Abstract
Vibrio cholerae is both an environmental bacterium and a human intestinal pathogen. The attachment of bacteria to surfaces in biofilms is thought to be an important feature of the survival of this bacterium both in the environment and within the human host. Biofilm formation occurs when cell-surface and cell-cell contacts are formed to make a three-dimensional structure characterized by pillars of bacteria interspersed with water channels. In monosaccharide-rich conditions, the formation of the V. cholerae biofilm requires synthesis of the VPS exopolysaccharide. MbaA (locus VC0703), an integral membrane protein containing a periplasmic domain as well as cytoplasmic GGDEF and EAL domains, has been previously identified as a repressor of V. cholerae biofilm formation. In this work, we have studied the role of the protein NspS (locus VC0704) in V. cholerae biofilm development. This protein is homologous to PotD, a periplasmic spermidine-binding protein of Escherichia coli. We show that the deletion of nspS decreases biofilm development and transcription of exopolysaccharide synthesis genes. Furthermore, we demonstrate that the polyamine norspermidine activates V. cholerae biofilm formation in an MbaA- and NspS-dependent manner. Based on these results, we propose that the interaction of the norspermidine-NspS complex with the periplasmic portion of MbaA diminishes the ability of MbaA to inhibit V. cholerae biofilm formation. Norspermidine has been detected in bacteria, archaea, plants, and bivalves. We suggest that norspermidine serves as an intercellular signaling molecule that mediates the attachment of V. cholerae to the biotic surfaces presented by one or more of these organisms.
In their natural habitats, bacteria are often found in surface-attached sessile communities known as biofilms (11). The mature biofilm is a three-dimensional structure composed of pillars of bacteria encased in a self-produced exopolysaccharide matrix surrounded by water channels (12). Compared to their planktonic or free-swimming counterparts, biofilm-associated bacteria are less susceptible to UV irradiation, host defense, and the toxic effects of antimicrobial agents (16, 46). Thus, the ability to form a biofilm is hypothesized to confer a survival advantage both in the environment and within a host.
Genetic and microscopic analyses of many bacterial species have shown that biofilm development proceeds through several distinct stages (13, 52, 56, 71). In the free-swimming or free-floating planktonic stage, bacteria exist as single cells that are not associated with a surface. When a surface is encountered, bacteria form transient associations with the surface, which may lead to permanent attachment, thus initiating the monolayer stage. In the monolayer, only cell-surface interactions are present. Specific environmental signals activate the synthesis of exopolysaccharide, which heralds the passage into the biofilm stage (14, 28, 72, 77). Environmental signals identified to date include surfaces, osmolarity, quorum sensing, and nutritional cues (60).
Vibrio cholerae, the causative agent of the diarrheal disease cholera, is a natural inhabitant of diverse aquatic environments, including lakes, rivers, estuaries, and oceans (9, 10). In its natural habitats, V. cholerae is thought to exist primarily in the surface-attached state (31). It has been identified in association with phytoplankton, zooplankton, crustaceans, and aquatic plants (32, 33, 59, 65). V. cholerae biofilm development on abiotic surfaces proceeds through stages similar to those described above (52, 72, 73). Attachment to surfaces is accelerated by cell surface appendages such as the polar flagellum and the mannose-sensitive hemagglutinin type IV pilus. Exopolysaccharide synthesis and the formation of the mature V. cholerae biofilm are highly regulated. Exopolysaccharide synthesis and secretion are carried out by a number of proteins, including those encoded by the vps (vibrio polysaccharide) genes, which reside in two operons approximately 8.3 kb apart. These operons are comprised of genes vpsA to vpsK (vpsA-vpsK) and vpsL-vpsQ, respectively (69, 72, 77). Transcription of the vps genes is under the control of the σ54-dependent two-component response regulator VpsR (75). As is the case for most two-component response regulators, VpsR is activated by phosphorylation (43). However, the signals that lead to the activation of VpsR are unknown, and its cognate phosphoryl group donor histidine kinase has not yet been identified. The transcription of the vps genes is also activated by VpsT, another two-component response regulator (4). VpsT and VpsR activate their own transcriptions, and each also activates that of the other (4). It is not known whether these regulators affect vps gene transcription directly or indirectly.
Several negative regulators of vps gene transcription have also been identified. CytR, first identified as a nucleoside-responsive transcriptional repressor of genes responsible for nucleoside uptake and catabolism, is also a repressor of vps gene transcription and biofilm formation in V. cholerae (25). HapR, which was initially identified as a transcriptional activator of the hap gene encoding hemagglutinin/protease, represses vps gene transcription and biofilm formation in response to high cell density in some strains of V. cholerae (24, 36, 43, 76, 79). Although the roles of nucleosides and quorum-sensing signals in the regulation of vps gene transcription and biofilm formation have been established, the signal transduction pathways that underlie this regulation have not yet been elucidated.
Genes encoding proteins belonging to the GGDEF and EAL families are highly abundant in the genomes of gram-negative organisms. Some of the functions regulated by GGDEF and EAL family proteins include cellulose production in Gluconacetobacter xylinus and Salmonella enterica serovar Typhimurium, autoaggregation and the wrinkled colony morphology in Pseudomonas aeruginosa, swarmer-to-stalked cell differentiation in Caulobacter crescentus, and biofilm formation in Yersinia pestis (15, 26, 41, 64). These domains are involved in nucleotide cyclization, specifically in the synthesis and degradation of bis-(2′,5′)-cyclic diguanylic acid (c-di-GMP).
It has been shown that GGDEF domains can function as diguanylate cyclases, enzymes that catalyze the conversion of GTP to c-di-GMP, while some EAL domains have been demonstrated to be phosphodiesterases, enzymes that degrade c-di-GMP (2, 57). However, there are also a number of diguanylate cyclases and phosphodiesterases that contain both of these domains (64). c-di-GMP has been shown to affect the function of some enzymes by an allosteric mechanism, and changes in the intracellular levels of c-di-GMP modulate gene transcription within the cell (35).
Recently, several proteins belonging to the GGDEF and EAL families have been shown to affect biofilm formation in V. cholerae (3, 58, 67). In a study by Tischler and Camilli, the overexpression of the putative diguanylate cyclase encoded by VCA956, a GGDEF family protein of unknown function, or the deletion of VieA, a putative phosphodiesterase containing an EAL domain, resulted in increased intracellular levels of the secondary messenger c-di-GMP, vps gene transcription, and biofilm formation in V. cholerae (67). Furthermore, MbaA, a putative integral membrane protein containing GGDEF and EAL domains, has also been identified as a repressor of biofilm formation (3). Because decreased biofilm formation has been correlated with the degradation of c-di-GMP, it is likely that MbaA functions as a phosphodiesterase. MbaA is encoded by the second gene in a putative three-gene operon that also includes genes coding for a predicted periplasmic protein with similarity to polyamine-binding proteins (locus VC0704) and a predicted cytoplasmic protein (locus VC0702).
