Abstract
Genes involved in magnetite biomineralization are clustered in the genome of the magnetotactic bacterium Magnetospirillum gryphiswaldense. We analyzed a 482-kb genomic fragment, in which we identified an approximately 130-kb region representing a putative genomic “magnetosome island” (MAI). In addition to all known magnetosome genes, the MAI contains genes putatively involved in magnetosome biomineralization and numerous genes with unknown functions, as well as pseudogenes, and it is particularly rich in insertion elements. Substantial sequence polymorphism of clones from different subcultures indicated that this region undergoes frequent rearrangements during serial subcultivation in the laboratory. Spontaneous mutants affected in magnetosome formation arise at a frequency of up to 10−2 after prolonged storage of cells at 4°C or exposure to oxidative stress. All nonmagnetic mutants exhibited extended and multiple deletions in the MAI and had lost either parts of or the entire mms and mam gene clusters encoding magnetosome proteins. The mutations were polymorphic with respect to the sites and extents of deletions, but all mutations were found to be associated with the loss of various copies of insertion elements, as revealed by Southern hybridization and PCR analysis. Insertions and deletions in the MAI were also found in different magnetosome-producing clones, indicating that parts of this region are not essential for the magnetic phenotype. Our data suggest that the genomic MAI undergoes frequent transposition events, which lead to subsequent deletion by homologous recombination under physiological stress conditions. This can be interpreted in terms of adaptation to physiological stress and might contribute to the genetic plasticity and mobilization of the magnetosome island.
Magnetotactic bacteria (MTB) form intracellular chains of magnetosomes, which are specific inorganic structures that serve as devices for magnetic navigation in the aquatic habitats of these organisms (6). Despite great interdisciplinary interest in magnetosome formation, the molecular mechanism of biomineralization and its genetic determination have remained poorly understood, mostly due to previous difficulties in genetic manipulation and in particular due to the lack of appropriate mutants for functional analysis, as well as the lack of complete genome data (6).
The microaerophilic alphaproteobacterium Magnetospirillum gryphiswaldense synthesizes magnetosomes, which consist of crystals of magnetite (Fe3O4) enclosed in intracytoplasmic vesicles of the magnetosome membrane (MM) (37). The MM is a unique compartment which provides spatial and physicochemical control over magnetite biomineralization and has a distinct biochemical composition. Besides phospholipids, the MM contains a complex, specific subset of magnetosome membrane proteins (MMPs) (18). The classes of MMPs in M. gryphiswaldense include MMPs with presumed functions in magnetosome-directed transport of iron, control of crystal growth, and assembly of magnetosome chains. The MMPs are encoded by the mam and mms genes, which are clustered in three operons that are close to each other (18, 19).
Expression of the magnetic phenotype in M. gryphiswaldense is under physiological control and depends on the availability of iron and the presence of microaerobic conditions (20, 38, 39). In addition, the magnetic phenotype seems to be genetically unstable, and spontaneous nonmagnetic mutants have been isolated repeatedly from various MTB during subcultivation in the laboratory (8, 12). In M. gryphiswaldense frequent spontaneous loss of the magnetic phenotype has been observed in long-term cultures during stationary growth (36). Nonmagnetic mutants accumulated at a frequency of 0.5 × 10−2 in aged cultures that were stored at 4°C, whereas these nonmagnetic mutants were virtually not detectable in growing cultures during repeated serial transfers. One nonmagnetic mutant designated strain MSR-1B was found to have a large chromosomal deletion which was estimated to extend over approximately 80 kb and encompasses the entire mamAB, mamGFDC, and mms6 operon-like gene clusters that encode all MMPs (18). Partial sequence analysis of the deleted region, as well as a region adjacent to the right boundary, revealed that this region contains a number of unknown genes and is remarkably rich in insertion elements (IS elements). The characteristics of this chromosomal region suggested that there is a genomic “magnetosome island” (MAI), which seems to be conserved in various MTB and might extend beyond the sequenced 35-kb region in M. gryphiswaldense (19, 36).
Genomic islands were first described as pathogenicity islands in pathogenic bacteria, but mobile and accessory genetic elements similar to pathogenicity islands have been identified in a range of nonpathogenic species isolated from the environment (11). Genomic islands are often found inserted near tRNA genes, typically contain direct and inverted repeat sequences and IS elements in the flanking regions, and exhibit genetic instability, which suggests that they can be acquired and transmitted via horizontal gene transfer.
In this study, we analyzed the putative MAI of M. gryphiswaldense. Sequence analysis of a genomic 482-kb contig revealed the presence of a conspicuous 130-kb region that, in addition to the previously identified magnetosome genes, contains genes putatively involved in magnetosome biomineralization. In addition, we investigated the occurrence of spontaneous magnetosome mutations that were associated with various types of deletion events in this region. Our results provide further evidence that there is a genomic MAI that undergoes frequent transposition and subsequent deletion under physiological stress conditions.
MATERIALS AND METHODS
Bacterial strains.
Different strains, subcultures, and mutants were derived from M. gryphiswaldense strain MSR-1 (= DSM 6361). A fresh subculture was obtained from the stock of MSR-1 deposited in the DSMZ strain collection and was designated the “archetype” strain (MSR-1A). A magnetic derivative of MSR-1 which had been routinely subcultured for an undefined number of passages in the laboratory was designated the “lab strain” (MSR-1L). Strain MSR-1B is a spontaneous nonmagnetic mutant of strain MSR-1 that has a large deletion comprising all known magnetosome genes (36).
Growth conditions.
