Abstract
Listeria monocytogenes strains belonging to phylogenetic lineage II (serotypes 1/2a, 1/2c, and 3a) carry a lineage-specific genome segment encoding a putative sigma subunit of RNA polymerase (lmo0423, herein referred to as sigC), a gene of unknown function (lmo0422) similar to the padR family of regulators, and a gene that is similar to the rodA-ftsW family of cell wall morphology genes (lmo0421). To understand the function of this set of genes, their expression patterns and the effects of null mutations in the lineage II L. monocytogenes strain 10403S were examined. The data are consistent with the three genes comprising an operon (the sigC operon) that is highly induced by temperature upshift. The operon is transcribed from three different promoters, the proximal of which (P1) depends upon sigC itself. Null mutations in sigC or lmo0422 increase the death rate at lethal temperatures and cause loss of thermal adaptive response, whereas the lmo0421 mutation causes only a loss of the adaptive response component. Only the sigC mutation affects transcription from the P1 promoter, whereas ectopic expression of lmo0422 from the PSPAC promoter complements the individual lmo0422 and sigC null mutations, showing that lmo0422 is the actual thermal resistance regulator or effector while sigC provides a mechanism for temperature-dependent transcription of lmo0422 from P1. Our genetic and phylogenetic analyses are consistent with lmo0422—renamed lstR (for lineage-specific thermal regulator)—and sigC comprising a system of thermal resistance that was ancestral to the genus Listeria and was subsequently lost during divergence of the lineage I L. monocytogenes population.
Although only a relatively small number of cases of human listeriosis occur each year in the United States (estimated at 2,500 per year), the associated rates of morbidity and mortality are substantial, with nearly 30% mortality in some outbreaks (40, 58). Because the causative agent of the disease, Listeria monocytogenes, is ubiquitous in nature and possesses durable physiological characteristics, the organism is one of the most significant food safety problems in the food production industry.
One of the best characterized physiological attributes of L. monocytogenes is its ability to grow at refrigeration temperatures, in conditions of high osmolarity, and at low pH. Several different stress adaptation systems have been defined for L. monocytogenes that contribute to growth characteristics under these conditions (2, 13, 33, 63). These characteristics facilitate contamination of foods and subsequent transmission to humans and have also been shown to contribute to virulence (18, 34, 42, 53, 62).
The role of the general stress regulator sigma B in facilitating growth under stress conditions and in facilitating virulence has recently been of interest with L. monocytogenes (2, 3, 11, 34, 41, 56, 63). In addition to sigma B, there are four other putative alternative sigma subunits in the L. monocytogenes EGDe genome sequence (21). So far, only one of these, the rpoN gene encoding sigma 54, has been studied (1), and its specific physiological role remains unclear.
Based on phylogenetic analysis of genome composition, we have recently shown that a previously unknown sigma factor-like gene, lmo0423 (herein referred to as sigC), is part of a three gene region that is carried only by L. monocytogenes strains comprising phylogenetic lineage II of the species (65). Lineage II includes serotype 1/2a, one of the most commonly found in foods (43, 47, 48, 59), and it is possible that the sigC region contributes to physiological characteristics important to survival in the food production environment. Analysis of compositional bias in sigC and the adjacent lmo0422-lmo0421 region is consistent with the region being ancestral to the species and subsequently lost during the divergence of phylogenetic lineage I (65). Indeed, orthologous genes are also found in the Listeria innocua genome at the same relative position, further underscoring the conclusion that it is ancestral to the genus.
Sequence alignments of the sigC-lmo0421 region indicate that sigC encodes a putative member of the extracytoplasmic function (ECF) family of sigma subunits that typically modulate regulons, responding to extracytoplasmic stress and/or mediating extracytoplasmic functions (25). Lmo0422 shows significant similarity to the PadR family of transcription regulators (23), while Lmo0421 encodes a member of the RodA/FtsW family of proteins, which modulate peptidoglycan biosynthesis during the elongation (RodA) and septation (FtsW) phases of cell division (4, 5, 8, 10, 16, 27, 46, 64).
To understand the characteristics conferred upon lineage II strains by the unique sigC-lmo0421 region, we examined the expression and function of these genes in the lineage II serotype 1/2a strain 10403S. Our data are consistent with the three genes comprising an operon that is induced in response to different types of environmental stress, including thermal stress and antibiotics. The operon is transcribed from three different promoters, with the primary heat-inducible promoter depending upon sigC itself. Mutations in sigC and lmo0422 genes both cause significant sensitivity to high temperature, but only the sigC deletion effects expression of the operon. When lmo0422 expression is placed under control of the PSPAC promoter, it complements both the Δlmo0422 and the ΔsigC deletions in an IPTG (isopropyl-β-d-thiogalactopyranoside)-dependent manner. Therefore, lmo0422 appears to be a lineage-specific regulator or effector of thermal resistance, while sigC comprises a mechanism for thermally regulated expression of lmo0422.
MATERIALS AND METHODS
Strains and plasmids.
The bacterial strains used and generated in this study are listed in Table 1. Listeria monocytogenes strain 10403S was the parental strain for all of the studies described (7). L. monocytogenes strains were propagated in brain heart infusion (BHI) broth at 37°C unless otherwise specified. Ampicillin was added to cultures of Eschierchia coli strains at 80 μg/ml. Antibiotic selection in L. monocytogenes was achieved with 10-μg/ml chloramphenicol, 50-μg/ml kanamycin, or 2-μg/ml erythromycin. The shuttle vector plasmid pKSV7 was used for allele replacements in L. monocytogenes (54). Plasmid pPL2 (38) was obtained from R. Calendar and used for site-specific integration of the lmo0422 gene. The PSPAC cassette was derived from pDEH21, a precursor of the pLIV-1 plasmid (14), which was obtained from N. Freitag.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and features | Reference or source |
|---|---|---|
| Strains | ||
| SM10 | E. coli conjugation donor; F−thi-1 thr-1 leuB6 recA tonA21 lacY1 supE44 (Muc+) λ−[RP4-2(Tc::Mu)] Kmr Tra+ | 52 |
| RM1602 | E. coli (dam recA negative) | W. Haldenwang |
| TOPO10 | E. coli host strain F−mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZ ΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (Strr)endA1 nupG | Invitrogen |
| 10403S | L. monocytogenes wild type | 7; N. Freitag |
| ΔsigC | L. monocytogenes ΔsigC derivative of 10403S | This study |
| Δlmo0422 | L. monocytogenes Δlmo0422 derivative of 10403S | This study |
| Δlmo0421 | L. monocytogenes Δlmo0421 derivative of 10403S | This study |
| ΔsigC/PSPAC-422E | pPD422E integrated in strain ΔsigC | This study |
| Δlmo0422/PSPAC-422E | pPD422E integrated in strain Δlmo0422 | This study |
| Plasmids | ||
| pDEH21 | PSPAC vector | N. Freitag |
| pPD422E | PSPAC-controlled lmo0422 | This study |
| pPL2 | Site-specific integration vector | 38 |
| pSKV7 | Temperature-sensitive integration vector | 54 |
| pKSV7Δlmo0421 | Δlmo0421 allele in the pKSV7 vector | This study |
| pKSV7Δlmo0422 | Δlmo0422 allele in the pKSV7 vector | This study |
| pKSV7ΔsigC | ΔsigC allele in the pKSV7 vector | This study |
| pCR4-TOPO | Cloning vector | Invitrogen |
Generation of the ΔsigC, Δlmo0422, and Δlmo0421 mutants.