Polyamines are organic polycations that are positively charged at physiological pH due to their protonated amine groups (63). They are involved in numerous cellular processes, including the modulation of DNA and RNA functions, protection against oxidative damage, and the regulation of bacterial porins and eukaryotic ion channels (34, 37, 45, 53, 78). In this work, we have studied the function of the predicted polyamine-binding protein encoded at locus VC0704. We show that this protein activates biofilm formation in response to increased environmental concentrations of the polyamine norspermidine. Thus, we have named this protein NspS (for norspermidine sensor). To our knowledge, this is the first report of the modulation of bacterial biofilms by environmental polyamines. Although studies are limited, norspermidine has been identified in many types of bacteria and in eukaryotes. Because signaling through NspS most likely does not require the entry of norspermidine into the cell, we suggest that we have identified a novel intercellular signaling pathway that enables V. cholerae to communicate with both its prokaryotic and its eukaryotic neighbors.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and reagents.
The bacterial strains and plasmids used in this study are listed in Table 1. pCR2.1-TOPO was propagated in the DH5α strain, and pWM91 was propagated in the DH5αλpir strain. All experiments were done in Luria-Bertani (LB) broth except where otherwise indicated. Norspermidine and spermidine were purchased from Sigma (St. Louis, MO). Restriction endonucleases were from New England Biolabs (Beverly, MA). Taq and Pfx polymerases and salmon sperm DNA were from Invitrogen (Carlsbad, CA). Primers used in this study are listed in Table 2. Primer synthesis and DNA sequencing were done by Tufts University Core Facility (Boston, MA).
TABLE 1.
Bacterial strains and plasmids
| Strain/plasmid | Genotype | Reference/source |
|---|---|---|
| E. coli strain SM10λpir | thi thr leu tonA lacY supE recA::RP4-2 Tc::MuλpirR6K; Kmr | 51 |
| V. cholerae strains | ||
| PW249 | MO10, clinical isolate of V. cholerae O139 from India, Smr | 70 |
| PW357 | MO10lacZ::vpsLp→lacZ, Smr | 25 |
| PW396 | MO10, ΔvpsA-vpsK, Smr | 40 |
| PW444 | MO10lacZ::vpsLp→lacZ, ΔmbaA, Smr | This study |
| PW512 | MO10lacZ::vpsLp→lacZ, ΔVC0702, Smr | This study |
| PW514 | MO10lacZ::vpsLp→lacZ, ΔVC0704, Smr | This study |
| PW522 | MO10lacZ::vpsLp→lacZ, ΔVC0704ΔmbaA, Smr | This study |
| Plasmids | ||
| pWM91 | oriR6K mobRP4 lacI Ptac tnp mini-Tn10Km; Kmr, Apr | 49 |
| pACYC184 | Kmr Cmr | 6 |
| pCR2.1-TOPO | Plasmid for TOPO cloning, Apr | Invitrogen |
| PK105 | pWM91 carrying an internal in-frame deletion of VC0704 | This study |
| PK104 | pWM91 carrying an internal in-frame deletion of VC0702 | This study |
| pNB4 | pWM91 carrying an internal in-frame deletion of mbaA | 3 |
TABLE 2.
Primers
| Construct | Primer | Description | Sequence |
|---|---|---|---|
| V. cholerae VC0702 deletion | P200 | Forward primer for upstream fragment 1 | 5′-GGAACTCTCGCGTCTTGGCTATG-3′ |
| P201 | Reverse primer for upstream fragment | 5′-GCCGCAGCGGCCGCAGGTGCTAAAGGCACTTCTAACG-3′ | |
| P202 | Forward primer for downstream fragment | 5′-TGCGGCCGCTCGGGCGCTCTGATCCCCTTTATTAATCCA-3′ | |
| P203 | Reverse primer for downstream fragment 1 | 5′-TCCTTCTTTTAGTGAGTGGCAGCAAG-3′ | |
| P208 | Forward primer for upstream fragment 2 | 5′-TCGAGGAACTCTCGCGTCTTGGCTATG-3′ | |
| P209 | Reverse primer for downstream fragment 2 | 5′-CTAGTCCTTCTTTTAGTGAGTGGCAGCAAG-3′ | |
| V. cholerae VC0704 deletion | P204 | Forward primer for upstream fragment 1 | 5′-GATATGACTAAGCTGGAATCGCGTC-3′ |
| P205 | Reverse primer for upstream fragment | 5′-GCCCGAGCGGCCGCACAGCGACAGAAAGCGTTTGATC-3′ | |
| P206 | Forward primer for downstream fragment | 5′-TGCGGCCGCTCGGGCCTCAGTCTGCGTGCCAAAATCATC-3′ | |
| P207 | Reverse primer for downstream fragment 1 | 5′-TTGGCCATTGAGGATCGATAACG-3′ | |
| P210 | Forward primer for upstream fragment 2 | 5′-TCGAGATATGACTAAGCTGGAATCGCGTC-3′ | |
| P211 | Reverse primer for downstream fragment 2 | 5′-CTAGTTGGCCATTGAGGATCGATAACG-3′ |
Construction of the deletion mutants.
In-frame deletions that removed 957 nucleotides from the 1,080-bp coding sequence at locus VC0704 and 469 nucleotides from the 552-bp coding sequence at locus VC0702 were constructed as follows. For the VC0704 deletion, a 392-bp region of chromosome I between positions 753277 and 753669 and a 404-bp region between positions 754624 and 755028 were amplified by PCR. For the deletion at locus VC0702, a 421-bp region of chromosome I between positions 750399 and 750820 and a 411-bp region between positions 751232 and 751643 were amplified by PCR. In both cases, the internal primers contained a 15-bp complementary region, which allowed the amplified fragments to be spliced together by overlap extension PCR (29). The spliced fragments were then amplified in two separate reactions for each gene by using primer pairs P200/P209 (reaction 1) and P203/P208 (reaction 2) for the VC0702 deletion and primer pairs P204/P211 (reaction 3) and P207/P210 (reaction 4) for the VC0704 deletion. The amplified products were heated to 100°C for 5 min for denaturation; mixtures for reactions 1 and 2 were combined and mixtures for reactions 3 and 4 were combined. They were then left to anneal by slow cooling to room temperature to yield sticky ends as described elsewhere (68). These were cloned directly into the suicide plasmid pWM91 digested with XhoI and SpeI to create pK104 and pK105. The correct assembly of the fragments was confirmed by sequencing. These plasmids were transformed into SM10λpir and conjugated into the relevant strains. The deletion mutants were created by double homologous recombination and sucrose selection as described previously (49). Similarly, pNB4 was used to create the ΔmbaA strain (3). All of the deletions were confirmed by PCR.
Fluorescence microscopy.