Cells grown on solid activated charcoal agar (ACA) were incubated at 28°C in anaerobic jars (Ochs, Bovenden-Lenglern, Germany) under an O2-CO2-N2 (0.5:5:94.5, vol/vol/vol) atmosphere (43). Liquid cultures of M. gryphiswaldense strains were routinely grown microaerobically in flask standard medium (FSM) as described previously (20) at 28°C with 50 μM ferric citrate as the iron source. Alternatively, iron was added at concentrations up to 2,000 μM for iron stress experiments, or ferric citrate was omitted from the growth medium and 10 μM 2,2′-dipyridyl was added for iron starvation experiments. The effect of aerobic conditions was determined with 50-ml cultures which were agitated for 24 h at 150 rpm (INNOVA 4330) with free gas exchange with air. Establishment of aerobic conditions was indicated by the repression of magnetosome formation in the cells. For nutritional deprivation experiments, carbon and nitrogen sources were omitted from the medium. The effect of hydrogen peroxide was tested similarly, as described previously (26). Overnight cultures were diluted into fresh FSM and grown to an optical density at 565 nm of 0.2. One-milliliter aliquots were exposed to 3% H2O2, and the cultures were shaken at 28°C for 10 min. Exposure was stopped by diluting the cultures 625-fold into FSM medium containing 130 U catalase/ml. For cold storage experiments, liquid cultures of M. gryphiswaldense strain MSR-1L were incubated for various times at 4°C under microaerobic or aerobic conditions before they were plated onto ACA.
Isolation of spontaneous nonmagnetic mutants.
Cells of MSR-1L from different incubation experiments were spread onto ACA (102 to 103 cells per plate) and incubated as described above. Colonies that developed after 5 to 7 days were visually examined for altered colony morphology. All colonies whose appearance differed from the appearance of the wild-type colonies were considered mutants. Compared to the wild type, which was dark brown due to the presence of fully developed magnetosome chains, clones with a reduced magnetosome content could be recognized by the lighter brown colonies, whereas magnetosome-free mutants were cream to whitish. After restreaking, mutant phenotypes were verified by microscopic inspection of the magnetic reaction, electron microscopy, and growth experiments. The average magnetic orientation of cell suspensions (“magnetism”) was assayed by an optical method as described previously (40). Briefly, cells were aligned at different angles relative to the light beam by means of an external magnetic field. The ratio of the resulting maximum and minimum scattering intensities (Cmag) was previously demonstrated to be correlated with the average number of magnetic particles and can be used for semiquantitative assessment of magnetite formation (for practical purposes, a Cmag value of 0 was assumed for nonmagnetic cells).
Approximately 500 stable mutants with aberrant morphology selected from a total of approximately 2.5 × 105 screened colonies were investigated further.
Generation and analysis of genome sequence data.
For BAC and whole-genome shotgun (WGS) cloning, genomic DNA was obtained from different independent subcultures of M. gryphiswaldense MSR-1A that were serially transferred for an undefined number of passages. Isolation of a BAC (bacterial artificial chromosome) clone harboring the magnetosome genes, generation of a whole-genome shotgun library, and sequencing were performed as described previously (36). In addition, a cosmid library (Epicenter Technologies, Madison, WI) was generated, and end sequences determined from 500 clones were used for WGS assembly. Sequencing gaps were eliminated and low-quality regions were improved by resequencing of selected plasmids/cosmids, primer walking, and sequencing of long-range PCR products. The quality of raw sequence data was checked with PHRED (13). Shotgun sequences were assembled with Phrap (http://www.genome.bnl.gov/Software/UW/), and Consed (version 14.00) (17) was used for final editing of the sequence. The final quality of the sequence data was less than 1 error in 20,000 bases. Glimmer 2.0 was used for prediction of open reading frames (ORFs) in the finished sequence (10). ORF predictions were manually refined using ARTEMIS (34). Similarity searches for annotation were carried out with BLASTP (2) by using the UniProt database and the translated amino acid sequences encoded by predicted ORFs as queries. Functional assignments were determined with the INTERPRO system (3), using the modules PROSITE, Pfam, PRINTS, ProDom, SMART, TIGRFAMs, and SIGNALP (5). These methods were implemented in the web-based platform HTGA (High Throughput Genome Annotation) (32) and were used for final annotation. tRNA genes were identified by the algorithm described at http://www.genetics.wustl.edu/eddy/tRNAscan-SE/ (24).
PCR amplification and sequence analysis.
Amplification of genetic markers within the 482-kb fragment was performed using standard procedures. The primers were purchased from MWG Biotech (Ebersberg, Germany). For direct sequencing the PCR products were purified (PCR purification kit; QIAGEN, Hilden, Germany) and sequenced using BigDye v3.1 (Applied Biosystems, Darmstadt, Germany) with an ABI 3700 capillary sequencer. Sequence data were analyzed with the Lasergene (DNAstar Inc., Madison, WI) and MacVector 7.0 (Oxford Molecular Ltd., Oxford, United Kingdom) programs. Other DNA manipulations were carried out by using standard methods (35).
Restriction fragment length polymorphism (RFLP) analysis by Southern hybridization.
Approximately 10 μg of chromosomal DNA was digested with EcoRI, EcoRV, MunI, SspI, and BamHI and electrophoresed in a 1.5% agarose gel. The DNA was blotted and hybridized by using standard procedures. Probes were labeled using an [α-33P]dATP HexaLabel DNA labeling kit (MBI Fermentas). Prehybridization was carried out in 20 ml of Church's phosphate buffer (0.25 M Na2HPO4 [pH 7.2], 1 mM EDTA, 1% sodium dodecyl sulfate) at 65°C for at least 1 h. Hybridization was performed in the same Church's buffer that was used for prehybridization containing the labeled probe at 60 to 65°C for at least 15 h. Washing was done twice at 65°C in wash buffer (0.1× SSC, 0.1% [wt/vol] sodium dodecyl sulfate) for 30 min (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). After the membranes were washed, they were exposed to a phosphor screen (Kodak storage phosphor screen; Molecular Dynamics, Krefeld, Germany) for 4 h to 1 day, and the hybridized signals were captured as image files by using a Typhoon 9400 scanner (Amersham Biosciences).
Electron microscopy.
Cells from concentrated suspensions were adsorbed onto 300-mesh carbon-coated copper grids (PLANO, Wetzlar, Germany) and rinsed twice with water. Samples were viewed and recorded without staining using an EM 10 transmission electron microscope (Zeiss, Germany) at an accelerating voltage of 70 kV.
Nucleotide sequence accession numbers.
The complete sequences of the 68-kb BAC clone and the 130-kb whole-genome shotgun assembly have been deposited in the GenBank, EMBL, and DDJB libraries under accession numbers BX571797 and AM085146, respectively.
RESULTS
A 482-kb genomic fragment harbors a conspicuous 130-kb region representing an MAI.
