmrH mutant than for a pmrA null strain, suggesting that the pmrH operon may be controlled in vivo by factors other than PmrA. To evaluate whether the in vivo expression of pmrH was dependent solely on phoP and pmrA, we transduced either a phoP::Tn10d-Cm or a pmrA::kan allele into strain JSG2428 pmrH-tnpRmut135. In experiments in vitro under PhoP- and PmrA-inducing conditions, expression in the pmrA background was consistently 0% (x = 100, n = 2), while in the phoP background the resolution of the tetAR reporter was 1 to 2.9% (x = 100, n = 2). Expression levels in vitro could be complemented by ectopic expression of the wild-type allele (data not shown). Similarly, in vivo expression of pmrH was fully dependent on the presence of functional phoP and pmrA (Fig. 4), suggesting that no other major activators of the pmrH operon exist.
FIG. 4.
In vivo expression of pmrH is fully dependent on both PhoP and PmrA. The phoP::Tn10d-Cm or pmrA::kan allele was transduced into S. enterica serovar Typhimurium JSG2428 (pmrH-tnpRmut135) to generate JSG2437 and JSG2436, respectively. The strains were grown in LB (pH 7.7) in the presence of Ap and Tc to prepare the inoculum. Two female BALB/c mice were infected with 108 CFU each by oral gavage. After 48 h, the mice were sacrificed and selected organs were dissected and homogenized. Aliquots were diluted and plated on LB-Ap, and up to 100 colonies were patched to LB-Tc from each sample. The proportion of Tets colonies was calculated, and the background resolution in the original inoculum was subtracted. Averages with the corresponding standard deviations are shown. WT, wild type.
Neutralization of stomach acidity does not affect lumenal pmrH and pagP induction.
Acidic pH and iron(III) are two recognized signals able to activate the PhoP and PmrA regulons in vitro. In vivo, upon oral inoculation, bacteria experience an acidic pH only in the stomach (pH range from 2 to 5) and the proximal portion of the duodenum (44). To rule out the stomach as the site of induction of pmrH-tnpRmut135, we treated the mice with relatively large volumes (100 μl) of 10% sodium bicarbonate 60 min before, at the time of oral inoculation of the bacterial suspension, and 30 min postinoculation. Transcription of pmrH in the lumen and in the spleen was followed over time (Fig. 5), and the results show that expression of the gene fusion was not affected by neutralizing treatments. Transient exposure to a residual acidic pH, due to insufficient buffering of the stomach, may be sufficient to induce the expression of TnpR at a level high enough to trigger resolution later on in the ileal lumen. To rule out this possibility, we exposed cells carrying the tnpR fusions to low-pH shocks (pH 5.6) in vitro for 15, 30, and 60 min followed by overnight growth in PhoP- and PmrA-repressing conditions (high Mg2+ NMM [pH 7.7]). A second promoter fusion was constructed to a PhoP-activated gene, pagP, also inducible by low pH. The expression of pagP, measured as the percentage of resolution, was increased from 0% to 2% after 60 min of exposure to low pH, while transcription of pmrH was measured at about 10.4% ± 3.7%, 19.4% ± 4.3%, and 32% ± 5.0% after 15-, 30-, and 60-min shocks, respectively. The non-acid-shocked control showed 6.9% ± 2.6% resolution after 16 h of growth. Therefore, these data show that the levels of TnpR-induced resolution observed in the lumen are unlikely to be caused by transient exposure to acidity due to the fact that, as discussed below, in fasting mice the half-emptying time of the stomach is on the order of a few minutes.
FIG. 5.