For fluorescence microscopy, biofilms were formed on borosilicate coverslips inserted into 50-ml Falcon tubes containing 7 ml of LB broth inoculated with V. cholerae to yield a starting optical density at 655 nm (OD655) of 0.02, as measured in a 96-well flat-bottomed plate by using a Bio-Rad model 680 microplate reader (Hercules, CA). After an 18-h incubation, the media and the planktonic cells were discarded, and biofilms were incubated in fresh media containing 2 μg of the fluorescent dye 4′,6-diamidino-2-phenylindole (DAPI) for 30 min at room temperature, washed with fresh LB, and placed on a concave microscope slide filled with sterile medium. An Eclipse TE-200 inverted fluorescence microscope (Nikon) equipped with an Orca digital charge-coupled-device camera (Hamamatsu) and a focus motor were used to collect a stack of transverse sections through the biofilm. The image stacks were subjected to nearest-neighbor deconvolution and three-dimensional image reconstruction using the Metamorph Imaging software (Universal Imaging).
Biofilm assays.
Borosilicate tubes were filled with 300 μl of growth medium. These tubes were inoculated with the strains of interest to yield an OD655 of 0.02. These cultures were incubated for 18 h at 27°C. Planktonic cells were removed, the remaining biofilm was washed with 300 μl of LB, and the biofilm was dispersed by vortexing for 10 s in the presence of 1-mm-diameter borosilicate glass beads (Biospec, Bartlesville, OK). The final planktonic and biofilm cell densities were measured by using 150 μl of the relevant cell suspension. All experiments were performed in triplicate and repeated multiple times to confirm reproducibility.
β-Galactosidase measurements.
β-Galactosidase assays were performed as previously described with the following modifications (25). Briefly, Erlenmeyer flasks were filled with 20 ml of LB broth with supplements as noted and inoculated with an overnight culture of the relevant strain at a 1:100 dilution. The culture was grown to logarithmic phase (OD655 of 0.3 to 0.4) at 27°C with shaking. One-ml aliquots were used for β-galactosidase measurements. Cells were washed twice and resuspended in 200 μl of Z buffer (50). Cells were lysed by three freeze-thaw cycles and incubated for 18 h at 37°C with O-nitrophenyl-β-d-galactopyranoside (Sigma). β-Galactosidase activity was determined by measuring the A415. Values were normalized using cell density measurements. All experiments were performed in triplicate and repeated multiple times.
RNA extraction.
RNA for use in microarray hybridizations was prepared as follows. Wild-type and ΔmbaA mutant cultures were grown as described above for β-galactosidase assays and grown to logarithmic phase. Half of the culture was pelleted by centrifugation at 5,000 rpm for 5 min at 4°C. RNA was extracted by lysing the cells in TRIzol reagent (Invitrogen), a procedure followed by chloroform extraction and isopropanol precipitation. The RNA was dissolved in 300 μl of RNase-free water, treated with DNase (Ambion, Austin, TX), and further purified for use in the microarray experiments by use of RNeasy columns following the manufacturer's instructions (QIAGEN, Valencia, CA).
cDNA synthesis.
Ten μg of purified RNA was reverse transcribed using random hexamers, a mixture of dNTPs and aminoallyl-labeled dUTP, and Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. After reverse transcription, 10-μl portions of 0.5 M EDTA and 1 N NaOH were added to inactivate the enzyme and hydrolyze the RNA, respectively, and the mixture was incubated at 65°C for 15 min. After the addition of 10μl 1N HCl and 40 μl water, cDNA was purified using a QIAGEN MinElute kit as described by the manufacturer with the following modification: cDNA bound to the column was washed with 75% ethanol rather than manufacturer-supplied PE buffer and eluted with 10 μl of water. PE buffer may contain free amines that would compete with the coupling of the Cy5 and Cy3 dyes to the aminoallyl-labeled nucleotides. The purified cDNA was quantified by measuring absorbance at 260 nm. We routinely obtained approximately 1 to 2 μg of cDNA by this method.
Coupling of Cy3 and Cy5 to cDNA.
All reactions with dyes were maintained in the dark when possible. cDNA was coupled to monoreactive Cy3 or Cy5 dye (Amersham) in the presence of 0.05 M NaHCO3 for 1 h at room temperature. The reaction was quenched by incubation with 1.2 M hydroxylamine, and the labeled cDNA was purified by use of Qiaquick columns (QIAGEN). The efficiencies of incorporation were determined by measuring absorbances at 260 and 650 nm for Cy5 and at 260 and 550 nm for Cy3. The Cy3- and Cy5-labeled cDNAs were combined such that they contained equal amounts of incorporated dyes. The mixture was dried using a SpeedVac system.
Array printing.
Microarrays were printed on UltraGAPS-coated slides (Corning, Acton, MA) using a Biorobotics MicroGrid II system. Each array consisted of 4,000 spots containing 70-mers designed to correspond to unique sequences in the open reading frames (ORFs) of the V. cholerae genome as well as to positive and negative controls (Illumina, San Diego, CA). Each slide contained two microarrays.
Preparation of cDNA for hybridization.
The labeled and dried cDNA was dissolved in 27 μl of hybridization solution (250 μl formamide, 250 μl 20× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 10 μl 10% sodium dodecyl sulfate [SDS], 490 μl water). After the addition of 3 μl of blocking solution (10 μl 10-mg/ml salmon sperm DNA, 20 μl 10-mg/ml tRNA, 20 μl water), the mixture was incubated in boiling water for 3 min, snap-chilled on ice for 30 s, and kept at room temperature. Before it was applied to the microarray for hybridization, it was centrifuged for 5 min at 13,000 rpm to remove any debris.
Microarray hybridization.
Arrays were prepared for hybridization by incubation in 50 ml prehybridization solution (12.5 ml 20× SSC, 500 μl 10% SDS, 1 g bovine serum albumin, and 37 ml water filtered through 0.22-μm syringe filters; Fisher) at 42°C for 45 min. The slides were rinsed with water and subsequently with isopropanol. Labeled cDNAs were applied to the microarrays and covered with a lifter slip. The microarrays were then incubated with the labeled cDNAs in hybridization chambers for 48 h in 42°C in a water bath. After hybridization, the slides were successively washed at room temperature for 5 min in solution 1 (2× SSC, 0.2% SDS), 10 min in solution 2 (0.1× SSC, 0.2% SDS), and three times for 1 min each in solution 3 (0.1× SSC) and scanned.
Image quantitation and data analysis.
Microarrays were scanned for Cy5 and Cy3 fluorescence intensities using a Packard ScanArray 4000 system. cDNAs synthesized from wild-type V. cholerae and the ΔmbaA mutant were labeled twice with Cy3 and twice with Cy5 each, so as not to introduce biases due to any possible differences between the dyes. Spot intensities and qualities were calculated using Imagene version 5.6.1 (BioDiscovery, El Segundo, CA). Gene transcription levels were calculated from background-subtracted fluorescence intensities. These data were normalized using GeneSpring version 6.1 (Silicon Genetics, Redwood City, CA) with the intensity-dependent Lowess normalization. Genes were scored as differentially regulated if the ratio of the mutant to wild-type transcripts was >1.5 and <0.67 in three out of four technical replicates.
RESULTS
Deletion of the chromosomal locus VC0704 decreases V. cholerae biofilm formation.