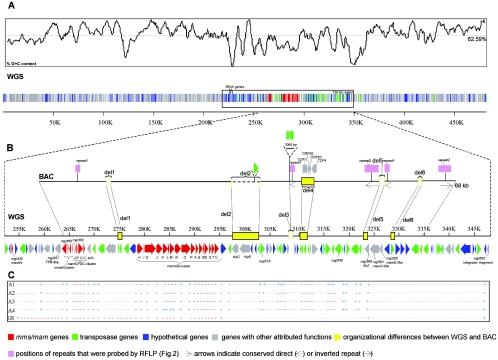
As previous work indicated that there is a large region resembling a genomic island which might extend beyond the previously analyzed 35-kb subsequence of the 68-kb BAC sequence region, we used two approaches to obtain extended and contiguous sequence information. First, a 482-kb contig was generated from the WGS assembly representing more than 10% of the whole genome (4.6 Mb) of M. gryphiswaldense. Second, sequence analysis of the BAC insert was completed, which generated an additional 33.6 kb of sequence information to the right of the previously analyzed 35-kb sequence. In addition to the known magnetosome genes, annotation of the 482-kb contig revealed numerous genes that encode diverse well-known metabolic functions that do not have any obvious relevance to magnetosome biomineralization, such as large operons for ribosome and flagellum synthesis, as well as operons for urea and phosphonate utilization and nitrate reduction. These genes are located predominantly in the left (approximately 1 to 220 kb) and right (350 to 482 kb) arms of the contig (Fig. 1A). These regions encompass a conspicuous 130-kb region (between approximately 220 and 350 kb), which has the following characteristics: (i) it harbors all previously identified magnetosome genes (mam and mms), (ii) it contains 42 of the 49 transposase-like genes present in the 482-kb region, and 23 of the transposase genes could be assigned to seven distinct groups of paralogs (Table 1), and (iii) most other ORFs (77 genes) are classified as hypothetical genes because of a lack of significant similarity to any known genes. Because of these characteristics, we suspected that the 130-kb region might encompass the entire magnetosome island and therefore focused our analysis on this region. The 130-kb region contains numerous direct and inverted repeats, most of which correspond to similar copies of transposase genes. Its G+C content (61.1%) is distinct from those of the 482-kb region (62.59%) and the WGS assembly (62.2%), and it has a more heterogeneous distribution (Fig. 1). For example, the mms6, mamGFDC, and mamAB operons have G+C contents of 63.8, 64.6, and 59.5%, respectively. Three tRNA genes are present in this region (tRNAAla,Ile,Met). An ORF (mgi605) encoding a putative phage-related integrase fragment is located at the right boundary of the 130-kb region, which exhibits 58% sequence similarity to phage integrases from the alphaproteobacteria Silicibacter sp. and Magnetospirillum magnetotacticum. Several other ORFs are also pseudogenes, including the idiA fragment (mgI469), which represents a remnant of a ferric iron transport system (27). Another notable feature of the region is the presence of several two-component systems and several ORFs with low levels of similarity to hemerythrin-like genes. We found several genes with potential relevance for magnetosome biomineralization, including mgI457 encoding a hypothetical TPR protein and mgI452 encoding an acidic pentapeptide repeat protein, as these motifs have been implicated in magnetosome formation and were identified in several MMPs (37). One ORF (mgI438) located about 11 kb to the left of mms6 encodes a protein containing the peptide sequence IAASPTASPIRK, which matches 100% a peptide sequence obtained by previous mass spectroscopic analysis of the magnetosome subproteome (18) but which could not been assigned to a known gene sequence. Therefore, we concluded that mgI438 encodes a magnetosome protein designated MamW, which is a 15.01-kDa protein with a basic pI (pI 12.8). Database searches failed to identify homologues, with the exception of a very similar hypothetical protein from M. magnetotacticum (ZP_00054421). Furthermore, we identified two additional ORFs in the putative mms6 operon (mgI460, 764 bp upstream of mms6; and mgI462, 722 bp downstream of mms6), which encode small basic proteins (for mgI460, a molecular weight of 8,756.55 and a pI of 12.3; for mgI462, a molecular weight of 9,729.75 and a pI of 11.8). Several of the newly identified ORFs outside the mam-mms cluster exhibited best hits to previously identified genes in the mamAB operon in BLAST searches. For example, the deduced gene product of mgI561, which is located 29.2 kb downstream of mamU, exhibits partial similarity (63%) to MamH belonging to the major facilitator superfamily. This gene is followed by mgI565, which encodes a protein with partial similarity to MamE (45%). Likewise, two additional ORFs (CDF3 and CDF4) encoding proteins with similarity to the magnetosome-associated putative iron transporters MamB and MamM are present in the 68-kb BAC sequence. However, these genes were not detected in the 482-kb WGS sequence derived from a different subculture of MSR-1, which prompted us to examine the differences between these two sequences more closely.
FIG. 1.
(A) Schematic representation of the 482-kb WGS genomic region. Magnetosome genes (mms and mam), hypothetical genes, tRNA genes, genes encoding other assigned functions, and transposase genes are indicated by different colors. The distribution of the G+C content is shown in the upper panel (the average value for the 482-kb region is 62.59%). The extent of the conspicuous 130-kb region described in the text is indicated by a box. (B) Major organizational differences between the BAC sequence and the homologous region in the WGS sequence (del1 to del6). Equivalent positions are connected by lines. The positions of selected features described in the text are indicated. (C) Identified deletions in different type A mutants (A1 to A4) and strain MSR-1B (1B). −, marker absent; +, marker present; dotted line, not tested. The positions refer to the WGS sequence shown in panel B.
TABLE 1.
Paralogous groups of IS element genes identified in the 130-kb MAIa
| Group | Paralogous ORFs |
|---|---|
| 1 | mgI398, mgI518, mgI546, mgI547, mgI556, mgI574 |
| 2 | mgI587, mgI400, mgI557, mgI575 |
| 3 | mgI428, mgI533, mgI535 |
| 4 | mgI441, mgI524, mgI578 |
| 5 | mgI436, mgI507, mgI593 |
| 6 | mgI474, mgI549 |
| 7 | mgI568, mgI602 |
Paralogous groups were formed from gene groups that had BLASTP hits to each other with e-values less than 10−6 and an alignment length of at least 60% of the deduced amino acid sequence. If the alignment length was not at least 60%, other parameters were taken into consideration with respect to the occurrence of fragmented genes (9).