Orally fed sodium bicarbonate does not affect the induction of pmrH in the gastrointestinal lumen. S. enterica serovar Typhimurium JSG2428 carrying a cointegrated pmrH-tnpRmut135 fusion was grown in LB (pH 7.7) in the presence of Ap and Tc to prepare the inoculum. Two female BALB/c mice were infected with 108 CFU each by oral gavage. Three oral doses of 100 μl 10% NaHCO3 were administered at −60, 0, and +30 min from the inoculation time. After 5, 24, and 72 h, the mice were sacrificed and selected organs were dissected and homogenized. Aliquots were diluted and plated on LB-Ap, and up to 100 colonies were patched to LB-Tc from each sample. The proportion of Tets colonies was calculated, and the background resolution in the original inoculum was subtracted. Averages with the corresponding standard deviations are shown.
Iron deprivation does not affect induction of pmrH.
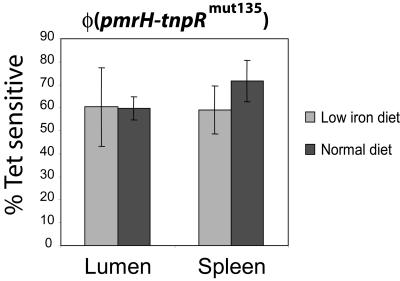
The previous experiments eliminated stomach acidic pH as the key signal inducing pmrH in the gastrointestinal tract under our experimental conditions. However, iron(III) at 0.1 mM has been shown to induce PmrA/PmrB in vitro and in the soil (9). In vivo, such elevated levels of bioavailable iron(III) can be observed only in the stomach and in the proximal duodenum as a function of the iron content in the diet (44). More distal regions of the gastrointestinal tract are characterized by higher pH, wherein the large majority of iron would be in insoluble, nonbioavailable forms (45). We proceeded to test directly whether the in vivo expression of pmrH was affected by dietary iron. Strain JSG2428 pmrH-tnpRmut135 was inoculated in mice which were fed for 21 days with an iron-deficient diet and treated for 24 h preinoculation and postinoculation with oral doses of DFO. The analysis of pmrH transcriptional levels at 48 h postinoculation showed no significant difference between the iron-deficient and control treatments (Fig. 6).
FIG. 6.
Expression of pmrH in the gastrointestinal lumen is not dependent on high concentrations of bioavailable iron. Female BALB/c mice were fed iron-deficient or normal diets for 3 weeks. S. enterica serovar Typhimurium strain JSG2428 (pmrH-tnpRmut135) was grown in LB (pH 7.7) in the presence of Ap and Tc. Mice were infected with 108 CFU each by oral gavage. The iron-deficient group was also administered water containing the iron(III) chelator desferrioxamine (DFO) 24 h before inoculation until the end of the experiment. After 48 h, the mice were sacrificed and selected organs were dissected and homogenized. Aliquots were diluted and plated on LB-Ap, and up to 100 colonies were patched to LB-Tc from each sample. The proportion of Tets colonies was calculated, and the background resolution in the original inoculum was subtracted. Bioavailable iron was measured using a Ferrozine protocol, and the concentrations ranged from 7.5 to 40 μmol/kg lumen content in the normal diet treatment to <0 to 1.4 μmol/kg lumen content in the low-iron diet treatment.
The iron levels in the ileal lumen were first evaluated indirectly using an fepA-tnpR RIVET strain. fepA codes for the enterobactin receptor, which is induced under low-iron conditions and repressed by Fur in conditions of high iron (53). In vitro, the fepA-tnpR fusion repression led to 0% resolution under high-iron growth (LB with 200 μM FeCl3 or 200 μM FeSO4) and 48% to 37% resolution under low-iron conditions due to DFO or 2,2-dipiridyl treatments (100 μM). In vivo 48 h postinoculation, fepA-tnpR was expressed in the lumen at slightly higher levels in iron-deficient mice than in mice fed with a normal diet (74.9% versus 65.6%), but this difference was not statistically significant (P = 0.102). Bioavailable iron levels in the ileal lumen of the same mice where transcriptional analysis of pmrH was performed were also measured directly using a Ferrozine spectrophotometric protocol. The results showed submicromolar concentration of iron (often below the detection threshold) in mice kept on an iron-deficient diet (Fig. 6), while concentrations ranging from 7.5 to ca. 40 μM were observed in “normal diet” mice. These results suggest that iron is not a relevant PmrA-PmrB-inducing signal in the mouse ileal lumen.