In-frame mutations were generated in the lmo0421, lmo0422, and sigC genes by using splicing by overlap extension (SOEing). The ΔsigC allele was generated with SOEing PCR primers sigCA1 and sigCB (Table 2), which amplify a 570-bp fragment comprising the 5′ end of sigC, and primers sigCBC and sigCD, which amplify a 782-bp fragment comprising the 3′ end of sigC. Recombinant PCR using the two fragments as templates and the sigCA1 and sigCD primers produced a 224-bp in-frame deletion of sigC extending from +753 to +977. In-frame deletions in the lmo0422 and lmo0421 genes were also constructed by a similar approach. The SOEing primers are summarized in Table 2. The Δlmo0422 allele has a 219-bp in-frame deletion and Δlmo0421 mutant has 963-bp in-frame deletion. The deletion junctions for all three alleles were confirmed by sequencing the SOEing PCR products cloned in plasmid pCR4-TOPO (Invitrogen, Carlsbad, CA). Once confirmed, the ΔsigC allele was excised as a BamHI-PstI fragment, and the Δlmo0422 and Δlmo0421 alleles were excised as EcoRI-HindIII fragments from the respective pCR4-TOPO vectors and cloned into the temperature-sensitive pKSV-7 shuttle vector (54) to generate plasmids pKSV7Δ421, pKSV7Δ422, and pKSV7ΔsigC.
TABLE 2.
Primers used in this study
| Probe | Sequence |
|---|---|
| Northern blot and S1 | |
| Lm423F1 | 5′-CAACTTCCCTGTACTTTACGGG-3′ |
| Lm423R1 | 5′-AGAATCGAACGACAGCGG-3′ |
| I421F | 5′-GCCATTAAAGTAGCACTGGG-3′ |
| I421R | 5′-CGAGTAGTGCAGCGTAAAC-3′ |
| sigCB | 5′-GCGTTAAATCTTCCGCTGTCG-3′ |
| SOEing primers | |
| sigCA1a,b | 5′-cgtCTGCAG-TGGTCAATCGCCAATTTAGGC-3′ |
| sigCB | 5′-GCGTTAAATCTTCCGCTGTCG-3′ |
| sigCBC | 5′-CGACAGCGGAAGATTTAACGCGCGCTGTTATGGATTTACCCG-3′ |
| SigC3Da,c | 5′-ggtGGATCCTCCCATTTCCTGCATCGC-3′ |
| Lm422Aa,d | 5′-gggAAGCTT-AAAGAATCGAACGACAGCGG-3′ |
| Lm422B | 5′-TTTGCTTCGTCTTGCAGTGC-3′ |
| Lm422BC | 5′-GCACTGCAAGACGAAGCAAAGACTCTAAGCAACACGGATCACTG-3′ |
| Lm422Da,e | 5′-tgcGAATTCTAGGATCAAGGCAGCCGC-3′ |
| Lm421Aa,d | 5′-gggAAGCTTACTGCAAGACGAAGCAAAGC-3′ |
| Lm421B | 5′-CGCAATAAGGAGCCAATCC-3′ |
| Lm421BC | 5′-GGATTGGCTCCTTATTGCGTTTACGCTGCACTACTCGGTC-3′ |
| Lm421Da,e | 5′-tgtGAATTCTTTATTGTTGCGAGCGGC-3′ |
| PSPAC-lmo0422 construct | |
| Lm422Ex1a,f | 5′-tggTCTAGA-GCGCTGTTATGGATTTACCCG-3′ |
| Lm422Ex2a,f | 5′tgcTCTAGA-CTCCCATTTCCTGCATCGCC-3′ |
Lowercase nucleotides are nongenomic sequences added to facilitate restriction endonuclease cleavage.
Recognition site for PstI underlined.
Recognition site for BamHI underlined.
Recognition site for HindIII underlined.
Recognition site for EcoRI underlined.
Recognition site for XbaI underlined.
To recombine ΔsigC, Δlmo0422, and Δlmo0421 deletions onto the L. monocytogenes chromosome, the pKSV7Δ421, pKSV7Δ422, and pKSV7ΔsigC plasmids were electroporated into L. monocytogenes 10403S as previously described (2, 11). Transformants were selected on BHI plates containing 10 μg of chloramphenicol/ml. The transformants were grown at briefly at 30°C and plated at 42°C in BHI plus chloramphenicol to select for integration of the plasmid by homologous recombination. Single colonies with a chromosomal integration were then serially transferred in BHI without chloramphenicol at 30°C to allow the excision and eventual loss of the plasmid. Single colonies were picked and replica plated on BHI and BHI plus chloramphenicol to identify those having undergone excision and loss of the plasmid. Chloramphenicol-sensitive colonies were picked to confirm allelic exchange by both PCR amplification and Southern blot analysis. PCR amplification was performed using the corresponding SOEing PCR primers. Southern blot analyses were performed using a sigC internal probe (293 bp) generated by primers Lm423F1 and Lm423R1 and an lmo0421 internal probe (516 bp) generated by primers I421F and I421R (Table 2). The deletion junctions were further confirmed by DNA sequence analysis of the PCR products.
Construction of the ΔsigC/PSPAC-lmo0422E and Δlmo0422/PSPAC-lmo0422E strains.
The lmo0422 gene was PCR amplified using primers Lm0422Ex1 (5′TGGTCTAGAGCGCTGTTATGGATTTACCCG3′; XbaI site underlined) and primer Lm0422Ex2 (5′TGCTCTAGACTCCCATTTCCTGCATCGCC3′; XbaI site underlined). The amplicon was digested with XbaI and then cloned into the XbaI site in the vector pDEH21. The pDEH21 is fundamentally the same as vector pLIV1 (14) except that a plcB gene was cloned in front of the orfXYZ locus. The entire PSPAC-lmo0422E cassette was excised from the recombinant pDEH21-422E with KpnI and cloned in the KpnI site of the integration vector pPL2, leading to the generation of pPD422E. The pPD422E plasmid was then transferred to E. coli SM10, and the transformants were used to mate pPD422E into L. monocytogenes as previously described (38). L. monocytogenes strain 10403S was originally selected as a streptomycin-resistant variant of strain 10403 (7), allowing us to directly counterselect against the SM10 E. coli donor strain on BHI plates with 50 μg of streptomycin/ml. Transconjugants were isolated on BHI agar containing 50-μg/ml streptomycin and 7.5-μg/ml chloramphenicol. The transconjugants were confirmed by Southern blot analysis, PCR, and sequencing.
RNA analysis by Northern blotting and S1 nuclease analysis.
Cells were grown overnight at 37°C in BHI and diluted 1:100 into fresh medium to initiate the experiments. The cultures were then grown to an optical density at 600 nm (OD600) of ∼0.4, subdivided into equal volumes, and shifted to the following conditions: 48°C for 25 min, addition of acetic acid to a pH of 4.5, addition of ethanol to a final concentration of 4%, addition of NaCl to 4%, addition of penicillin to 50 μg/ml, addition of bile salts mixture (BBL, Maryland) to 0.08%, and addition of nisin to 62.5 IU/ml. After 25 min of each treatment, the cells were harvested and resuspended in 1 ml of TRI-REAGENT (MRC, Cincinnati, OH). The cells were then disrupted by homogenization with glass beads for four 1-min intervals at 4,000 rpm with a Mini-Bead Beater. Total RNA was then extracted from the disrupted cells according to the manufacturer's directions.