Clones from different magnetic subcultures exhibit extensive sequence polymorphism in the magnetosome island.
Alignment of the 68-kb BAC sequence and the equivalent subsequence from the 482-kb WGS assembly, which were obtained from different subcultures of the magnetic archetype strain MSR-1A, revealed that the homologous regions were interrupted by insertions, deletions, or substitutions. Besides several minor conflicts, such as single nucleotide mismatches and short deletions, six major organizational differences between the two sequences (designated del1 to del6) were detected (Fig. 1B). del1 is a 1,049-bp insertion harboring two transposase genes in the WGS sequence, which are missing in the BAC sequence. In del2, a 196-bp transposase fragment in the BAC sequence is replaced by a 5,459-bp insertion in the WGS sequence, which contains a putative hemolysine transport operon that is bracketed by transposase genes. The excision site in the BAC sequence is flanked by two imperfect inverted repeats, which are present in four or more inverse or direct copies within both sequences. del3, a 1,066-bp fragment harboring two transposase genes in the BAC sequence, is deleted in the WGS sequence. del4, a 2,307-bp fragment harboring two CDF genes in the BAC sequence, is replaced by a 1,710-bp fragment harboring three transposase genes. del5, a 1,065-bp fragment harboring two transposase genes, is deleted in the BAC sequence. del 6, a 924-bp transposase fragment, is inserted into the WGS sequence but is not present in the BAC sequence. All organizational differences are associated with insertion elements or repeats or a combination of insertion elements and repeats. In two of the deletions (del2 and del6), a conserved sequence (CCGCCT) is present immediately to the left of the excision site, whereas the sequence CTAR is adjacent to the right excision site in four of the six deletions (Table 2).
TABLE 2.
Nucleotide sequences of regions flanking the excision sites found for the organizational differences between the BAC and WGS sequences and the excision site of the large deletion in strain MSR-1B
| Region | Sequencea |
|---|---|
| del1 | 274621-GGCAGG....CTAGAG-275711 |
| del2 | 297431-CCGCCT...TCAGCCC-302932 |
| del3 | 308996-AAATCA....TAAGCA-309034 |
| del4 | 310060-CAAAGG....CTAGAA-3110807 |
| del5 | 322778-GGGAAG....CTAACA-322816 |
| del6 | 328270-CCGCCT....CTAATA-329235 |
| MSR-1B | 254558-CTCGCA....CATGGA-294955 |
The nucleotide positions are the positions in the WGS sequence. Conserved residues are underlined.
Isolation of spontaneous magnetosome mutants under various conditions.
The considerable sequence polymorphisms observed for two different subcultures and the previously observed spontaneous mutability of the magnetic phenotype prompted us to investigate the induction, mechanism, and targets of the suspected genetic instability in more detail. Invariantly, the abundance of nonmagnetic clones was below the level of detection (<10−5 mutations per cell) in serially transferred cultures growing under standard conditions. In contrast, stationary cultures stored in the cold (4°C) for 3 to 5 weeks reproducibly gave rise to numerous white colonies. These colonies appeared at frequencies between about 2 × 10−2 under aerobic storage conditions (2 mM iron) and 10−3 under microaerobic storage conditions (1 μM iron). In order to mimic stress conditions presumably encountered during prolonged storage and to determine if a particular stress factor was responsible for the induction of mutants, we determined mutant frequencies in cells cultivated under various defined growth conditions. Growth at a high temperature (37°C), growth at a low temperature (12°C), freezing and thawing, or deprivation of a nutrient (carbon, nitrogen, or iron) did not result in significantly increased mutation rates compared to the rates observed after serial subcultivation. However, high iron concentrations (>500 μM) significantly increased the frequency of mutants (4 × 10−4). Also, exposure to increased oxygen concentrations, which were already inhibitory for growth and magnetosome formation, resulted in mutant frequencies of 6 × 10−4. Treatment of cells with H2O2 for 10 min had the most drastic effect and resulted in an increase in mutant frequencies up to nearly 10−2.
Spontaneous magnetosome mutants have various distinct phenotypes.
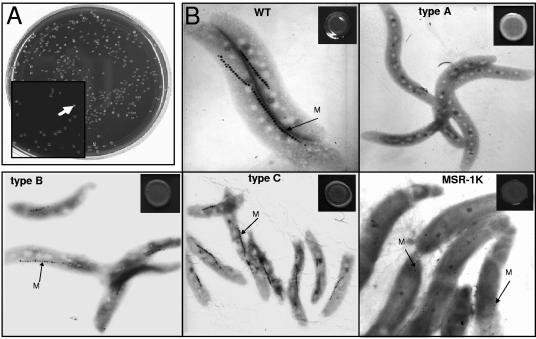
During the initial mutant screening, colonies with a whitish appearance compared with the dark brown wild-type clones were not differentiated further. However, upon closer inspection of approximately 500 restreaked colonies from various experiments, four mutant classes could be distinguished (Fig. 2). Type A accounted for 36% of the mutants, and the colonies were bright white. The cells failed to align in magnetic fields, which was due to the total lack of magnetosome particles. Type B mutants (61% of the mutants) formed darker gray colonies, and the cells exhibited a magnetic response that was detectable but was weaker (0.1 to 1.6) than that of the wild type (1.8 to 2.0), as detected by differential light-scattering measurements (40). This was consistent with the reduced number of magnetosomes (18 to 29 particles per cell) that were aligned in shorter and less regular chains (Fig. 2). The color of colonies of type C mutants was only slightly lighter than the color of colonies of the wild type, but the colonies had a heterogeneous “fried-egg” appearance, with a convex peripheral region surrounding a flat central region. The cells were otherwise indistinguishable from the wild-type cells in terms of the number and appearance of magnetosomes, as determined by electron microscopy. Type C mutants accounted for less than 3% of the clones. A further mutant-type phenotype was found only once in a clone (MSR-1K) from a culture after storage at 4°C. MSR-1K formed light brown colonies, and the cells contained fewer magnetosomes, which had a less regular chain-like appearance. Unlike type B mutants, the particles also were smaller (diameter, 15 to 25 nm). All mutants exhibited very similar growth characteristics under high- and low-iron conditions (data not shown).