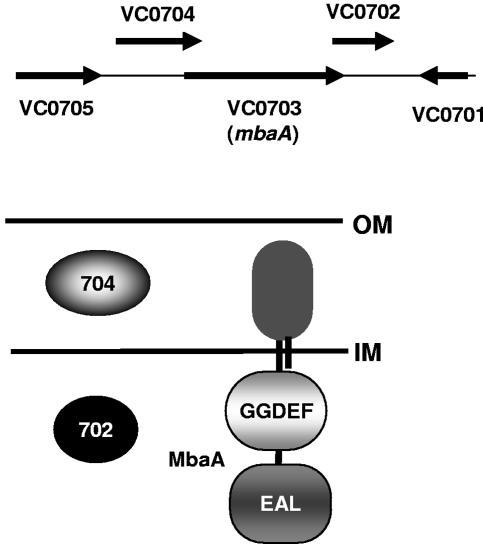
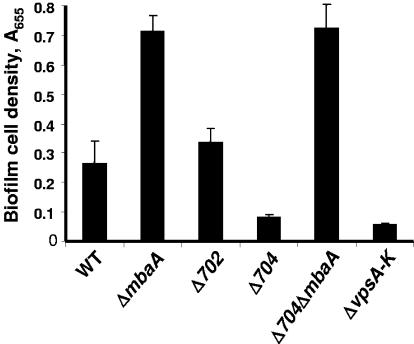
MbaA, a protein named for its role in the maintenance of biofilm architecture, was previously identified as a repressor of biofilm formation in a transposon mutagenesis screen designed to select biofilm-altered V. cholerae O1 El Tor mutants (3). mbaA, which is located on the large chromosome of V. cholerae at locus VC0703, is predicted to code for an integral membrane protein comprised of a periplasmic domain as well as cytoplasmic GGDEF and EAL domains. The ORF of mbaA is flanked by overlapping ORFs at loci VC0702 and VC0704. These genes are predicted to encode a cytoplasmic protein and a periplasmic protein, respectively (Fig. 1). The genomic arrangement of these three genes suggested to us that they might function together to regulate biofilm formation. To test this hypothesis, we constructed V. cholerae O139 mutants carrying in-frame deletions in each gene of this putative operon and quantified the growth rates of these mutants as well as their propensities to attach to surfaces and accumulate in biofilms. The growth rate of each mutant as well as its ability to attach to a surface and form a monolayer was similar to that of wild-type V. cholerae (data not shown); however, differences in biofilm formation were observed. (Fig. 2). As had previously been reported for the V. cholerae O1 El Tor strain N16961, the V. cholerae O139 ΔmbaA mutant demonstrated an increased propensity to accumulate in a biofilm. In contrast, surface accumulation by the ΔVC0704 mutant was greatly reduced compared to that of wild-type V. cholerae and was just slightly more than that of a ΔvpsA-vpsK deletion mutant. Surface accumulation by the ΔVC0702 deletion mutant was consistently slightly more than that of wild-type V. cholerae, but this difference was not always statistically significant (P = 0.1). We hypothesize that the environmental conditions utilized in these experiments may not highlight the role of VC0702 in the regulation of biofilm formation.
FIG. 1.
Genomic arrangement of VC0702, mbaA, and VC0704 and predicted cellular locations of the proteins these genes encode. Horizontal lines represent the bacterial inner membrane (IM) and outer membrane (OM).
FIG. 2.
Quantification of wild-type and mutant V. cholerae biofilms. Wild-type V. cholerae (WT) as well as ΔmbaA, ΔVC0702, ΔVC0704, ΔVC0704ΔmbaA, and ΔvpsA-vpsK (ΔvpsA-K) mutant biofilms were formed in borosilicate tubes overnight in LB broth and quantified as described in Materials and Methods. Error bars show standard deviations of three replicates.
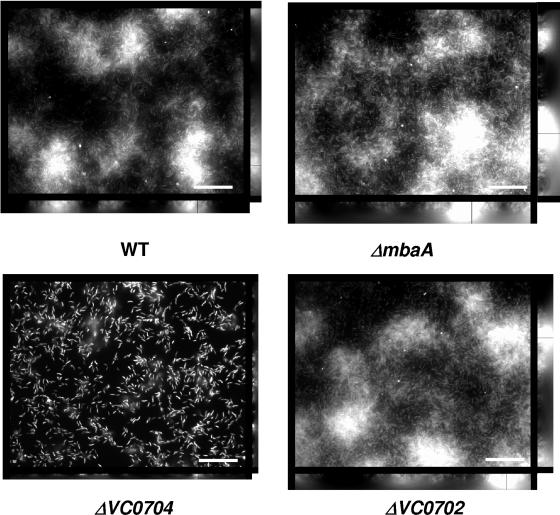
Fluorescence microscopy was used to visualize the architectures of biofilms made by wild-type V. cholerae as well as by the three mutants described above (Fig. 3). As previously demonstrated in the El Tor background, the height of the ΔmbaA mutant biofilm was greater than that of the biofilm formed by wild-type V. cholerae. In contrast, the ΔVC0704 mutant biofilm was only a few cells in depth. The height of the ΔVC0702 mutant biofilm was similar to that of the wild-type V. cholerae biofilm. These results confirm that MbaA is a repressor of biofilm formation in V. cholerae O139 and suggest that the protein encoded at locus VC0704 is an activator of biofilm formation.
FIG. 3.
Architecture of wild-type V. cholerae and mutant biofilms. Transverse and vertical cross sections through DAPI-stained wild-type V. cholerae (WT) as well as ΔmbaA, ΔVC0702, and ΔVC0704 mutant biofilms formed after overnight incubation in LB broth. The transverse sections were obtained at the level of the substratum. (Bar ≈ 10 μm.)
We hypothesized that the protein encoded at locus VC0704, a putative periplasmic protein, might interact with the periplasmic domain of MbaA to block the repression of biofilm formation by MbaA. We predicted that if this were the case, then the deletion of VC0704 would have no effect on biofilm accumulation in the absence of MbaA. To test this prediction, we constructed a ΔmbaAΔVC0704 double mutant and quantified its ability to accumulate in a biofilm. As shown in Fig. 2, the ΔmbaAΔVC0704 double mutant displayed a phenotype similar to that of the ΔmbaA mutant. This is consistent with the hypothesis that protein encoded at locus VC0704 modulates the regulation of biofilm development by MbaA.
Norspermidine enhances biofilm formation in a VC0704-dependent manner.