Northern blot analyses were carried out using 30 μg of total RNA for each sample. The RNA was electrophoresed in 1% agarose gels with 0.66 M formaldehyde in MOPS buffer (20 mM morpholinepropanesulfonic acid [MOPS], 8 mM sodium acetate, 1 mM EDTA, pH 7.0). The RNA was then transferred to nylon membranes in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). After UV cross-linking, the membranes were prehybridized and then hybridized in solution as described by Church and Gilbert (12) at 70°C and 65°C, respectively. The 293-bp sigC internal probe generated by primers Lm423F1 and Lm423R1 (Table 2) and the 516-bp lmo0421 internal probe generated by primers I421F and I421R (Table 2) were used for the Northern blot analysis. Both of the probes span the deleted region of the corresponding mutants. The probes were labeled by incorporation of [γ-32P]dATP. The membranes were washed twice at 65°C, each time for 45 min in 0.1 M Na2HPO4, and twice at 55°C, each time for 30 min in 0.2× SSC and 0.15% sodium dodecyl sulfate, before being subjected to autoradiography.
S1 nuclease reactions were performed using single-stranded probes that were synthesized by elongation of primer sigCB on a single-strand template. The primer was end labeled with [32P]ATP and annealed in 1× sequenase buffer (USB, Cleveland, Ohio) to single-strand DNA prepared from an M13mp19 clone carrying a 540-bp fragment of the sigC region extending from +151 of the sigC coding region to −389 bp upstream of sigC. The sigCB primer was extended on the template with 1 U of Sequenase and 1 mM final concentration of deoxynucleoside triphosphates for 20 min at 42°C. The extended primer-template hybrids were then digested with HindIII, which cuts at −315 relative to sigC, giving a final single-strand fragment of 466 bp that extends from −315 to +151. The single-stranded, end-labeled probe molecules were then purified by electrophoresis in a 4.5% denaturing gel using autoradiography to locate the band. The gel slices were crushed and soaked overnight in diffusion buffer (0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA [pH 8.0], and 0.1% sodium dodecyl sulfate), followed by phenol extraction and ethanol precipitation. Precipitated probe molecules were redissolved in diethyl pyrocarbonate-treated H2O and annealed to 50 μg of RNA by being heated to 95°C for 5 min and incubated at 65°C for 1 h. After annealing, 300 μl of S1 nuclease buffer (50 mM sodium acetate [pH 4.5], 280 mM NaCl, 4.5 mM ZnSO4) and 300 U of S1 nuclease were added, and the reaction was incubated at 37°C for 30 min. The reaction products were phenol extracted, ethanol precipitated, redissolved in distilled H2O, and electrophoresed in 6% denaturing polyacrylamide gels alongside sequencing reactions primed with the same primer.
Thermal sensitivity.
Wild-type and mutant strains were compared for their ability to survive lethal temperatures by subjecting mid-logarithmic-phase cultures to sublethal and lethal temperatures and enumerating survivors over a time course. Overnight cultures were grown in BHI at 37°C and diluted 1:200 into fresh BHI. The cells were then grown to OD600 of ∼0.4, and the culture was shifted to a 60°C shaking water bath. Samples were removed before and after temperature upshift, serially diluted, and plated onto BHI. The number of colonies from duplicate plates of each dilution was enumerated and averaged after 24 h of growth at 37°C. To test for adaptive responses to sublethal upshifts, the diluted overnight cultures were grown to an OD600 of ∼0.4, followed by transfer of the culture to 48°C for 30 min prior to shifting to 60°C. Samples were removed and enumerated immediately before upshift to 60°C and at 15-min intervals thereafter. Each experiment was repeated at least three times, and the curves shown are representative of the trend in all three experiments.
Thermal sensitivity of PSPAC-lmo0422E strains.
Overnight cultures of the 10403S, ΔsigC, Δlmo0422, ΔsigC/PSPAC-lmo0422E, and Δlmo0422/PSPAC-lmo0422E strains were grown in BHI broth at 37°C (with 7.5-μg/ml chloramphenicol added for the ΔsigC/PSPAC-lmo0422E and Δlmo0422/PSPAC-lmo0422E strains). The overnight cultures were diluted 1:200 into fresh BHI without antibiotic and grown to mid-logarithmic phase (OD600, ∼0.4). The cultures were then subdivided into equal volumes, and IPTG was added to some of the cultures at concentrations ranging from 1 μM to 2.5 μM as indicated; the cells were incubated an additional 30 min at 37°C. The cultures were then transferred to a 60°C shaking water bath. Samples were removed immediately before upshift and at 30 min intervals after the upshift, diluted, and plated onto BHI plates. Colonies were enumerated from duplicate plates after overnight incubation at 37°C and averaged. The experiments were repeated at least three times, and the results shown are representative of the trend in all experiments.
RESULTS
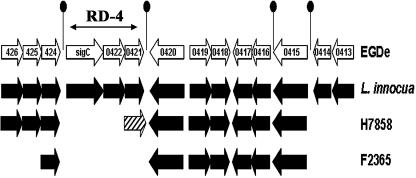
Organization of the sigC operon. The sigC-lmo0421region (Fig. 1) was originally identified in Listeria monocytogenes as a region of genome difference (RD-4) that is specific to lineage II strains (65) and is absent in all lineage I and III strains that have been examined (65; C. Zhang and and A. K. Benson, unpublished data). Comparison of the available L. monocytogenes and L. innocua genome sequences (21, 45) further reveals that this region is conserved in the L. monocytogenes EGDe and L. innocua CLIP strains, suggesting it is ancestral to the Listeria species and was likely lost early during divergence of lineage I strains. Indeed, genome sequences of the L. monocytogenes serotype 4b strains H7858 and F2365 show that the region has undergone further differentiation between different populations of lineage I strains (Fig. 1).
FIG. 1.
Organization and synteny of the RD4 region in genomes of L. monocytogenes and L. innocua strains. Representation of the alignment between the L. monocytogenes strain EGDe, L. innocua, and L. monocytogenes strains H7858 (serotype 4b) and F2365 (serotype 4b) genomes in the RD-4 region is illustrated. Genes are indicated by arrows showing their relative orientations (not drawn to scale). Putative transcription terminators are indicated by a line and filled ellipse. Black arrows represent orthologous genes, stippled arrows indicate nonhomologous genes, and white space indicates absence of the orthologous genes.
The genes within RD-4 encode a protein that is homologous to the ECF family of sigma subunits of RNA polymerase (Lmo0423, referred to herein as sigma C), a PadR-like protein (Lmo0422), and a protein belonging to the RodA-FtsW family (Lmo0421). In the strain EGD genome sequence, these genes appear to comprise an operon based on two characteristics. First, they are positioned close together; sigC and lmo0422 are separated by 2 bp, and the lmo0422 and lmo0421 open reading frames overlap by 4 bp. Second, putative transcription terminators are positioned downstream of lmo0421 (the terminal gene of the operon) and upstream of the initial gene of the operon, sigC (Fig. 1). The latter terminator likely terminates upstream transcription from the lmo0424 transcription unit.
To determine experimentally if sigC, lmo0422, and lmo0421 comprise an operon and to determine the conditions under which these genes are expressed, the accumulation pattern and size distribution of transcripts containing sigC and lmo0421 genes in L. monocytogenes strain 10403S were examined by Northern blotting. Probes for these experiments were generated by PCR amplification of internal segments from the lmo0421 and sigC open reading frames. RNA was prepared from logarithmically growing cells (OD600, ∼0.4) grown at 37°C, as well as logarithmically growing cells that had been subjected to temperature upshift or downshift, osmotic upshift, addition of bile, addition of ethanol, addition of nisin, or addition of penicillin G. As shown in Fig. 2, transcripts were detected under each of the conditions except for the logarithmically growing cells. By visual inspection of the band intensities, the most significant accumulation of RNA occurred in cells that had been subjected to temperature upshift. The lmo0421 and sigC probes both detected bands of 2.3 kb each, and the blots showed identical patterns of accumulation under the different environmental conditions (data not shown). Along with the organization of these genes in the genome, the Northern hybridization experiments support the conclusion that sigC, lmo0422, and lmo0421 comprise a stress-induced operon.
FIG. 2.