In vivo expression of ssaB and the PhoP-regulated genes pagP and prgH.
To further examine the in vivo spatiotemporal expression of virulence genes, including those belonging to the PhoP regulon, we constructed additional fusions to the tnpR reporter. pagP is a gene encoding a palmitoyl transferase believed to be induced inside phagocytic cells by PhoP/PhoQ. Activation of the PhoP/PhoQ regulon would also lead to expression of the PmrA/PmrB regulon, including the pmrH operon. Given the strong dependence on PhoP shown by pmrH in vivo (Fig. 4), we would expect activation of the PhoP/PhoQ regulon also in the ileal lumen, not just inside cells of the reticuloendothelial system. A pagP-tnpRmut135 fusion showed the expected behavior in vitro (i.e., repressed by high pH and high magnesium concentrations and induced by low pH and low Mg; data not shown) and was also upregulated 20-fold in S. enterica serovar Typhimurium cells recovered from the lumen of orally infected mice compared to that of repressing conditions in vitro (Fig. 7). A fusion to the SPI-1 gene prgH was also expressed in lumenal bacteria, upregulated ca. twofold in Peyer's patches compared to an SPI-1 repressing low osmolarity acidic medium (NMM pH 5.6) but not further activated in organs of the reticuloendothelial system. Lumenal expression of prgH is consistent with its role in enterocyte and M-cell invasion. Finally, a fusion to the SPI-2 structural gene ssaB was tested as a control based on its known site of expression in vivo. The ssaB fusion was not induced in the lumen but was upregulated in bacteria isolated from Peyer's patches and spleen, consistent with its essential role in intramacrophage survival and systemic spread (Fig. 7). This fusion also served as a control to rule out nonspecific induction of tnpR fusions in the intestine.
FIG. 7.
In vivo transcription of the PhoP-activated gene pagP, the PhoP-repressed gene prgH, and the SPI-2 gene ssaB. S. enterica serovar Typhimurium strains carrying cointegrated fusions to tnpRmut135 were grown in LB (pH 7.7) in the presence of Ap and Tc. Female BALB/c mice were infected with 108 CFU each by oral gavage. At 48 h postinoculation, two mice per treatment were sacrificed, and the luminal contents, Peyer's patches, and spleens were homogenized. Aliquots were diluted and plated on LB-Ap, and up to 100 colonies were patched to LB-Tc from each sample. The proportion of Tets colonies was calculated, and the background resolution in the original inoculum was subtracted. Averages with the corresponding standard deviations are shown. JSG2502, pagP-tnpRmut135; JSG2504, prgH-tnpRmut135; JSG2524, ssaB-tnpRmut135. ND, not determined.
DISCUSSION
In this study we developed RIVET for use in S. enterica serovar Typhimurium (Fig. S1) and showed its application to the study of in vivo patterns of transcriptional expression of virulence genes belonging to the PmrA and PhoP regulons. In particular we demonstrated that, contrary to what was expected, both pagP and pmrH, two genes involved in LPS modification and antimicrobial peptide resistance, are upregulated in bacteria located extracellularly in the ileum of BALB/c mice. Prior to this work, the model had been that, in vivo, the PmrA and PhoP regulons were induced exclusively intracellularly. Indeed, consistent with the published literature (1), expression was even higher in organs of the reticuloendothelial system, where the bacteria are localized in phagocytic cells.