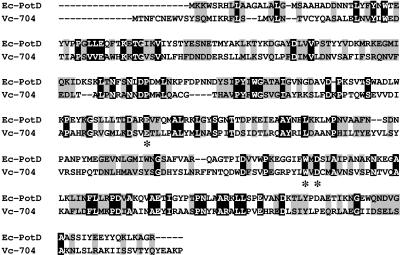
Numerous studies have shown that V. cholerae biofilm formation is an environmentally regulated process. We hypothesized that the VC0704-encoded protein might serve as the ligand-binding sensory component of the MbaA regulatory system. In order to identify potential ligands for the VC0704-encoded protein through homologies with other bacterial proteins, a BLAST search was performed. This demonstrated 22% sequence identity and 44% similarity at the amino acid level between the VC0704-encoded protein and PotD, the periplasmic component of the Escherichia coli ABC-type spermidine transport system (Fig. 4). Furthermore, three amino acids that have been shown to be essential for spermidine binding by PotD (E171, W255, and D257) are conserved in the VC0704-encoded protein as E173, W261, and D263 (38).
FIG. 4.
Multiple sequence alignment of E. coli PotD and the protein encoded by V. cholerae VC0704. Identical residues are boxed in black, conserved residues are boxed in gray, and nonconserved residues are shown on a white background. The positions of three conserved amino acids (E173, W261, D263) are depicted with asterisks below the sequences. Sequence analysis and alignment was done using the online tools at Biology Workbench (http://workbench.sdsc.edu). Ec, E. coli; Vc, V. cholerae.
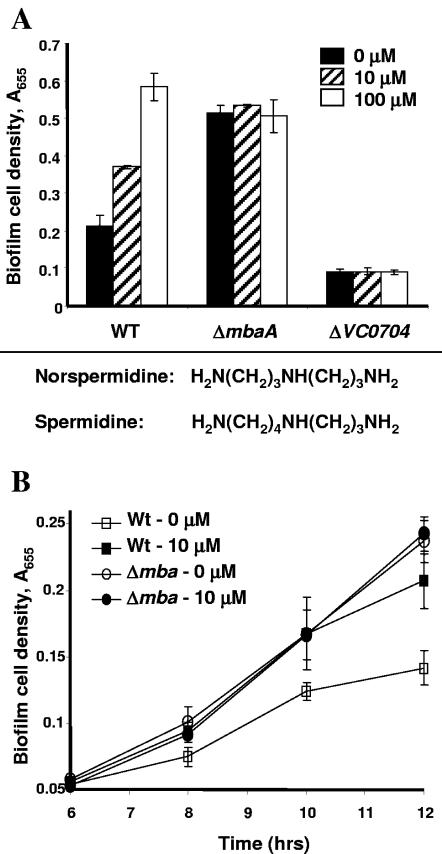
We first tested the effect of spermidine, the ligand of PotD, on V. cholerae biofilm formation. In wild-type V. cholerae and the ΔmbaA mutant, the addition of exogenous spermidine at concentrations greater than or equal to 1 mM resulted in decreases in biofilm formation (data not shown). No effect was seen at lower concentrations of spermidine. We were unable to document an effect of spermidine on the ΔVC0704 mutant biofilm, most likely because the biofilm formed by this mutant is already quite minimal. The high exogenous concentration of spermidine required to cause an effect on biofilm formation led us to hypothesize that a related polyamine might alter V. cholerae biofilm formation at lower concentrations. Because norspermidine is very similar to spermidine, differing only by one carbon residue, we tested the effect of norspermidine on V. cholerae biofilm formation. As shown in Fig. 5A, wild-type V. cholerae biofilm formation was enhanced at concentrations of norspermidine as low as 10 μM, and, at 100-μM concentrations, a threefold increase in biofilm formation was observed. In contrast, the addition of norspermidine to the growth medium had no effect on the biofilms formed by the ΔVC0704 and ΔmbaA mutants. We questioned whether norspermidine might affect biofilm formation by a ΔmbaA mutant at earlier times in biofilm development. To address this possibility, we quantified biofilm development by wild-type V. cholerae and a ΔmbaA mutant in the presence and absence of 10 μM norspermidine after 6, 8, 10, and 12 h of incubation (Fig. 5B). In these experiments, the activation of wild-type V. cholerae biofilm formation by norspermidine was observed after only 8 h. In contrast, even at early times, norspermidine had no effect on biofilm formation by the ΔmbaA mutant. Planktonic cell densities were unaffected by the addition of norspermidine in all the experiments performed (data not shown). These results suggest that the protein encoded at locus VC0704 specifically binds norspermidine and that this interaction results in the activation of biofilm formation. For this reason, we have named the gene at locus VC0704 nspS (for norspermidine sensor). Furthermore, because norspermidine had no effect on the biofilm formed by ΔmbaA, we hypothesize that MbaA also forms part of this regulatory pathway.
FIG. 5.
Effect of norspermidine on biofilm cell densities. A. Effect of norspermidine on surface accumulation by wild-type V. cholerae (WT) as well as on that by ΔmbaA and ΔVC0704 mutants. Biofilms were formed in borosilicate tubes in LB broth for 20 h with indicated quantities of norspermidine and quantified as described in Materials and Methods. Error bars show standard deviations of three replicates. The chemical formulas of norspermidine and spermidine are shown below the graph. B. Time course of wild-type V. cholerae and ΔmbaA mutant biofilm formation in the presence and absence of norspermidine. Wild-type or ΔmbaA mutant biofilms were formed in borosilicate tubes in LB broth alone or supplemented with 10 μM norspermidine and quantified after 6, 8, 10, or 12 h. Error bars show standard deviations of three replicates.
Even in LB broth alone, the NspS mutant biofilm was decreased in comparison to the wild-type biofilm. One possibility is that LB broth itself may contain enough norspermidine to activate this signal transduction pathway. Alternatively, while norspermidine may enhance the function of NspS in activating biofilm formation, it is possible that NspS has a basal level of function even in the absence of norspermidine.
Modulation of vps gene transcription contributes to the biofilm phenotypes of the ΔmbaA and ΔnspS mutants.
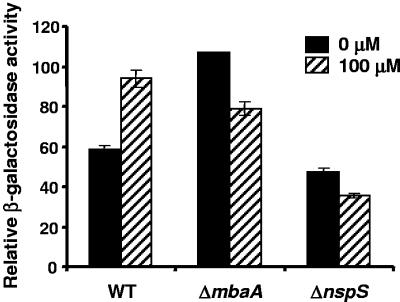
Wild-type V. cholerae biofilm formation in freshwater, monosaccharide-supplemented minimal medium, and LB is dependent on vps gene transcription (40, 52). Furthermore, most transcriptional regulators that alter V. cholerae biofilm formation effect this change through the regulation of vps gene transcription (4, 24, 25). Thus, we hypothesized that ΔmbaA and ΔnspS mutants would also show altered levels of vps gene transcription. To test this hypothesis, ΔmbaA and ΔnspS mutations were inserted into the chromosome of a previously constructed V. cholerae strain carrying a chromosomal promoter fusion of the vpsL operon to the E. coli lacZ gene, and transcription at the vpsL promoter was measured in the resulting strains (25). As shown in Fig. 6, the level of vpsL transcription in the ΔmbaA mutant was twice that observed in wild-type V. cholerae. The level of vpsL transcription in the ΔnspS mutant showed a small but statistically significant decrease (P = 0.01). This result is consistent with our hypothesis that ΔnspS and ΔmbaA mutants have altered vps gene transcription levels. The addition of norspermidine increased vpsL transcription in wild-type V. cholerae almost to the level observed in the ΔmbaA mutant. This result suggests that norspermidine interacts with NspS to relieve the repression of vpsL transcription by MbaA. Interestingly, norspermidine repressed vpsL transcription in the ΔmbaA and ΔnspS mutants.