Northern blot analysis of RNA from the sigC operon. RNA was prepared from L. monocytogenes strain 10403S grown to mid-logarithmic phase at 37°C in brain heart infusion and 30 min after temperature upshift or downshift (48°C or 4°C), addition of NaCl (to 4%), ethanol (to 4%), nisin (to 62.5 U/ml), penicillin G (50 μg/ml), or bile (0.08%). Total RNA was extracted from the treated cells and 50-μg samples of RNA were loaded in the appropriate lanes. The blots are derived from independent experiments with 37°C and 48°C included on both blots as a point of reference. Both blots shown were probed with a 293-bp segment of the sigC gene amplified using the Lm423F1 and Lm423R1 primers (the amplicon extends from +121 to +414 of the lmo0423 coding region).
Sensitivity of the ΔsigC, Δlmo0422, and Δlmo0421 mutants to high temperature.
Our discovery that the sigC operon is expressed under several conditions of environmental stress prompted us to test whether the sigC, lmo0422, and lmo0421 genes contribute to survival under different environmental stress conditions. To measure their contribution, in-frame deletions were introduced into the sigC, lmo0422, and lmo0421 coding regions; the resulting ΔsigC, Δlmo0422, and Δlmo0421 mutants were then subjected to several different stress conditions. In the first set of experiments, the parental and mutant strains were grown at 37°C to mid-logarithmic phase and then shifted to medium containing 10% ethanol, 4% NaCl, or 0.1% bile salts or to medium with a pH of 4.5. Additional cultures of the strains were also grown at 37°C to mod-logarithmic phase and subjected to temperature downshift to 4°C or upshift to 60°C. Survival of the different strains was monitored by enumeration of viable cells at various times after each of the shifts. The parental (wild-type) strain and the ΔsigC, Δlmo0422, and Δlmo0421 derivatives all showed similar susceptibilities to high osmolarity, low pH, temperature downshift, ethanol, and bile salt treatments (data not shown).
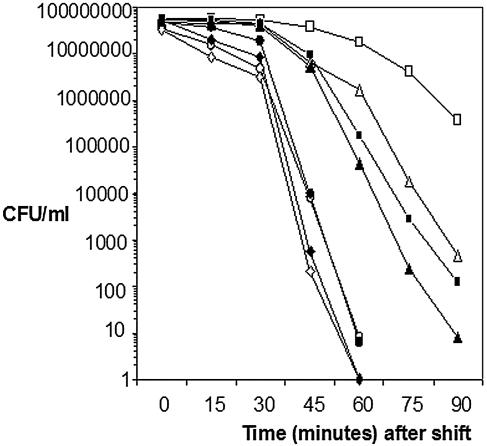
In contrast to osmolarity, pH, ethanol, or bile, the ΔsigC and Δlmo0422 strains showed substantially enhanced death rates when mid-logarithmic cells were upshifted from 37°C to the lethal temperature of 60°C. As shown in Fig. 3, viability of the parental strain assumes a logarithmic decline that began about 45 min after upshift from 37°C to 60°C. If cells of the parental strain were preadapted by a shift to 48°C prior to the 60°C upshift, the parental strain displayed a characteristic adaptive response, assuming a much slower decline in viability and achieving a nearly 5,000-fold enhancement of survival at the later timepoints. Unlike the wild-type strain, viability of the Δlmo0422 and ΔsigC strains decreased at a much more rapid rate after the upshift to 60°C. Moreover, these strains also showed no adaptive response when the cells were preadapted to 48°C prior to the upshift to 60°C. The Δlmo0421 mutant showed a nearly indistinguishable death rate from the wild type when upshifted from 37°C to 60°C, but it lost a portion of the adaptive response when the cells were preadapted at 48°C prior to the upshift to 60°C. The level of adaptation of the Δlmo0421 mutant was only 100 fold compared to the nearly 5,000-fold adaptation achieved with the wild type.
FIG. 3.
Survival of sigC operon mutants at 60°C. Overnight cultures of the parental strain 10403S and mutant derivatives Δlmo0421, Δlmo0422, and ΔsigC were grown in BHI and inoculated 1:200 into fresh BHI and grown to mid-log phase (OD600, ∼0.4) at 37°C. Each mid-log phase culture was then divided in half, with one-half remaining at 37°C and the other half transferred to a 48°C water bath (preadaptation) for 20 min. After the 20-min period, both halves were then shifted to a 60°C water bath. Immediately before the shift to 60°C (time zero) and at intervals after the shift, samples from the cultures were withdrawn, serially diluted, and enumerated by plating onto BHI agar. The results shown here are representative of three independent experiments. Symbols: ▪, 10403S; □, 10403S preadapted; ▴, Δlmo0421; ▵, Δlmo0421 preadapted; •, Δlmo0422; ○, Δlmo0422 preadapted; ⧫, ΔsigC; ⋄, ΔsigC preadapted.
Based on the temperature-induced expression of the sigC operon, the thermal sensitivity of the ΔsigC and Δlmo0422 mutants, and the similarity of sigC and lmo0422 to the ECF sigma factors and the padR family of transcription regulators (23), respectively, we conclude that sigC and lmo0422 likely contribute to regulation of a unique heat shock regulon(s). Because the Δlmo0421 mutation only affects the adaptive response, its role must be less critical than that of sigC or lmo0422, at least under the conditions tested.
The sigC operon is autoregulated in L. monocytogenes.
Our discovery that the sigC operon is thermally regulated and that sigC, lmo0422, and lmo0421 each contribute to thermal resistance prompted us to more closely examine transcription of the sigC operon. Alternative sigma factors, particularly members belonging to the ECF family, are often autoregulatory and positioned adjacent to genes that control their function (25). To determine if sigC contributes to expression of the operon under conditions of temperature upshift, Northern blots were performed to measure transcript accumulation in the mutant backgrounds before and after upshift to 48°C. Probes for the experiment were prepared from both lmo0421 and sigC genes, since transcripts from the operon in the Δlmo0421 mutant will not hybridize to a lmo0421-derived probe and vice versa for sigC.
As shown in Fig. 4, transcripts from the sigC operon were detected in all of the mutant strains after thermal upshift. The hybridizing bands in the mutant strains are shorter in length, due to the deletions. However, inspection of the band intensity in the ΔsigC strain (probed with lmo0421) showed substantially reduced signal after upshift to 48°C. The reduced intensity is particularly notable compared to the signal generated by stripping and reprobing the blot with the dapE gene probe, which is not known to be heat shock regulated (39). The reduced transcript accumulation observed in the ΔsigC background is consistent with the operon being transcribed from multiple promoters, at least one of which is σC dependent and accumulates after temperature upshift.
FIG. 4.
Northern blot analysis of sigC operon RNA from the wild type and Δlmo0421, Δlmo0422, and ΔsigC mutants. Overnight cultures of the wild-type 10403S and derived Δlmo0421, Δlmo0422, and ΔsigC mutants were diluted 1:200 into fresh BHI, grown to mid-log phase (OD600, ∼0.4), and shifted to 48°C. RNA was extracted from samples harvested before (37) and 30 min after (48) the shift, and 50 μg of RNA was electrophoresed in formaldehyde-agarose gels and transferred to nylon membranes. The membranes were probed with labeled PCR products derived from internal segments of the sigC (+121 to +414 of the lmo0423 coding region) or lmo0421 (+735 to +1191 of the lmo0421 coding region) genes. The observed hybridizing transcripts are shorter in length in the mutant strains due to the length of the individual deletions (sigC deletion = 224 bp; lmo0422 deletion = 219 bp; lmo0421 deletion = 963 bp). To confirm equal loading of RNA, the blots were stripped and reprobed with a probe from a temperature-independent housekeeping gene, lmo0265 (dapE) gene (extending from +190 to +319), which encodes succinyl-diaminopimelate desuccinylase (39).