FIG. 2.
(A) Appearance of nonmagnetic mutants (white colonies) (arrow) among magnetic clones of the wild type (dark brown colonies). (B) Phenotypes (electron micrographs and colony morphology) of wild-type M. gryphiswaldense (WT) and type A, type B, type C, and MSR-1K mutants. M, magnetosome chain.
Genotypic analysis of mutants.
As the region flanking the genes had a particularly high number of repeated IS elements belonging to the IS21 and IS66 families, we hypothesized that these elements were involved in the instability of this genomic region. We therefore performed a Southern (RFLP hybridization) analysis to determine the copy numbers of these IS elements in the genomes of mutants in which magnetosome synthesis was affected (Fig. 3). Probes were derived from selected ORFs corresponding to an IS21 (mgI549) element and an IS66 (mgI518) element (Fig. 1B). Hybridizations revealed the presence of three DNA fragments that exhibited similarity with IS21 and 11 fragments that exhibited similarity with IS66 in the archetype strain (MSR-1A). Unexpectedly, MSR-1L, which had been used as the parental strain for the isolation of mutants, exhibited only two IS21-hybridizing fragments and five IS66-hybridizing fragments. While all representatives of the type B mutants tested and strain MSR-1K produced hybridization patterns indistinguishable from that of parental strain MSR-1L, all type A mutants, including strain MSR-1B, displayed reduced copy numbers of IS elements, with one IS21-hybridizing band and three to five IS66-hybridizing bands. Type A mutants not only differed in terms of copy numbers of IS elements but also displayed RFLP, indicating that there were insertions or deletions in adjacent genomic regions. Many reports have shown that repeated elements can often be recombinogenic, facilitating the formation of deletions (1, 14). We therefore investigated the genomic distribution of an IS21-related repeat (repeat1) and an IS66-related repeat (repeat2) that are present in the MAI at levels of multiple copies (Fig. 3; for localization of repeats see Fig. 1). While MSR-1A yielded 10 repeat1-hybridizing fragments, there was one fragment in MSR-1L and type B mutants and there was no fragment in type A mutants. Compared to the four repeat2-hybridizing repeats of the archetype strain (MSR-1A), three fragments were detected in strain MSR-1L and type A mutants; however in the A3 and A4 mutants hybridizing fragments that were different lengths were observed after EcoRI digestion. Three of seventeen isolated type B mutants lacked a 2.3-kb fragment hybridizing with the repeat2 probe.
FIG. 3.
RFLP analysis of EcoRI, EcoRV, and MunI enzyme-digested genomic DNA from various mutants. Restriction fragments were blotted and hybridized with [α-33P]ATP-labeled IS66, IS21, repeat1, and repeat2 probes. Lane A, MSR-1A; lane L, MSR-1L; lane K, MSR-1K; lane B, MSR-1B; lanes A1, A2, A3, and A4, type A mutants; lanes B1, B2, B3, and B4, type B mutants.
We found an increase in the copy number, which would have been indicative of recent transposition events, in none of the nonmagnetic strains isolated during mutant screening. While nonmagnetic mutants had the lowest numbers of IS elements, loss of multiple copies apparently resulted in no detectable phenotype for the magnetic parental strain (MSR-1L). Therefore, we asked whether the observed polymorphism was specifically associated with the loss of mam genes observed in isolated mutants or whether consecutive loss of copies occurred independently of the coincident loss of mam genes. If deletion of IS elements was a general consequence of high genetic instability of the entire genome, then IS element polymorphism with comparable frequency should have been detectable also in magnetic clones, as additional copies of IS elements were also detected outside the magnetosome island. To answer this question, 10 magnetic clones were arbitrarily selected from independent mutant screening experiments and analyzed as described above. All clones tested produced RFLP patterns indistinguishable from that of parental strain MSR-1L, suggesting that loss of IS elements was a relatively rare event in magnetic clones, while nonmagnetic mutants always exhibited loss of IS elements.
All mutants were tested by PCR for the presence of genetic markers located in two of the known magnetosome operons (mamC, mamF, mamB, mamE). As in the wild type, all markers tested could be amplified from the type B, type C, and MSR-1K mutants. In contrast, all type A mutants were negative for all of these markers, indicating that as in strain MSR-1B, genomic deletions in the magnetosome operons had occurred. To further characterize the extent of the deletions, we examined the presence of additional markers in the mam and mms operons, as well as in adjacent and remote regions of the magnetosome island, in selected type A mutants by hybridization and PCR. If they were present, the PCR products of various markers were shorter than the expected sizes, indicating that the regions adjacent to the IS element were also affected by partial deletions. The results of these experiments indicated that there were diverse mosaic-like deletion patterns in the different mutants, all of which were confined to an approximately 64-kb section in the 130-kb region (Fig. 1C). Mutant A1 had at least four different deletions. The entire mamGFDC operon and the idiA fragment were deleted. In addition, two large parts of the mamAB operon (mamHIEJKLMN and mamAQRBSTU) were absent, whereas the mamO and mamP genes were present. In addition, a copy of IS21 (mgI554) located 24.4 kb to the right of mamU was missing due to a short deletion. Mutant A2 had at least four different deletions. One large deletion seemed to encompass the entire mms6, mamGFDC, and mamAB operons. Three shorter deletions were present further to the right. Mutant A3 had at least three different deletions. Again, one large deletion seemed to comprise the entire mms6, mamGFDC, and mamAB operons, and two shorter deletions were present further to the right. At least five different deletions were detected in A4, which resulted in the loss of the entire mms6 and mamGFDC operons and two fractions of the mamAB operon, while the mamKLMNOP genes were still present. Three shorter deletions were present to the right of the mamAB operon. In MSR-1B, a single large 40.385-kb deletion extended from nucleotide 254564 to nucleotide 294949 (Table 2).