FIG. 6.
vpsL transcription in wild-type and mutant V. cholerae strains. Wild-type V. cholerae (WT) as well as ΔmbaA and ΔnspS mutants containing a chromosomal fusion of the E. coli lacZ gene to the vpsL promoter were grown to logarithmic phase with shaking in the absence (black bars) or presence (striped bars) of 100 μM norspermidine. β-Galactosidase measurements were done on the planktonic cultures as described in Materials and Methods. Error bars show standard deviations.
The NspS-MbaA regulatory pathway specifically regulates V. cholerae biofilm formation.
Proteins in the GGDEF and EAL protein families, such as MbaA, have been shown to affect intracellular levels of c-di-GMP, which is thought to be a secondary messenger (35). Secondary messengers often have global effects on gene transcription. Thus, we questioned whether other V. cholerae genes might be regulated by MbaA. To identify additional genes regulated by MbaA, we compared the whole-genome transcriptional profiles of wild-type V. cholerae and of a ΔmbaA mutant grown to logarithmic phase in shaking cultures. Data were collected from four separate microarray experiments on RNA extracted on different days (four technical replicates). In an analysis of the microarray data, the gene transcription in the ΔmbaA mutant was used as the test and the gene transcription in wild-type V. cholerae was used as the reference. We identified 23 genes with 1.5-fold-or-greater increases in transcription in the ΔmbaA mutant and 6 genes with 1.5-fold-or-greater decreases in transcription (Table 3). As predicted from our vpsL reporter fusion experiments, 14 of the genes demonstrating transcriptional activation in the ΔmbaA mutant were located in or between the two vps operons. Genes at loci VC1585 (katB), VC1888, VCA0849, and VCA0952 (vpsT), whose transcriptions were also found to beincreased in previous microarray studies of a V. cholerae rugose-phase variant (76), demonstrated increased transcription in the ΔmbaA mutant. VC0930 and VC1888, both of which demonstrated transcriptional activation in our microarray study, are annotated as encoding putative hemolysins. Because these genes encode secreted proteins, we hypothesize that they may contribute to the formation of the biofilm matrix. VC1585, which is 59% identical and 72% similar to the P. aeruginosa katB gene, was also activated in our microarray studies. The P. aeruginosa katB gene has been shown to be induced in biofilms (17). In this work, the catalase KatA was shown to be mainly responsible for the resistance of P. aeruginosa biofilms to H2O2. Because V. cholerae does not appear to encode KatA, we propose that KatB may account for the resistance of the rugose V. cholerae variant to H2O2 (69, 77). Two response regulators, VpsR and VpsT, regulate biofilm formation (4, 75). Transcription of vpsT but not of vpsR was found to be increased in the ΔmbaA mutant. VC1216, a gene predicted to encode a GGDEF protein with an N-terminal hemerythrin histidine-histidine-glutamate cation-binding domain, was also upregulated in the ΔmbaA mutant. These domains, which usually bind oxygen, have been shown to be important when low oxygen concentrations are encountered (66). The induction of VC1216 suggests that low oxygen tensions in the environment may also direct biofilm formation. The majority of genes identified in our microarray study had increased transcription levels. Furthermore, approximately 84% of these genes are likely to be directly involved in biofilm formation. Thus, we conclude that the NspS-MbaA regulatory pathway does not have global effects on gene transcription but more likely is specifically regulating V. cholerae biofilm formation.
TABLE 3.
Genes differentially regulated in ΔmbaA relative to wild-type V. cholerae
| Locus (gene name[s])a | Predicted function | Fold change |
|---|---|---|
| VC0916 | Phosphotyrosine protein phosphatase | 1.7-5 |
| VC0918 (vpsB, epsD) | UDP-N-acetyl-d-mannosaminuronic acid dehydrogenase | 2.7-5.4 |
| VC0919 (vpsC) | Serine acetyltransferase-related protein | 1.6-6.3 |
| VC0921 (vpsE) | Polysaccharide export protein, putative | 1.7-4.8 |
| VC0922* (vpsF) | Hypothetical protein | 1.5-4 |
| VC0926 (vpsJ) | Hypothetical protein | 1.5-5.6 |
| VC0927 (vpsK, cpsF) | UDP-N-acetyl-d-mannosamine transferase | 1.5-6.1 |
| VC0928* | Hypothetical protein | 1.9-6.2 |
| VC0930* | Hemolysin-related protein | 2.1-2.6 |
| VC0932* | Hypothetical protein | 1.5-15.6 |
| VC0933* | Hypothetical protein | 1.8-16.2 |
| VC0934 (vpsL) | Capsular polysaccharide biosynthesis glycosyltransferase, putative | 1.5-7 |
| VC0935* (vpsM) | Hypothetical protein | 2.1-18.6 |
| VC0936 (vpsN) | Polysaccharide export-related protein | 2.2-6.1 |
| VC1216 | GGDEF family protein, heme-erythrin domain | 1.6-2.2 |
| VC1585 (katB) | Catalase | 2-2.5 |
| VC1888* | Hemolysin-related protein | 1.8-6.4 |
| VC2134 (fliE) | Flagellar hook-basal body complex protein FliE | 1.7-2.3 |
| VC2455 | Hypothetical protein | 1.5-1.7 |
| VC2456 | Hypothetical protein | 1.8-2.2 |
| VCA0470 | Acetyltransferase, putative | 1.6-2.4 |
| VCA0480* | Hypothetical protein | 1.5-2.1 |
| VCA0849a | Hypothetical protein, bp 6531-9792 | 2.1-2.7 |
| VCA0952* (vpsT) | Transcriptional regulator, LuxR family | 1.5-5.5 |
| VC2046 | Hypothetical protein | 0.4-0.49 |
| VC2735 | Conserved hypothetical protein | 0.53-0.64 |
| VCA0188 | Hypothetical protein | 0.55-0.65 |
| VCA0689 | Conserved hypothetical protein | 0.51-0.57 |
| VCA0734 | Hypothetical protein | 0.39-0.6 |
*, differentially regulated in all four experiments.
DISCUSSION
In bacteria, changes in gene transcription are usually the result of the sensing of a signal in the environment. The genome of V. cholerae codes for 31 proteins with GGDEF domains, 12 proteins with EAL domains, and 10 that contain both GGDEF and EAL domains. More than half of these are linked to sensory or signaling domains. Despite the growing number of functionally characterized proteins in the GGDEF and EAL families, the signals that they process remain largely unknown.