The transcription start site(s) of the sigC operon were next mapped using S1 nuclease assays to identify the 5′ end of transcripts originating upstream of the sigC gene under inducing (48°C) and noninducing (37°C) conditions. As shown in Fig. 5A, S1 nuclease protection assays detected transcripts with start sites positioned at −19, −58, and −92 relative to the sigC start codon (termed P1, P2, and P3, respectively). Alignment of the sequences upstream of these start sites (Fig. 5E) did not reveal similarity to any known promoter elements of L. monocytogenes. To determine if any of the transcripts depend on sigC, lmo0422, or lmo0421, S1 nuclease protection assays were used to measure the relative concentrations of transcripts from the wild type and the mutant strain before and after upshift to 48°C. As shown in Fig. 5B, the ΔsigC mutation eliminates only the proximal transcript at −19, indicating that this transcript is σC dependent. The other two transcripts remained intact with the ΔsigC mutant, indicating that only the proximal P1 promoter depends on sigC. These results are consistent with the Northern blot data of Fig. 4 and show that the sigC operon is transcribed from multiple promoters, one of which is σC dependent.
FIG. 5.
S1 nuclease protection mapping of transcripts from the sigC operon. (A to C) A single-stranded S1 nuclease probe was generated by elongation of the CEX 1 primer (+172 to +151 of sigC) that had been end labeled with [γ-32P]ATP using T4 kinase. The primer was extended on a single-strand template of an M13 mp19 clone carrying the promoter region using Sequenase. The extension products were digested with HindIII at position −316 relative to the transcription start site, and the end-labeled single-stranded probe molecules (extending from +172 to −316 relative to the sigC start codon) were purified by electrophoresis in a denaturing gel. The purified probe was then mixed with 50 μg of RNA from wild-type or mutant cells, heated to 95°C, annealed at 65°C, and treated with S1 nuclease as previously described (35). The digestion products were dissolved in loading buffer and electrophoresed alongside a sequencing ladder primed with the CEX1 primer. (A) RNA was derived from mid-logarithmic-phase wild-type cells that had been shifted from 37°C to 48°C for 20 min. The positions of the three prominent bands relative to the start codon of the sigC gene are indicated to the right. (B) S1 nuclease reactions were performed on RNA extracted from mid-logarithmic-phase cells of the wild-type and ΔsigC strains that had been shifted from 37°C to 48°C for 20 min. The lengths of the three transcripts are indicated to the left of the image. (C) RNA was extracted from cultures of mid-logarithmic-phase wild-type, Δlmo0421, Δlmo0422, and ΔsigC cells at 37°C (37) and 20 min after upshift to 48°C (48). Only the P1 transcript is shown. (D) Relative map of the operon with the three transcription start sites indicated upstream of the sigC gene. The map is not to scale. (E) Putative promoter sequences upstream of the transcription start sites. Putative −10 and −35 region sequences are underlined.
In contrast to the ΔsigC mutation, the Δlmo0422 and Δlmo0421 deletions did not negatively effect accumulation of the sigC-dependent transcript from the P1 promoter (Fig. 5C). This finding was somewhat surprising, given the similarity in phenotypes of the ΔsigC and Δlmo0422 mutants. Thus, if σC is directly responsible for recognition of the P1 promoter, then σC activity itself is not substantially negatively affected by the Δlmo0422 or Δlmo0421 mutation.
Ectopic expression of lmo0422 can complement the sigC thermal resistance phenotype.
The fact that the temperature-inducible, sigC-dependent transcript from P1 accumulates normally in the Δlmo0422 mutant implies that sigC function is intact in the lmo0422 mutant background. Therefore, a simple explanation for the similarity in thermal death phenotypes of the ΔsigC and Δlmo0422 mutants is that lmo0422 is the actual thermal resistance effector-regulator and that sigC is only necessary because it is required for induction of lmo0422 under conditions of temperature upshift. To test this hypothesis, we introduced an IPTG-inducible copy of lmo0422 into the ΔsigC and Δlmo0422 mutants to determine if ectopic expression of lmo0422 could complement the temperature sensitivity of the ΔsigC mutant. An intact copy of the lmo0422 gene was placed under the control of the IPTG-inducible PSPAC promoter as described in Materials and Methods and inserted at a unique location in the genome using the pPL2 system of Lauer et al. (38). Dot blot analysis, shown in Fig. 6, confirmed that expression of lmo0422 in the Δlmo0422/PSPAC-lmo0422E and the ΔsigC/PSPAC-lmo0422E strains was IPTG inducible.
FIG. 6.
Dot blot analysis of lmo0422 transcripts from PSPAC-422E strains. Cultures of the Δlmo0422/PSPAC-422E and ΔsigC/PSPAC-422E were grown at 37°C to mid-logarithmic phase (OD600, ∼0.4), and RNA was extracted from cells harvested prior to or 30 min after addition of IPTG to a final concentration of 1 mM. As a control, RNA was also extracted from mid-logarithmic-phase cells of the Δlmo0422 strain 30 min after upshift of the culture to 48°C. Samples of 20 μg of RNA (each) were then spotted onto nylon, cross-linked by UV light, and then hybridized with a 32P-labeled PCR product derived from the lmo0422 coding region, which includes 180 bp of the 3′ end of sigC, the entire lmo0422 coding region, and 182 bp of the 5′ end of lmo0421.
When the thermal resistance profiles of the Δlmo0422 and Δlmo0422/PSPAC-lmo0422E strains were compared, we observed that, as expected, addition of IPTG allowed the PSPAC-lmo0422E construct to complement the Δlmo0422 deletion (Fig. 7A). Importantly, titration experiments showed that full complementation only occurred at low IPTG concentrations (1 μM), implying that tightly coordinated expression of lmo0422 is important for its function. As with the Δlmo0422/PSPAC-lmo0422E strain, we also observed that IPTG-dependent induction of lmo0422 expression in the ΔsigC/PSPAC-lmo0422 strain restored its thermal resistance phenotype to wild-type levels (Fig. 7B). Moreover, complementation of the ΔsigC thermal resistance phenotype was also dependent on IPTG concentration; full complementation was only observed at 2.5 μM and was not complete at concentrations of >5 μM or <2.5 μM.
FIG. 7.
Ectopic expression of lmo0422 rescues the Δlmo0422 and ΔsigC thermal sensitivity phenotypes. Cells of the wild-type, Δlmo0422, ΔsigC, Δlmo0422/PSPAC-422E, and ΔsigC/PSPAC-422E strains were grown overnight in BHI, diluted 1:200 in fresh BHI, and grown to mid-logarithmic phase (OD600, ∼0.4). The cultures were then subdivided into several equal portions, and IPTG was then added at 0, 0.5 μM, 1 μM, 2.5 μM, and 5 μM concentrations to the cultures of the PSPAC-lmo0422E strains. After 30 min, samples of the cultures were removed and enumerated by serial dilution and plating. The remaining portions of the cultures were then transferred to 60°C. Samples were removed at various times after temperature upshift and enumerated by serial dilution and plating. (A) ○, 10403S; □, Δlmo0422; ⧫, Δlmo0422/PSPAC-422E; ▪, Δlmo0422/PSPAC-422E plus 0.5 μM IPTG; ▴, Δlmo0422/PSPAC-422E plus 1 μM IPTG; •, Δlmo0422/PSPAC-422E plus 2.5 μM IPTG. (B) ○, 10403S; □, ΔsigC; ⧫, ΔsigC/PSPAC-422E; ▴, ΔsigC/PSPAC-422E plus 1 μM IPTG; •, ΔsigC/PSPAC-422E plus 2.5 μM IPTG. Data shown are representative of three or more independent experiments.