DISCUSSION
In this paper, we present further evidence that there is a genomic MAI in M. gryphiswaldense. The 482-kb sequence analyzed in this work is the most extended contiguous genomic sequence from a magnetotactic bacterium and is likely to comprise all gene functions involved in magnetosome formation. In addition to previously identified structural genes, we identified a gene (mamW) encoding a magnetosome membrane protein in this region and found two new genes in the mms6 operon. The conspicuous 130-kb region is further characterized by the presence of numerous genes having unknown functions. Several of these genes exhibit similarity to previously identified mam genes (mgI561 and mgI565), are colocalized in an operon along with other magnetosome proteins (mgI460, mgI462), or contain conspicuous sequence motifs (mgI457, mgI452) and thus might be candidates for involvement in magnetosome formation. The roles of other hypothetical gene products encoded in this region remain cryptic. The observed loss of genetic material during serial subculturing, such as the loss of two mamB-like CDF genes, was not always accompanied by a nonmagnetic phenotype, which indicates that the putative MAI contains sections that are not essential for magnetosome formation. The presence of many pseudogenes suggests that parts of the region might simply represent a “junkyard” of genes derived from previous rearrangements or horizontal gene transfer events. The occurrence of multiple representatives of magnetosome-specific gene families also seems to indicate that there is some redundancy of genetic determination of magnetosome formation.
Typically, genomic islands are integrated into or near tRNA genes (30). We found three tRNA genes close to the left terminus of the 130-kb region, but it is not clear if these genes were involved in the insertion of the region. The G+C content of the 130-kb region is heterogeneous, suggesting that the MAI may have a mosaic structure. Our sequence and experimental analyses of the MAI revealed that this region is unusually prone to insertion and excision events. Hypervariable regions of other bacteria, such as Salmonella enterica and Escherichia coli (7), are characterized by the presence of phage-related genes, such as genes encoding integrases, and IS elements, which has led to the assumption that there are genomic hot spots for insertion and excision of mobile elements. Likewise, a striking characteristic of the region in M. gryphiswaldense analyzed is the great abundance of multiple copies of transposase genes belonging to different families of IS elements. Between position 220000 and position 350000 IS elements account for 22.7% of the coding region, suggesting that these mobile genetic elements played a major role in driving the variability of the putative MAI. The detected organizational differences between the different magnetic clones are consistent with recent activity of insertion elements, and all deletions in nonmagnetic mutants are associated with IS elements. These data show that transposition and subsequent deletions are dominant mechanisms of mutations in the magnetosome island of M. gryphiswaldense and that IS elements are responsible for the genetic instability and plasticity observed in this region. Insertion elements are frequently also involved in the mobilization of intervening genomic regions, which results in composite transposons (25). Thus, the numerous IS elements might promote the mobilization and acquisition of the magnetosome island. Together, the presence of IS elements, the presence of phage-associated genes, the presence of an integrase remnant, and the presence of tRNAs in and at the extremities of this region, as well as the distinct G+C content, suggest that the region might have been acquired via horizontal gene transfer from different taxa, potentially by a phage-mediated horizontal gene transfer event. Although comparable data are not yet available for other MTB, a preliminary analysis of the genome assembly of M. magnetotacticum MS-1 and the magnetotactic coccus MC-1 (available at http://www.jgi.doe.gov/) revealed similar characteristics of the homologous regions. Interestingly, the synteny within the homologous regions is not strictly conserved in related MTB, but genetic elements appear to be shuffled in these regions, probably due to transposition-mediated reorganization (data not shown). Further research could reveal whether this region can in fact be mobilized between different strains of bacteria.
Genomic islands often tend to be deleted en bloc owing to the presence of functional integrase genes, which are involved in site-specific insertion and excision of the DNA region via recombination between specific sequences situated on either side of the element (11, 30). In some genomic islands the integrase gene may be deleted or nonfunctional, resulting in permanently anchored islands (4, 23, 30). The integrase gene fragment present at the right boundary of the 130-kb region is unlikely to be functional. Consistently, we did not observe spontaneous precise excision of the MAI at predetermined unique sites; rather, multiple consecutive deletions occurred at various different sites, leading to a mosaic-like structure in type A mutants. This indicates that there is a different, integrase-independent mechanism of deletion. Faure et al. (14) described genomic arrangements that arose in long-term cultures of E. coli K-12. In the stationary phase, large-scale deletions spanning many kilobases of the genome arose. Some cultures were observed to be polymorphic and to contain up to three different types of deletions. Most deletions were bordered, either precisely or within a 5-kb range, by sequences associated with IS elements. This led to the assumption that there is a two-step process for generation of deletions in which the insertion of a new IS element copy near a preexisting IS element is followed by homologous recombination between the two copies. As all deletions that we observed in the magnetosome island were associated with IS elements, a similar mechanism for deletion can be assumed for the MAI.
Although a genetic system has been established recently for M. gryphiswaldense (41-43), genetic analysis by directed mutagenesis has remained laborious and time-consuming. Thus, the analysis of a diverse set of spontaneous magnetosome mutants provides a powerful tool for genetic dissection of biomineralization, particularly for determination and mapping of the set of essential genes required for magnetosome formation. Mutants isolated in this study are heterogeneous with respect to their pheno- and genotypes. In addition to nonmagnetic type A mutants, mutants that were aberrant sizes and mutants that had different numbers of magnetosomes (type B, MSR-1K) or a different colony morphology (type C) were identified. While the genotypes of the latter mutants could not be determined clearly, mutations in all type A mutants, including strain MSR-1B, exhibited deletions in 64 kb of the MAI, which in all cases affected the mms and mam operons. This further substantiates the assumption that this set of genes is crucial for magnetosome synthesis.
Spontaneous loss of the ability to synthesize magnetosomes seems to be a characteristic trait common to diverse MTB and has been reported for different species (8, 12). For instance, spontaneous nonmagnetic mutants were regularly observed in marine magnetotactic vibrio MV-1. All mutants failed to express a major copper-containing periplasmic protein (ChpA) presumably involved in iron uptake (12). Although the mutants were genetically heterogeneous, all of the mutants had two point mutations (transversions) at identical wobble positions in chpA, which apparently prevented translation of the transcript by an unknown mechanism. This finding suggests that there is no universal explanation for the observed genetic instability of the magnetic phenotype and that different mechanisms are responsible for the appearance of spontaneous nonmagnetic mutants in different MTB.