In previous work, Bomchil and colleagues demonstrated that the protein MbaA is a repressor of V. cholerae biofilm formation (3). However, the environmental signal and the sensory component of the MbaA signal transduction system were not identified. In this work, we have identified norspermidine as the environmental signal sensed by the MbaA regulatory cascade. Furthermore, we present evidence that the protein NspS is the sensory component of this signal transduction system. We propose that NspS interacts with the periplasmic domain of MbaA to regulate its enzymatic activity and that this interaction is modulated by binding of norspermidine to NspS. MbaA contains both GGDEF and EAL domains. Both of these domains have been shown to dimerize, and dimerization is thought to be required for the activity of the GGDEF domain (2, 5). Interestingly, PotD, a protein that is homologous to NspS, forms a dimer that is disrupted in the presence of its ligand, spermidine (62). Thus, we hypothesize that one possible mechanism by which the norspermidine-NspS complex decreases the repression of biofilm formation by MbaA may be through the destabilization of the MbaA dimer. Alternatively, the binding of norspermidine to NspS may induce a conformational change in MbaA that reduces its function. Experiments to test these various possibilities directly are under way.
We have also demonstrated that exogenous norspermidine results in an increase of vpsL gene expression in wild-type V. cholerae but a decrease in vpsL gene expression in the absence of NspS or MbaA. We hypothesize that norspermidine affects biofilm formation through multiple pathways. While the NspS-MbaA pathway is dominant in wild-type V. cholerae, alternative pathways may predominate in the absence of a functional NspS-MbaA pathway.
Polyamines such as spermidine, putrescine, and spermine are abundant in nature, and their roles in a wide variety of cellular processes have been studied extensively. Polyamines have been previously shown to act as intercellular signaling molecules both in prokaryotes and eukaryotes. For example, putrescine, a diamine, can act as an extracellular signal that is required for the swarming cell differentiation and migration ability of Proteus mirabilis (61). In eukaryotes, extracellular polyamines can increase fluid secretion in rat distal colonic crypts via their interactions with the calcium-sensing receptors, which in turn affects levels of cyclic secondary messengers in the cell (8). Furthermore, the intestinal polyamine pool, which may be modulated by diet and the commensal bacterial population, affects epithelial cell development and apoptosis (44, 48). Norspermidine has also been shown to have inhibitory effects on immunoglobulin M production by lipopolysaccharide-stimulated murine splenocytes by interfering with the polyamine metabolism pathway (42). Thus, commensal or pathogenic gut bacteria may influence gut epithelial development and immunity through the synthesis and export of polyamines.
V. cholerae contains very small amounts of spermidine, a polyamine found in species as diverse as humans, yeast (Saccharomyces cerevisiae), and E. coli. Instead, the major polyamine in Vibrio species is norspermidine (74). The genes for norspermidine synthesis are present in the genomes of V. cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus as well as in those of many other members of the proteobacteria and of a few members of the deinococci, bacteroidetes, cyanobacteria, and gram-positive bacteria (7, 27, 47). Furthermore, norspermidine has been identified as a major polyamine in a number of species of archaea and thermophilic bacteria (19, 20, 30). Norspermidine has also been identified as a major polyamine in a number of aquatic plants and in other aquatic organisms such as algae, bivalve mollusks, sea urchins, and sea cucumbers as well as in terrestrial plants and insects (18, 21-23). These studies suggest that norspermidine is present in a wide range of prokaryotes and eukaryotes.
Relatively little is known about the role of norspermidine in Vibrio species. Vibriobactin and vulnibactin, the iron-scavenging molecules of V. cholerae and V. vulnificus, respectively, contain norspermidine, suggesting an essential role for this molecule in iron-limiting environments (39, 55). It is also likely that norspermidine is essential for normal growth in these organisms, as is the case for spermidine or spermine in Saccharomyces cerevisiae (1).
Our results suggest that norspermidine promotes V. cholerae surface accumulation. In seawater, the concentration of norspermidine is in the nanomolar range (54). Therefore, global pools of norspermidine in the marine environment would not be sufficient to promote the association of V. cholerae with biotic or abiotic surfaces. However, it is likely that V. cholerae associates with species of bacteria, archaea, plants, and animals that maintain norspermidine gradients at their surfaces. Thus, we suggest that polyamines serve as a form of communication that enhances close associations between V. cholerae and its prokaryotic and eukaryotic neighbors.
Acknowledgments
We thank Natalia Bomchil and Roberto Kolter for generous sharing of plasmid pNB4, which was used to make the ΔmbaA mutant. We thank Kate Pflughoeft for constructing the ΔmbaA mutant. We thank Anne Kane of the Tufts-NEMC GRASP Center and her staff for their expert preparation of many reagents. We thank Lan Wei of the Tufts Microarray Core Facility for help with the printing and scanning of microarray slides, Dagmar Kapfhammer for optimization of microarray experiments, and Emily Lyettefi for assistance with analysis of microarray data.
This work was supported by an award from the Ellison Medical Foundation, the Tufts-NEMC GRASP Center NIH/NIDDK, P30 DK34928 and NIH R01 AI50032 to P.I.W.
REFERENCES
- 1.Balasundaram, D., C. W. Tabor, and H. Tabor. 1991. Spermidine or spermine is essential for the aerobic growth of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88:5872-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bobrov, A. G., O. Kirillina, and R. D. Perry. 2005. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247:123-130. [DOI] [PubMed] [Google Scholar]
- 3.Bomchil, N., P. Watnick, and R. Kolter. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 185:1384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 186:1574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, C., R. Paul, D. Samoray, N. C. Amiot, B. Giese, U. Jenal, and T. Schirmer. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. USA 101:17084-17089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Y., S. Vujcic, P. Liang, P. Diegelman, D. L. Kramer, and C. W. Porter. 2003. Genomic identification and biochemical characterization of a second spermidine/spermine N1-acetyltransferase. Biochem. J. 373:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, S. X., J. P. Geibel, and S. C. Hebert. 2004. Extracellular polyamines regulate fluid secretion in rat colonic crypts via the extracellular calcium-sensing receptor. Gastroenterology 126:148-158. [DOI] [PubMed] [Google Scholar]
- 9.Colwell, R. R., and A. Huq. 1994. Environmental reservoir of Vibrio cholerae. The causative agent of cholera. Ann. N. Y. Acad. Sci. 740:44-54. [DOI] [PubMed] [Google Scholar]
- 10.Colwell, R. R., and W. M. Spira. 1992. The ecology of Vibrio cholerae, p. 107-127. In D. Barua and W. B. I. Greenough (ed.), Cholera. Plenum, New York, N.Y.