Because IPTG-dependent expression of lmo0422 can overcome the temperature sensitivity of the ΔsigC mutation, the role of sigC in thermal resistance must be to provide temperature-inducible expression of lmo0422. The fact that overexpression of either sigC or lmo0422 was detrimental to the thermal resistance phenotype underscores the importance of appropriate control of lmo0422 expression by sigC. Thus, we conclude that lmo0422 is the actual effector or regulator of the heat resistance regulon and we therefore propose to name this gene lstR (for lineage-specific thermal regulator).
DISCUSSION
Several independent studies have observed that L. monocytogenes strains from the three phylogenetic lineages are isolated at different frequencies from food, environmental, and clinical samples (9, 19, 22, 32, 61, 66). These findings have been regarded as evidence that the three evolutionary lineages (and indeed the serotypes which are distributed in a monophyletic fashion among the lineages) possess unique virulence or transmissibility characteristics. Recent studies of genome diversity among populations of Listeria monocytogenes have further shown that transcription factors and genes associated with the cell surface (encoding cell surface proteins or biosynthetic pathways of extracellular material) represent the largest classes of lineage-specific genes (17, 28, 31, 45, 65), implying that these genes could be responsible for lineage-specific traits.
In this study, we have shown that the lineage II-specific sigC, lstR, and lmo0421 genes comprise an operon and that lstR is important for thermal resistance characteristics of the lineage II serotype 1/2a strain 10403S. Thermal resistance characteristics are primarily conferred by temperature-inducible sets of genes, collectively referred to as heat shock genes. There are three major heat shock regulatory pathways that appear to be conserved in L. monocytogenes and the related species Bacillus subtilis. The primary pathway is controlled by HrcA. The hrcA gene is positioned as the promoter-proximal member of the dnaK operon in B. subtilis and L. monocytogenes (24, 50), and HrcA functions to reduce transcription of heat shock genes at moderate growth temperatures by binding to CIRCRE elements in the promoter regions of its target genes (50). A secondary pathway (referred to as class II genes) is controlled by an alternative sigma factor, σB, which, like the putative σC, is induced under a wide variety of environmental and physical stress conditions including heat shock (2, 6, 34, 60, 63). The third pathway is controlled by CtsR, the proximal gene of the clp operon in B. subtilis and L. monocytogenes (33, 44). Like HrcA, CtsR is a DNA-binding protein that acts to negatively regulate expression of its target genes in a temperature-dependent manner (36). In both L. monocytogenes and B. subtilis, at least one additional class of thermally regulated genes exist, exemplified by the lon protease gene, for which no regulatory system has been identified (51).
Our results from this report have now clearly defined yet another regulatory system, σC-LstR, which participates in thermally regulated gene expression in phylogenetic lineage II L. monocytogenes populations. The lstR gene encodes a PadR-like protein whose function is necessary for full thermal resistance, presumably by controlling transcription of a fifth class of heat shock genes. lstR is embedded within an operon that carries a positive regulator of its transcription (sigC) and a gene whose function contributes to the adaptive phase of the response (lmo0421). Although σC is the primary positive regulator of the operon via the thermally regulated P1 promoter, the sigC gene appears to be dispensable for thermal resistance apart from its role in lstR expression. Therefore, σC may comprise a specialized device for controlling lstR expression. As shown by ectopic expression of lstR (Fig. 7), appropriate expression of lstR is absolutely critical for its function. Thus, σC activity is likely to be tightly governed in the cell.
The contribution of σC-LstR to thermal resistance in the lineage II strain 10403S background is substantial. We are in the process of measuring the relative contribution of the different heat shock responses by comparing the thermal sensitivity of isogenic strains carrying deletions in lstR, sigC, and sigB and strains carrying IPTG-inducible versions of hrcA and ctsR in the 10403S background. Preliminary studies of the ΔsigC and ΔsigB single and double mutants indicate that ΔsigC mutants are much more susceptible to thermal death than sigB:Kmr mutants (Zhang and Benson, unpublished). Moreover, ΔsigC sigB:Kmr double mutants are even more sensitive than either single mutant, underscoring that the σC-LstR and σB pathways are independent. No formal, systematic comparisons of thermal resistance characteristics and pathways of lineage I and lineage II strains have been reported. Therefore, it remains to be determined how lineage I strains mount adaptive responses to high temperatures.
Distribution of lstR and the sigC operon in Listeria and the bacilli.
LstR is a member of a family of proteins sharing similarity to PadR. Originally discovered in Lactobacillus plantarum, PadR negatively controls transcription of genes involved in utilization of phenolic acids, presumably by acting directly at cis-acting sequences (23). Although several paralogous members of the PadR family can be found in the genomes of Listeria and Bacillus species, only lineage II Listeria monocytogenes, Listeria innocua, Bacillus cereus, Bacillus anthracis, Bacillus thuringiensis, and Bacillus halodurans strains carry an orthologous lstR gene. In each instance, lstR is positioned between sigC and lmo0421 orthologues, indicating that the operon is a conserved element.
In the L. monocytogenes and L. innocua genomes, the sigC operon is positioned between a glucose permease and a conserved hypothetical protein with hydrolase-like domains. The synteny implies that the operon was likely ancestral to the genus Listeria, while absence of the operon in lineage I and lineage III L. monocytogenes strains is a consequence of gene loss during divergence of these populations. In the sensu lato members of the B. cereus subgroup (B. cereus, B. anthracis, and B. thuringiensis), the operon is positioned adjacent to the glpD and glpK genes while in B. halodurans, the operon is positioned in a third genomic region, suggesting that it has undergone rearrangement in the genome or that it was acquired independently in Listeria and the two different Bacillus lineages. It is interesting to note that like the situation with L. monocytogenes phylogenetic lineages, the sigC region in the B. cereus ATCC14579 strain has apparently been lost, but it is conserved in the B. cereus ATCC10987 and ZK strains, suggesting that there may be some propensity for the region to be lost in certain subpopulations of a species.
Lmo0421 and lineage-specific expansion of rodA-ftsW genes.
The function of lmo0421, the distal gene of the sigC operon, remains enigmatic. The Δlmo0421 mutant had a normal death rate after direct upshift to lethal temperature but had lost a portion of its adaptive response when first upshifted to a sublethal temperature (Fig. 3). This phenotype is consistent with Lmo0421 representing a unique pathway to controlling lstR expression or function. In addition to its unique effect on temperature resistance, the Lmo0421 protein has some rather remarkable structural characteristics. Lmo0421 shares substantial similarity to the RodA-FtsW family of proteins, which modulates cell wall biosynthesis during the elongation phase (RodA) and the septation phase (FtsW) of cell division (4, 5, 15, 27). Because of the similarity to this family, we initially examined the Δlmo0421 strain for morphological defects but were unable to observe defects under any of the conditions tested (room temperature, 37°C, or 45°C). One reason for the absence of morphological phenotype could be the redundancy of the RodA-FtsW family of proteins in L. monocytogenes. Most rod-shaped species carry a single copy of the rodA and ftsW genes, and they are essential for normal morphology in E. coli and B. subtilis (27, 55). B subtilis carries an additional member of this family, spoVE, which appears to be dedicated to wall biosynthesis in the developing endospore (26). BLAST analyses of the L. monocytogenes genome sequences show that Lineage II L. monocytogenes and L. innocua carry six paralogus members of the RodA-FtsW family (including Lmo0421 and its orthologue in L. innocua, Lin0441). Remarkably, B. anthracis, B. cereus, B. thuringiensis, B. halodurans, and Bacillus licheniformis also share this phenomenon of carrying multiple copies of RodA-FtsW genes. With the exception of B. licheniformis, these are also the only genomes which carry orthologous sigC operons, begging the question of whether functional linkage exists between these systems. Although B. licheniformis does not contain a sigC operon orthologue, it is interesting to note that it carries a cell wall peptidase-like gene adjacent to the sigX operon and that sigX is known to confer thermal resistance (30).