As both iron ions (58) and acidic pH (48) have been shown to activate the PhoP/PhoQ and/or the PmrA/PmrB regulons in vitro, we sought to determine if these environmental factors were responsible for luminal pagP and pmrH expression. We did not consider magnesium as an inducing signal in the lumen, because the normal concentration in the plasma is in the millimolar range and we would expect similar or higher levels in the intestine. However, direct magnesium measurements in the intestine have not been performed. The neutralization of stomach acidity, as well as dietary and treatment regimens minimizing the levels of bioavailable iron in the ileum, did not affect the levels of intestinal induction of the pmrH fusion. Lacking a solely pH-regulated fusion, our controls for the neutralization of stomach pH relied on indirect assays. Specifically, almost 30 mg of sodium bicarbonate per mouse was used to buffer the stomach acidity, which is three times the standard amount used in such assays. Moreover, in vitro experiments with transient pH pulses did not show marked induction of pagP or pmrH. Based on data from the literature, the emptying kinetics of the stomach in young mice follows an exponential decay function, and the half-emptying time with phenol red in saline solution is about 8 min (28), while it is less than 5 min for a BaSO4 meal (37). Similarly, in the duodenum, transit time is approximately 1 min (39). These transition times are well below the levels required for a significant induction of pmrH, even in the unlikely case that the neutralizing treatment had been incomplete. In experiments with mice kept on an iron-deficient diet, we exploited the observation that intestinal iron levels are determined solely by dietary iron, with the duodenal iron transport being vectorial and directed toward the bloodstream (49). Moreover, oral treatments with iron chelators were concurrently performed to minimize the free, bioavailable iron. The level of iron directly measured in iron-deficient mice was well below the inducing levels necessary for activation of the PmrA/PmrB system. The phagosome of Salmonella-infected macrophages probably contains relatively low concentrations of iron, given that genes in the Fur regulon were isolated in in vivo-induced screenings (25). Such low levels may not allow full PmrA/PmrB induction, and other signals, such as low pH, may play a more important role. This would be consistent with the absence of any repressing effect on pmrH-tnpR expression caused by 100 μM desferrioxamine (DFO), an iron(III) chelator, in cultured mouse macrophages (data not shown). We conclude that iron is not a likely signal for the induction of the PmrA/PmrB regulon either in the ileum or in the reticuloendothelial organs of mice.
In control experiments, prgH, a SPI-1 secretion gene required for invasion of enterocytes and M cells of the gastrointestinal tract, was expressed in the intestinal lumen, consistent with its role in cell invasion, but expression did not increase in progressive sites of invasion (Peyer's patches and spleen). Control strains carrying fusions of tnpR to ssaB, a structural SPI-2 gene necessary for intramacrophage survival and systemic spread, showed a reciprocal expression pattern compared to that of prgH, consistent with previous data showing macrophage upregulation of SPI-2 genes (11). The induction of both prgH and pagP in the intestine may seem contradictory, given that the former is a PhoP-repressed gene and the second is a PhoP-activated gene by functional definition. Several hypotheses may be formulated to explain this observation, including differential promoter affinity for PhoP and/or the involvement of multiple environmental signals acting at suboptimal threshold levels that lead to partial activation of PhoP/PhoQ in the intestinal lumen environment. Such partial activation would likely result in some bacteria with activated PhoP and some in which PhoP is not activated. In addition, PhoP/PhoQ could be activated by signals in the lumen but repressed upon association with intestinal epithelial cells, leading to expression of both prg and pag genes when measured by RIVET.
Our study does not identify the signal for intestinal expression of the PhoP and PmrA regulons, but recent work using in vitro sublethal concentrations of antimicrobial peptides showed that these defense molecules may act as a signal for PhoP/PhoQ (3). Consistent with this line of thought was the finding that, in vivo, pmrH was not expressed to measurable levels in either a PhoP null background or a PmrA null mutant. These observations, and the fact that PhoP/PhoQ and PmrA/PmrB expression are connected by a regulatory cascade, also suggest that, in vivo, signals are primarily perceived by PhoQ and not PmrB. If perceived by PmrB, partial or complete resolution would have been observed in a phoP mutant. It is tempting to speculate that in vivo induction by mucosal antimicrobial peptides may occur. Experiments using knockout mice unable to produce mature alpha defensins may help to test this hypothesis.