IS elements have been shown to be ubiquitously distributed in bacterial genomes (25), and abnormally high mutation rates in many other bacteria are also often due to transposition events. There have been a number of reports which have described increases in the transposition frequency under specific growth conditions. Prolonged nutritional deprivation led to increased levels of transposition of a range of IS elements in stab cultures of E. coli K-12 (29). Spontaneous gas vesicle mutants caused by transposition appeared frequently in cultures of several cyanobacteria and archaea after prolonged maintenance of cells, particularly after physiological stress, such as cryopreservation (28). A transposition burst of the Halobacterium halobium element ISH27 was observed after storage of the cells at 4°C (31). It has also been demonstrated that large numbers of mutants caused by IS element transposition can accumulate in stationary-phase cultures of Xanthomonas oryzae, and it has been suggested that IS elements play prominent roles in the adaptation of bacteria to life in the stationary phase (33). We also observed that magnetosome mutants were abundant in stationary cultures during prolonged storage in the cold, while their levels were was below the level of detection under standard growth conditions. However, conditions encountered during cold storage are rather undefined and likely involve complex environmental changes, such as nutrient deprivation and oxidative stress due to decreased respiratory activity of the cells. Of the potential stress factors tested, only increased oxygenation and high iron concentrations led to significantly increased mutation frequencies. Like oxygenation, excess iron also may result in oxidative stress and is known to increase the vulnerability of cells to damage by H2O2 because of Fenton chemistry (21). Likewise, direct exposure to the strong oxidant H2O2 was most effective for induction of nonmagnetic mutants. In Mycobacterium smegmatis transposition was stimulated by exposure to microaerobic conditions or a subsequent oxygen shock (16). Induction of hypertranspositional activity by the presence of diverse inhibitors, including cyanide and hydrogen peroxide, was observed in Corynebacterium glutamicum (15). Hydrogen peroxide is known to be a potent inducer of the SOS response which enhances recombinational activity. As a consequence, phage induction, mutagenesis, and filamentation are induced at sublethal H2O2 concentrations in E. coli (22). Hence, the existence of a similar mechanism could also explain the observed effect of oxidative stress in M. gryphiswaldense.
Results of this study demonstrated that parts of the genome of M. gryphiswaldense may undergo rapid rearrangement upon subculture in the laboratory. In fact, we observed several other indications of microevolution or “domestication,” such as increased oxygen tolerance of the lab strain compared to the archetype, which might have been caused by accidental selection of mutants during repeated subculture, eventually leading to improved adaptation to growth under prolonged laboratory conditions. Besides practical implications, such as the need for proper maintenance and regular selection to prevent accumulation of mutants, the question of whether the induced mutability of the magnetic phenotype might reflect adaptations to stress conditions under environmental conditions arises. We were unable to detect a discernible growth advantage for the nonmagnetic mutants compared to the magnetic parental strain under standard conditions when growth rates were used. Yet stationary cultures were repeatedly found to be completely taken over by nonmagnetic mutants of a single type, which displaced the parental strain. The ubiquity of such deletions in independent cultures and their increase in frequency under physiological stress conditions support the hypothesis that they have a selective advantage. A selective advantage was also reported for large chromosomal deletions in E. coli by Faure et al. (14). Likewise, Zambrano et al. (44) and Zinser et al. (45) have shown that mutants carrying loss-of-function alleles predominate in stationary-phase cultures of E. coli (44, 45). Further experimentation is required to determine if a selective advantage for survival during the stationary phase and under oxidative stress conditions is conferred by loss of the magnetic phenotype. Alternatively, the frequent loss of large parts of the MAI may simply reflect the energy costs of producing a large multiprotein organelle under conditions in which it provides no selective advantage.
Acknowledgments
This study was supported by the BMBF Biofuture Program and the Max Planck Society.
We thank Ekaterina Schmidt (MPI Bremen) and Ines Müller (MPI Berlin) for excellent technical assistance.
REFERENCES
- 1.Albertini, A. M., M. Hofer, M. P. Calos, and J. H. Miller. 1982. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell 29:319-328. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apweiler, R., T. K. Attwood, A. Bairoch, A. Bateman, E. Birney, M. Biswas, P. Bucher, L. Cerutti, F. Corpet, M. D. Croning, R. Durbin, L. Falquet, W. Fleischmann, J. Gouzy, H. Hermjakob, N. Hulo, I. Jonassen, D. Kahn, A. Kanapin, Y. Karavidopoulou, R. Lopez, B. Marx, N. J. Mulder, T. M. Oinn, M. Pagni, and F. Servant. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29:37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bach, S., C. Buchrieser, M. Prentice, A. Guiyoule, T. Msadek, and E. Carniel. 1999. The high-pathogenicity island of Yersinia enterocolitica Ye8081 undergoes low-frequency deletion but not precise excision, suggesting recent stabilization in the genome. Infect. Immun. 67:5091-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baxevanis, A. D. 2003. The Molecular Biology Database Collection: 2003 update. Nucleic Acids Res. 31:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bazylinski, D. A., and R. B. Frankel. 2004. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2:217-230. [DOI] [PubMed] [Google Scholar]
- 7.Bishop, A. L., S. Baker, S. Jenks, M. Fookes, P. O. Gaora, D. Pickard, M. Anjum, J. Farrar, T. T. Hien, A. Ivens, and G. Dougan. 2005. Analysis of the hypervariable region of the Salmonella enterica genome associated with tRNAleuX. J. Bacteriol. 187:2469-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blakemore, R., D. Maratea, and R. S. Wolfe. 1979. Isolation and pure culture of a freshwater magnetic spirillum in chemically defined medium. J. Bacteriol. 140:720-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien, M. C., I. Morozova, S. Shi, H. Sheng, J. Chen, S. M. Gomez, G. Asamani, K. Hill, J. Nuara, M. Feder, J. Rineer, J. J. Greenberg, V. Steshenko, S. H. Park, B. Zhao, E. Teplitskaya, J. R. Edwards, S. Pampou, A. Georghiou, I. C. Chou, W. Iannuccilli, M. E. Ulz, D. H. Kim, A. Geringer-Sameth, C. Goldsberry, P. Morozov, S. G. Fischer, G. Segal, X. Qu, A. Rzhetsky, P. Zhang, E. Cayanis, P. J. De Jong, J. Ju, S. Kalachikov, H. A. Shuman, and J. J. Russo. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966-1968. [DOI] [PubMed] [Google Scholar]