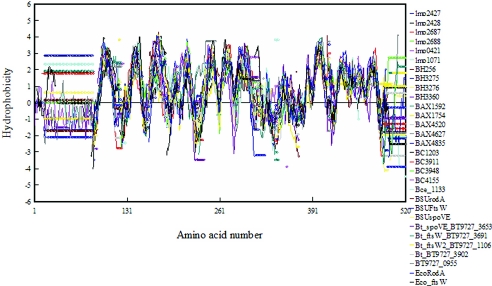
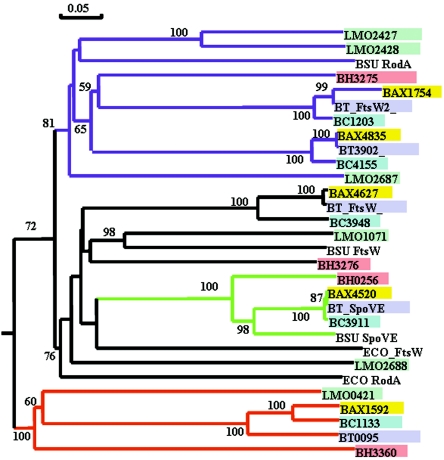
Is Lmo0421 a unique member of the RodA-FtsW family that somehow links gene expression and cell wall biosynthesis? Multiple alignment of the individual hydrophobicity plots of the RodA-FtsW proteins from Listeria and Bacillus species reveals that indeed Lmo0421 and all of the additional RodA-FtsW proteins share striking topological characteristics with the known B. subtilis and E. coli RodA and FtsW proteins (Fig. 8). Moreover, cluster analysis (Fig. 9) shows that Lmo0421 and the orthologous proteins Lin0441, BH3360, BA1592, BT0955, and BC1133 (derived from the sigC operons of these organisms) comprise a single cluster supported by a significant bootstrap score. Thus, the structural characteristics of Lmo0421 are consistent with it being a unique member of the RodA-FtsW family. The phenotype, operon organization, and structural characteristics of Lmo0421 make it tempting to speculate that the Lmo0421 provides a unique mechanism for linking cell wall biosynthesis to gene expression, perhaps through σC and LstR. It will be important to further characterize function of σC, LstR, and Lmo0421 and to understand the characteristics this specialized set of proteins confers on the phylogenetic lineage II populations of Listeria monocytogenes.
FIG. 8.
Multiple hydropathy alignment of RodA-FtsW-like proteins from L. monocytogenes and related species. Proteins belonging to the RodA-FtsW family were identified from the genomes of L. monocytogenes strain EGDe (Lmo), Bacillus subtilis 168 (BSU), Bacillus cereus ATCC10987 (BC), Bacillus anthracis Sterne (BAX), Bacillus thuringiensis serovar konkukian strain 97-27 (BT), Bacillus halodurans C-125 (BH), and Escherichia coli MG1655 (ECO). Multiple alignment was performed using CLUSTAL W (57) using a Blosum scoring matrix, and multiple hydrophobicity plots based on the alignment were generated based on the methods of Hopp and Woods (29, 37) as implemented in the DNAman package. The plot shown was developed using a window size of eight amino acid residues. Each colored plot and the corresponding protein are shown to the right of the plot. Circles in the plot indicate gaps in the alignment or start-stop positions.
FIG. 9.
Phylogenetic analysis of Lmo0421 and other RodA-FtsW-like proteins from L. monocytogenes and related species. Proteins belonging to the RodA-FtsW family were identified from the genomes of L. monocytogenes strain EGDe (Lmo), Bacillus subtilis 168 (BSU), Bacillus cereus ATCC10987 (BC), Bacillus anthracis Sterne (BAX), Bacillus thuringiensis serovar konkukian strain 97-27 (BT), and Bacillus halodurans C-125 (BH) using BLAST analyses. The set of proteins was then subjected to bootstrap analysis (20) using a neighbor-joining search (49). Significant bootstrap values (>50%) from 10,000 repetitions are indicated on the branches of the relevant nodes. Proteins from the genome of each species are filled with the same color. Nodes corresponding to the B. subtilis RodA (purple), B. subtilis SpoVE (green), and the L. monocytogenes Lmo0421 proteins (red) are colored to highlight putatively functionally similar proteins.
Acknowledgments
We thank R. Hutkins for many valuable discussions and for critical reading of the manuscript.
This work was funded by the USDA National Research Initiative Competitive Grants Program, award no. 2002-35201-12649 to A.K.B.
Footnotes
A contribution of the University of Nebraska Agricultural Research Division, Lincoln (journal series no. 14675).
REFERENCES
- 1.Arous, S., C. Buchrieser, P. Folio, P. Glaser, A. Namane, M. Hebraud, and Y. Hechard. 2004. Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiology 150:1581-1590. [DOI] [PubMed] [Google Scholar]
- 2.Becker, L. A., M. S. Cetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, L. A., S. N. Evans, R. W. Hutkins, and A. K. Benson. 2000. Role of σB in adaptation of Listeria monocytogenes to growth at low temperature. J. Bacteriol. 182:7083-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Begg, K. J., and W. D. Donachie. 1985. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J. Bacteriol. 163:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begg, K. J., B. G. Spratt, and W. D. Donachie. 1986. Interaction between membrane proteins PBP3 and RodA is required for normal cell shape and division in Escherichia coli. J. Bacteriol. 167:1004-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, A. K., and W. G. Haldenwang. 1993. The σB-dependent promoter of the Bacillus subtilis sigB operon is induced by heat shock. J. Bacteriol. 175:1929-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. [PubMed] [Google Scholar]
- 8.Botta, G. A., and J. T. Park. 1981. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J. Bacteriol. 145:333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruhn, J. B., B. F. Vogel, and L. Gram. 2005. Bias in the Listeria monocytogenes enrichment procedure: lineage 2 strains outcompete lineage 1 strains in University of Vermont selective enrichments. Appl. Environ. Microbiol. 71:961-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canepari, P., C. Signoretto, M. Boaretti, and M. Del Mar Lleo. 1997. Cell elongation and septation are two mutually exclusive processes in Escherichia coli. Arch. Microbiol. 168:152-159. [DOI] [PubMed] [Google Scholar]
- 11.Cetin, M. S., C. Zhang, R. W. Hutkins, and A. K. Benson. 2004. Regulation of transcription of compatible solute transporters by the general stress sigma factor, σB, in Listeria monocytogenes. J. Bacteriol. 186:794-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotter, P. D., N. Emerson, C. G. Gahan, and C. Hill. 1999. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J. Bacteriol. 181:6840-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dancz, C. E., A. Haraga, D. A. Portnoy, and D. E. Higgins. 2002. Inducible control of virulence gene expression in Listeria monocytogenes: temporal requirement of listeriolysin O during intracellular infection. J. Bacteriol. 184:5935-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Pedro, M. A., W. D. Donachie, J. V. Holtje, and H. Schwarz. 2001. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J. Bacteriol. 183:4115-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Pedro, M. A., J. C. Quintela, J. V. Holtje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doumith, M., C. Cazalet, N. Simoes, L. Frangeul, C. Jacquet, F. Kunst, P. Martin, P. Cossart, P. Glaser, and C. Buchrieser. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dussurget, O., D. Cabanes, P. Dehoux, M. Lecuit, C. Buchrieser, P. Glaser, and P. Cossart. 2002. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45:1095-1106. [DOI] [PubMed] [Google Scholar]
- 19.Evans, M. R., B. Swaminathan, L. M. Graves, E. Altermann, T. R. Klaenhammer, R. C. Fink, S. Kernodle, and S. Kathariou. 2004. Genetic markers unique to Listeria monocytogenes serotype 4b differentiate epidemic clone II (hot dog outbreak strains) from other lineages. Appl. Environ. Microbiol. 70:2383-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felsenstein, J. 1988. Phylogenies from molecular sequences: inference and reliability. Annu. Rev. Genet. 22:521-565. [DOI] [PubMed] [Google Scholar]