Based on the fact that a pmrA null strain was more virulent than a pmrH operon mutant by peroral infection, we hypothesized that the pmrH operon may be expressed in vivo in a PmrA-independent manner (24). Our data suggest that this hypothesis is most likely false, as pmrH was not expressed in vivo in PmrA or PhoP null backgrounds. However, it is possible, but unlikely, that the pmrH operon could be expressed at a level sufficient for LPS modification but insufficient for resolution of the Tetr cassette. An alternative hypothesis for the original finding (24) is that the loss of PmrA affects determinants that both positively and negatively affect virulence, while loss of aminoarabinose only negatively affects virulence. More work is necessary to explain this phenomenon.
Few studies have applied RIVET to the analysis of bacterial in vivo gene expression since the seminal work done on V. cholerae (7, 31). The only application of the kind is a recent work where Bordetella pertussis in vivo expression of several BvgSA-regulated genes (cya, fha, prn, and ptx) was analyzed using a modification of RIVET (56). Other applications have commonly used RIVET as a screening procedure (30, 38, 42). Our work adds this approach to the arsenal of genetic tools available for the study of Salmonella virulence in vivo.
Supplementary Material
Acknowledgments
Support was provided by National Institutes of Health grants AI043521 (J.S.G.) and AI63230 and 00-25 of the Roy J. Carver Charitable Trust (J.M.S.).
We thank Andrew Camilli for providing pIVET5n derivative plasmids before publication and for his technical advice.
Footnotes
Supplemental material for this article is available at http://jb.asm.org/.
REFERENCES
- 1.Alpuche Aranda, C. M., J. A. Swanson, W. P. Loomis, and S. I. Miller. 1992. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc. Natl. Acad. Sci. USA 89:10079-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology, vol. 1. John Wiley & Sons, New York, N.Y.
- 3.Bader, M. W., W. W. Navarre, W. Shiau, H. Nikaido, J. G. Frye, M. McClelland, F. C. Fang, and S. I. Miller. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol. Microbiol. 50:219-230. [DOI] [PubMed] [Google Scholar]
- 4.Belden, W. J., and S. I. Miller. 1994. Further characterization of the PhoP regulon: identification of new PhoP-activated virulence loci. Infect. Immun. 62:5095-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brogden, K. A., M. Ackermann, P. B. McCray, Jr., and B. F. Tack. 2003. Antimicrobial peptides in animals and their role in host defences. Int. J. Antimicrob. Agents 22:465-478. [DOI] [PubMed] [Google Scholar]
- 6.Bron, P. A., C. Grangette, A. Mercenier, W. M. de Vos, and M. Kleerebezem. 2004. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 186:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camilli, A., D. T. Beattie, and J. J. Mekalanos. 1994. Use of genetic recombination as a reporter of gene expression. Proc. Natl. Acad. Sci. USA 91:2634-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camilli, A., and J. J. Mekalanos. 1995. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol. Microbiol. 18:671-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamnongpol, S., W. Dodson, M. J. Cromie, Z. L. Harris, and E. A. Groisman. 2002. Fe(III)-mediated cellular toxicity. Mol. Microbiol. 45: 711-719. [DOI] [PubMed] [Google Scholar]
- 10.Chamnongpol, S., and E. A. Groisman. 2002. Mg2+ homeostasis and avoidance of metal toxicity. Mol. Microbiol. 44:561-571. [DOI] [PubMed] [Google Scholar]
- 11.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 12.Clark, M. A., K. A. Reed, J. Lodge, J. Stephen, B. H. Hirst, and M. A. Jepson. 1996. Invasion of murine intestinal M cells by Salmonella typhimurium inv mutants severely deficient for invasion of cultured cells. Infect. Immun. 64:4363-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 16.Galan, J. E., and R. Curtiss III. 1990. Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect. Immun. 58:1879-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galan, J. E., C. Ginocchio, and P. Costeas. 1992. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J. Bacteriol. 174:4338-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710-720. [DOI] [PubMed] [Google Scholar]