- 10.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 12.Dubbels, B. L., A. A. DiSpirito, J. D. Morton, J. D. Semrau, J. N. Neto, and D. A. Bazylinski. 2004. Evidence for a copper-dependent iron transport system in the marine, magnetotactic bacterium strain MV-1. Microbiology 150:2931-2945. [DOI] [PubMed] [Google Scholar]
- 13.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 14.Faure, D., R. Frederick, D. Wloch, P. Portier, M. Blot, and J. Adams. 2004. Genomic changes arising in long-term stab cultures of Escherichia coli. J. Bacteriol. 186:6437-6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garbe, T. R., N. Suzuki, M. Inui, and H. Yukawa. 2004. Inhibitor-associated transposition events in Corynebacterium glutamicum. Mol. Genet. Genomics 271:729-741. [DOI] [PubMed] [Google Scholar]
- 16.Ghanekar, K., A. McBride, O. Dellagostin, S. Thorne, R. Mooney, and J. McFadden. 1999. Stimulation of transposition of the Mycobacterium tuberculosis insertion sequence IS6110 by exposure to a microaerobic environment. Mol. Microbiol. 33:982-993. [DOI] [PubMed] [Google Scholar]
- 17.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 18.Grünberg, K., E. C. Müller, A. Otto, R. Reszka, D. Linder, M. Kube, R. Reinhardt, and D. Schüler. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 70:1040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grünberg, K., C. Wawer, B. M. Tebo, and D. Schüler. 2001. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 67:4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heyen, U., and D. Schüler. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 61:536-544. [DOI] [PubMed] [Google Scholar]
- 21.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 22.Imlay, J. A., and S. Linn. 1987. Mutagenesis and stress response induced in Escherichia coli by hydrogen peroxide. J. Bacteriol. 169:2967-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lesic, B., S. Bach, J. M. Ghigo, U. Dobrindt, J. Hacker, and E. Carniel. 2004. Excision of the high-pathogenicity island of Yersinia pseudotuberculosis requires the combined actions of its cognate integrase and Hef, a new recombination directionality factor. Mol. Microbiol. 52:1337-1348. [DOI] [PubMed] [Google Scholar]
- 24.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maringanti, S., and J. A. Imlay. 1999. An intracellular iron chelator pleiotropically suppresses enzymatic and growth defects of superoxide dismutase-deficient Escherichia coli. J. Bacteriol. 181:3792-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michel, K. P., and E. K. Pistorius. 2004. Adaptation of the photosynthetic electron transport chain in cyanobacteria to iron deficiency: the function of IdiA and IsiA. Physiol. Plant. 120:36-50. [DOI] [PubMed] [Google Scholar]
- 28.Mlouka, A., K. Comte, A.-M. Castets, C. Bouchier, and N. T. de Marsac. 2004. The gas vesicle gene cluster from Microcystis aeruginosa and DNA rearrangements that lead to loss of cell buoyancy. J. Bacteriol. 186:2355-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naas, T., M. Blot, W. M. Fitch, and W. Arber. 1994. Insertion sequence-related genetic variation in resting Escherichia coli K-12. Genetics 136:721-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osborn, A. M., and D. Böltner. 2002. When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid. 48:202-212. [DOI] [PubMed] [Google Scholar]
- 31.Pfeifer, F., and U. Blaseio. 1990. Transposition burst of the ISH27 insertion element family in Halobacterium halobium. Nucleic Acids Res. 18:6921-6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabus, R., M. Kube, J. Heider, A. Beck, K. Heitmann, F. Widdel, and R. Reinhardt. 2005. The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch. Microbiol. 183:27-36. [DOI] [PubMed] [Google Scholar]
- 33.Rajeshwari, R., and R. V. Sonti. 2000. Stationary-phase variation due to transposition of novel insertion elements in Xanthomonas oryzae pv. oryzae. J. Bacteriol. 182:4797-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M.-A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schübbe, S., M. Kube, A. Scheffel, C. Wawer, U. Heyen, A. Meyerdierks, M. Madkour, F. Mayer, R. Reinhardt, and D. Schüler. 2003. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J. Bacteriol. 185:5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schüler, D. 2004. Molecular analysis of a subcellular compartment: the magnetosome membrane in Magnetospirillum gryphiswaldense. Arch. Microbiol. 181:1-7. [DOI] [PubMed] [Google Scholar]
- 38.Schüler, D., and E. Baeuerlein. 1998. Dynamics of iron uptake and Fe3O4 biomineralization during aerobic and microaerobic growth of Magnetospirillum gryphiswaldense. J. Bacteriol. 180:159-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schüler, D., and E. Baeuerlein. 1996. Iron-limited growth and kinetics of iron uptake in Magnetospirillum gryphiswaldense. Arch. Microbiol. 166:301-307. [DOI] [PubMed] [Google Scholar]
- 40.Schüler, D., R. Uhl, and E. Baeuerlein. 1995. A simple light-scattering method to assay magnetism in Magnetospirillum gryphiswaldense. FEMS Microbiol. Lett. 132:139-145. [Google Scholar]
- 41.Schultheiss, D., R. Handrick, D. Jendrossek, M. Hanzlik, and D. Schüler. 2005. The presumptive magnetosome protein Mms16 is a poly(3-hydroxybutyrate) granule-bound protein (phasin) in Magnetospirillum gryphiswaldense. J. Bacteriol. 187:2416-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultheiss, D., M. Kube, and D. Schüler. 2004. Inactivation of the flagellin gene flaA in Magnetospirillum gryphiswaldense results in nonmagnetotactic mutants lacking flagellar filaments. Appl. Environ. Microbiol. 70:3624-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schultheiss, D., and D. Schüler. 2003. Development of a genetic system for Magnetospirillum gryphiswaldense. Arch. Microbiol. 179:89-94. [DOI] [PubMed] [Google Scholar]
- 44.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757-1760. [DOI] [PubMed] [Google Scholar]
- 45.Zinser, E. R., D. Schneider, M. Blot, and R. Kolter. 2003. Bacterial evolution through the selective loss of beneficial genes. Trade-offs in expression involving two loci. Genetics 164:1271-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]