- 21.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 22.Gray, M. J., R. N. Zadoks, E. D. Fortes, B. Dogan, S. Cai, Y. Chen, V. N. Scott, D. E. Gombas, K. J. Boor, and M. Wiedmann. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gury, J., L. Barthelmebs, N. P. Tran, C. Divies, and J. F. Cavin. 2004. Cloning, deletion, and characterization of PadR, the transcriptional repressor of the phenolic acid decarboxylase-encoding padA gene of Lactobacillus plantarum. Appl. Environ. Microbiol. 70:2146-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanawa, T., M. Kai, S. Kamiya, and T. Yamamoto. 2000. Cloning, sequencing, and transcriptional analysis of the dnaK heat shock operon of Listeria monocytogenes. Cell Stress Chaperones 5:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 26.Henriques, A. O., H. de Lencastre, and P. J. Piggot. 1992. A Bacillus subtilis morphogene cluster that includes spoVE is homologous to the mra region of Escherichia coli. Biochimie 74:735-748. [DOI] [PubMed] [Google Scholar]
- 27.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235-247. [DOI] [PubMed] [Google Scholar]
- 28.Herd, M., and C. Kocks. 2001. Gene fragments distinguishing an epidemic-associated strain from a virulent prototype strain of Listeria monocytogenes belong to a distinct functional subset of genes and partially cross-hybridize with other Listeria species. Infect. Immun. 69:3972-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hopp, T. P., and K. R. Woods. 1981. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. USA 78:3824-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, X., A. Decatur, A. Sorokin, and J. D. Helmann. 1997. The Bacillus subtilis σX protein is an extracytoplasmic function sigma factor contributing to survival at high temperature. J. Bacteriol. 179:2915-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacquet, C., E. Gouin, D. Jeannel, P. Cossart, and J. Rocourt. 2002. Expression of ActA, Ami, InlB, and listeriolysin O in Listeria monocytogenes of human and food origin. Appl. Environ. Microbiol. 68:616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeffers, G. T., J. L. Bruce, P. L. McDonough, J. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 33.Karatzas, K. A., J. A. Wouters, C. G. Gahan, C. Hill, T. Abee, and M. H. Bennik. 2003. The CtsR regulator of Listeria monocytogenes contains a variant glycine repeat region that affects piezotolerance, stress resistance, motility and virulence. Mol. Microbiol. 49:1227-1238. [DOI] [PubMed] [Google Scholar]
- 34.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konyecsni, W. M., and V. Deretic. 1990. DNA sequence and expression analysis of algP and algQ, components of the multigene system transcriptionally regulating mucoidy in Pseudomonas aeruginosa: algP contains multiple direct repeats. J. Bacteriol. 172:2511-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kruger, E., D. Zuhlke, E. Witt, H. Ludwig, and M. Hecker. 2001. Clp-mediated proteolysis in gram-positive bacteria is autoregulated by the stability of a repressor. EMBO J. 20:852-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 38.Lauer, P., M. Y. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, S., J. E. Graham, L. Bigelow, P. D. Morse II, and B. J. Wilkinson. 2002. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microbiol. 68:1697-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milohanic, E., P. Glaser, J. Y. Coppee, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 42.Nadon, C. A., B. M. Bowen, M. Wiedmann, and K. J. Boor. 2002. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70:3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nadon, C. A., D. L. Woodward, C. Young, F. G. Rodgers, and M. Wiedmann. 2001. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 39:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nair, S., I. Derre, T. Msadek, O. Gaillot, and P. Berche. 2000. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol. Microbiol. 35:800-811. [DOI] [PubMed] [Google Scholar]
- 45.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park, J. T., and L. G. Burman. 1985. Elongation of the murein sacculus of Escherichia coli. Ann. Inst. Pasteur Microbiol. 136A:51-58. [DOI] [PubMed] [Google Scholar]
- 47.Piffaretti, J. C., H. Kressebuch, M. Aeschbacher, J. Bille, E. Bannerman, J. M. Musser, R. K. Selander, and J. Rocourt. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. USA 86:3818-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasmussen, O. F., P. Skouboe, L. Dons, L. Rossen, and J. E. Olsen. 1995. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 141:2053-2061. [DOI] [PubMed] [Google Scholar]
- 49.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 50.Schulz, A., and W. Schumann. 1996. hrcA, the first gene of the Bacillus subtilis dnaK operon encodes a negative regulator of class I heat shock genes. J. Bacteriol. 178:1088-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schumann, W. 2003. The Bacillus subtilis heat shock stimulon. Cell Stress Chaperones 8:207-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simon, R., U. Priefer, and A. Puhler. 1982. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 53.Sleator, R. D., J. Wouters, C. G. Gahan, T. Abee, and C. Hill. 2001. Analysis of the role of OpuC, an osmolyte transport system, in salt tolerance and virulence potential of Listeria monocytogenes. Appl. Environ. Microbiol. 67:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705-711. [DOI] [PubMed] [Google Scholar]
- 55.Spratt, B. G. 1975. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 72:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sue, D., K. J. Boor, and M. Wiedmann. 2003. σB-dependent expression patterns of compatible solute transporter genes opuCA and lmo1421 and the conjugated bile salt hydrolase gene bsh in Listeria monocytogenes. Microbiology 149:3247-3256. [DOI] [PubMed] [Google Scholar]
- 57.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Todd, E. 1990. Epidemiology of foodborne illness: North America. Lancet 336:788-790. [DOI] [PubMed] [Google Scholar]
- 59.Vines, A., M. W. Reeves, S. Hunter, and B. Swaminathan. 1992. Restriction fragment length polymorphism in four virulence-associated genes of Listeria monocytogenes. Res. Microbiol. 143:281-294. [DOI] [PubMed] [Google Scholar]
- 60.Volker, U., B. Maul, and M. Hecker. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ward, T. J., L. Gorski, M. K. Borucki, R. E. Mandrell, J. Hutchins, and K. Pupedis. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 186:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wemekamp-Kamphuis, H. H., J. A. Wouters, R. D. Sleator, C. G. Gahan, C. Hill, and T. Abee. 2002. Multiple deletions of the osmolyte transporters BetL, Gbu, and OpuC of Listeria monocytogenes affect virulence and growth at high osmolarity. Appl. Environ. Microbiol. 68:4710-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wiedmann, M., T. J. Arvik, R. J. Hurley, and K. J. Boor. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180:3650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wientjes, F. B., and N. Nanninga. 1989. Rate and topography of peptidoglycan synthesis during cell division in Escherichia coli: concept of a leading edge. J. Bacteriol. 171:3412-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, C., M. Zhang, J. Ju, J. Nietfeldt, J. Wise, P. M. Terry, M. Olson, S. D. Kachman, M. Wiedmann, M. Samadpour, and A. K. Benson. 2003. Genome diversification in phylogenetic lineages I and II of Listeria monocytogenes: identification of segments unique to lineage II populations. J. Bacteriol. 185:5573-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng, W., and S. Kathariou. 1995. Differentiation of epidemic-associated strains of Listeria monocytogenes by restriction fragment length polymorphism in a gene region essential for growth at low temperatures (4°C). Appl. Environ. Microbiol. 61:4310-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]