- 19.Garcia Vescovi, E., F. C. Soncini, and E. A. Groisman. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165-174. [DOI] [PubMed] [Google Scholar]
- 20.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groisman, E. A., E. Chiao, C. J. Lipps, and F. Heffron. 1989. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc. Natl. Acad. Sci. USA 86:7077-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guina, T., E. C. Yi, H. Wang, M. Hackett, and S. I. Miller. 2000. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J. Bacteriol. 182:4077-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunn, J. S., and S. I. Miller. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunn, J. S., S. S. Ryan, J. C. Van Velkinburgh, R. K. Ernst, and S. I. Miller. 2000. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 68:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heithoff, D. M., C. P. Conner, U. Hentschel, F. Govantes, P. C. Hanna, and M. J. Mahan. 1999. Coordinate intracellular expression of Salmonella genes induced during infection. J. Bacteriol. 181:799-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes, K. T., and J. R. Roth. 1985. Directed formation of deletions and duplications using Mud(Ap, lac). Genetics 109:263-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kox, L. F., M. M. Wosten, and E. A. Groisman. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19:1861-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo, W. H., K. S. Wadwa, and C. D. Ferris. 1998. Cephalosporin antibiotics accelerate gastric emptying in mice. Digest. Dis. Sci. 43:1690-1694. [DOI] [PubMed] [Google Scholar]
- 29.Lee, H., F. F. Hsu, J. Turk, and E. A. Groisman. 2004. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J. Bacteriol. 186:4124-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, S. H., S. M. Butler, and A. Camilli. 2001. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. USA 98:6889-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, S. H., D. L. Hava, M. K. Waldor, and A. Camilli. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625-634. [DOI] [PubMed] [Google Scholar]
- 32.Mann, B. A., and J. M. Slauch. 1997. Transduction of low-copy number plasmids by bacteriophage P22. Genetics 146:447-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Miller, S. I. 1991. PhoP/PhoQ: macrophage-specific modulators of Salmonella virulence? Mol. Microbiol. 5:2073-2078. [DOI] [PubMed] [Google Scholar]
- 35.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, S. I., and J. J. Mekalanos. 1990. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J. Bacteriol. 172:2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreto, M., L. Guzman, and A. Diez. 1982. A pattern for gastric emptying in mice. Am. J. Physiol. 242:G333-G336. [DOI] [PubMed] [Google Scholar]
- 38.Osorio, C. G., J. A. Crawford, J. Michalski, H. Martinez-Wilson, J. B. Kaper, and A. Camilli. 2005. Second-generation recombination-based in vivo expression technology for large-scale screening for Vibrio cholerae genes induced during infection of the mouse small intestine. Infect. Immun. 73: 972-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raja, K. B., R. J. Simpson, and T. J. Peters. 1987. Comparison of 59Fe3+ uptake in vitro and in vivo by mouse duodenum. Biochim. Biophys. Acta 901:52-60. [DOI] [PubMed] [Google Scholar]
- 40.Rosenberger, C. M., R. L. Gallo, and B. B. Finlay. 2004. Interplay between antibacterial effectors: a macrophage antimicrobial peptide impairs intracellular Salmonella replication. Proc. Natl. Acad. Sci. USA 101:2422-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salzman, N. H., D. Ghosh, K. M. Huttner, Y. Paterson, and C. L. Bevins. 2003. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422:522-526. [DOI] [PubMed] [Google Scholar]
- 42.Saviola, B., S. C. Woolwine, and W. R. Bishai. 2003. Isolation of acid-inducible genes of Mycobacterium tuberculosis with the use of recombinase-based in vivo expression technology. Infect. Immun. 71:1379-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmid, M. B., and J. R. Roth. 1983. Genetic methods for analysis and manipulation of inversion mutations in bacteria. Genetics 105:517-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson, R. J., and T. J. Peters. 1990. Forms of soluble iron in mouse stomach and duodenal lumen: significance for mucosal uptake. Br. J. Nutr. 63:79-89. [DOI] [PubMed] [Google Scholar]
- 45.Simpson, R. J., S. Sidhar, and T. J. Peters. 1992. Application of selective extraction to the study of iron species present in diet and rat gastrointestinal tract contents. Br. J. Nutr. 67:437-444. [DOI] [PubMed] [Google Scholar]
- 46.Slauch, J. M., and A. Camilli. 2000. IVET and RIVET: use of gene fusions to identify bacterial virulence factors specifically induced in host tissues. Methods Enzymol. 326:73-96. [DOI] [PubMed] [Google Scholar]
- 47.Soncini, F. C., E. Garcia Vescovi, F. Solomon, and E. A. Groisman. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soncini, F. C., and E. A. Groisman. 1996. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 178:6796-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Srai, S. K., A. Bomford, and H. J. McArdle. 2002. Iron transport across cell membranes: molecular understanding of duodenal and placental iron uptake. Best Pract. Res. Clin. Haematol. 15:243-259. [DOI] [PubMed] [Google Scholar]
- 50.Stanley, T. L., C. D. Ellermeier, and J. M. Slauch. 2000. Tissue-specific gene expression identifies a gene in the lysogenic phage Gifsy-1 that affects Salmonella enterica serovar Typhimurium survival in Peyer's patches. J. Bacteriol. 182:4406-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stark, W. M., N. D. Grindley, G. F. Hatfull, and M. R. Boocock. 1991. Resolvase-catalysed reactions between res sites differing in the central dinucleotide of subsite I. EMBO J. 10:3541-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamayo, R., B. Choudhury, A. Septer, M. Merighi, R. Carlson, and J. S. Gunn. 2005. Identification of cptA, a PmrA-regulated locus required for phosphoethanolamine modification of the Salmonella enterica serovar Typhimurium lipopolysaccharide core. J. Bacteriol. 187:3391-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsolis, R. M., A. J. Baumler, I. Stojiljkovic, and F. Heffron. 1995. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J. Bacteriol. 177:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Velkinburgh, J. C., and J. S. Gunn. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 67:1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vazquez-Torres, A., J. Jones-Carson, A. J. Baumler, S. Falkow, R. Valdivia, W. Brown, M. Le, R. Berggren, W. T. Parks, and F. C. Fang. 1999. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401:804-808. [DOI] [PubMed] [Google Scholar]
- 56.Veal-Carr, W. L., and S. Stibitz. 2005. Demonstration of differential virulence gene promoter activation in vivo in Bordetella pertussis using RIVET. Mol. Microbiol. 55:788-798. [DOI] [PubMed] [Google Scholar]
- 57.Vescovi, E. G., Y. M. Ayala, E. Di Cera, and E. A. Groisman. 1997. Characterization of the bacterial sensor protein PhoQ. Evidence for distinct binding sites for Mg2+ and Ca2+. J. Biol. Chem. 272:1440-1443. [DOI] [PubMed] [Google Scholar]
- 58.Wosten, M. M., L. F. Kox, S. Chamnongpol, F. C. Soncini, and E. A. Groisman. 2000. A signal transduction system that responds to extracellular iron. Cell 103:113-125. [DOI] [PubMed] [Google Scholar]
- 59.Zhou, Z., A. A. Ribeiro, S. Lin, R. J. Cotter, S. I. Miller, and C. R. Raetz. 2001. Lipid A modifications in polymyxin-resistant Salmonella typhimurium: PmrA-dependent 4-amino-4-deoxy-L-arabinose, and phosphoethanolamine incorporation. J. Biol. Chem. 276:43111-43121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.