- 11.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 12.Costerton, J. W., Z. Lewandowski, D. DeBeer, D. Caldwell, D. Korber, and G. James. 1994. Biofilms, the customized microniche. J. Bacteriol. 176: 2137-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danese, P. N., L. A. Pratt, and R. Kolter. 2001. Biofilm formation as a developmental process. Methods Enzymol. 336:19-26. [DOI] [PubMed] [Google Scholar]
- 14.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Argenio, D. A., M. W. Calfee, P. B. Rainey, and E. C. Pesci. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184:6481-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elasri, M. O., and R. V. Miller. 1999. Study of the response of a biofilm bacterial community to UV radiation. Appl. Environ. Microbiol. 65: 2025-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elkins, J. G., D. J. Hassett, P. S. Stewart, H. P. Schweizer, and T. R. McDermott. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl. Environ. Microbiol. 65: 4594-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamana, K., T. Aizaki, E. Arai, A. Saito, K. Uchikata, and H. Ohnishi. 2004. Distribution of norspermidine as a cellular polyamine within micro green algae including non-photosynthetic achlorophyllous Polytoma, Polytomella, Prototheca and Helicosporidium. J. Gen. Appl. Microbiol. 50:289-295. [DOI] [PubMed] [Google Scholar]
- 19.Hamana, K., and T. Itoh. 2001. Polyamines of the hyperthermophilic archaebacteria belonging to the genera Thermococcus and Methanothermus and two new genera Caldivirga and Palaeococcus. Microbios 104:105-114. [PubMed] [Google Scholar]
- 20.Hamana, K., M. Niitsu, K. Samejima, and T. Itoh. 2001. Polyamines of the thermophilic eubacteria belonging to the genera Thermosipho, Thermaerobacter and Caldicellulosiruptor. Microbios 104:177-185. [PubMed] [Google Scholar]
- 21.Hamana, K., M. Niitsu, K. Samejima, and S. Matsuzaki. 1991. Novel tetraamines, pentaamines, and hexaamines in sea-urchin, sea-cucumber, sea squirt, and bivalves. Comp. Biochem. Physiol. B 100:59-62. [Google Scholar]
- 22.Hamana, K., M. Niitsu, and K. Samejima. 1998. Unusual polyamines in aquatic plants: the occurrence of homospermidine, norspermidine, thermospermine, norspermine, aminopropylhomospermine, bis(aminopropyl)ethanediamine, methylspermidine. Can. J. Bot. 76:130-133. [Google Scholar]
- 23.Hamana, K., H. Uemiya, and M. Niitsu. 2004. Polyamines of primitive apterygotan insects: springtails, silverfish and a bristletail. Comp. Biochem. Physiol. B 137:75-79. [DOI] [PubMed] [Google Scholar]
- 24.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 25.Haugo, A. J., and P. I. Watnick. 2002. Vibrio cholerae CytR is a repressor of biofilm development. Mol. Microbiol. 45:471-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hecht, G. B., and A. Newton. 1995. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J. Bacteriol. 177:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 29.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 30.Hosoya, R., K. Hamana, M. Niitsu, and T. Itoh. 2004. Polyamine analysis for chemotaxonomy of thermophilic eubacteria: polyamine distribution profiles within the orders Aquificales, Thermotogales, Thermodesulfobacteriales, Thermales, Thermoanaerobacteriales, Clostridiales and Bacillales. J. Gen. Appl. Microbiol. 50:271-287. [DOI] [PubMed] [Google Scholar]
- 31.Huo, A., B. Xu, M. A. Chowdhury, M. S. Islam, R. Montilla, and R. R. Colwell. 1996. A simple filtration method to remove plankton-associated Vibrio cholerae in raw water supplies in developing countries. Appl. Environ. Microbiol. 62:2508-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huq, A., E. B. Small, P. A. West, M. I. Huq, R. Rahman, and R. R. Colwell. 1983. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 45:275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Islam, M. S., B. S. Drasark, and D. J. Bradley. 1989. Attachment of toxigenic Vibrio cholerae O1 to various freshwater plants and survival with a filamentous green alga, Rhizoclonium fontanum. J. Trop. Med. Hyg. 92:396-401. [PubMed] [Google Scholar]
- 34.Iyer, R., and A. H. Delcour. 1997. Complex inhibition of OmpF and OmpC bacterial porins by polyamines. J. Biol. Chem. 272:18595-18601. [DOI] [PubMed] [Google Scholar]
- 35.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185-191. [DOI] [PubMed] [Google Scholar]
- 36.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023-1034. [DOI] [PubMed] [Google Scholar]
- 37.Jung, I. L., and I. G. Kim. 2003. Transcription of ahpC, katG, and katE genes in Escherichia coli is regulated by polyamines: polyamine-deficient mutant sensitive to H2O2-induced oxidative damage. Biochem. Biophys. Res. Commun. 301:915-922. [DOI] [PubMed] [Google Scholar]
- 38.Kashiwagi, K., R. Pistocchi, S. Shibuya, S. Sugiyama, K. Morikawa, and K. Igarashi. 1996. Spermidine-preferential uptake system in Escherichia coli. Identification of amino acids involved in polyamine binding in PotD protein. J. Biol. Chem. 271:12205-12208. [DOI] [PubMed] [Google Scholar]
- 39.Keating, T. A., C. G. Marshall, and C. T. Walsh. 2000. Vibriobactin biosynthesis in Vibrio cholerae: VibH is an amide synthase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry 39:15513-15521. [DOI] [PubMed] [Google Scholar]
- 40.Kierek, K., and P. I. Watnick. 2003. Environmental determinants of Vibrio cholerae biofilm development. Appl. Environ. Microbiol. 69:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirillina, O., J. D. Fetherston, A. G. Bobrov, J. Abney, and R. D. Perry. 2004. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 54:1134. [DOI] [PubMed] [Google Scholar]
- 42.Komori, T., and Y. Ohsugi. 1991. Norspermidine inhibits LPS-induced immunoglobulin production in an FCS-independent mechanism different from spermidine and spermine. Int. J. Immunopharmacol. 13:67-73. [DOI] [PubMed] [Google Scholar]
- 43.Lauriano, C. M., C. Ghosh, N. E. Correa, and K. E. Klose. 2004. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J. Bacteriol. 186:4864-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li, L., J. N. Rao, B. L. Bass, and J. Y. Wang. 2001. NF-kappaB activation and susceptibility to apoptosis after polyamine depletion in intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 280:G992-G1004. [DOI] [PubMed] [Google Scholar]
- 45.Lopatin, A. N., E. N. Makhina, and C. G. Nichols. 1994. Potassium channel block by cytoplasmic polyamines as the mechanism of intrinsic rectification. Nature 372:366-369. [DOI] [PubMed] [Google Scholar]
- 46.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 47.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 48.McCormack, S. A., and L. R. Johnson. 1991. Role of polyamines in gastrointestinal mucosal growth. Am. J. Physiol. 260:G795-G806. [DOI] [PubMed] [Google Scholar]
- 49.Metcalf, W. W., W. Jiang, L. L. Daniels, S. K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZ a for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 50.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 51.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moorthy, S., and P. I. Watnick. 2004. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol. Microbiol. 52:573-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morgan, J. E., J. W. Blankenship, and H. R. Matthews. 1987. Polyamines and acetylpolyamines increase the stability and alter the conformation of nucleosome core particles. Biochemistry 26:3643-3649. [DOI] [PubMed] [Google Scholar]
- 54.Nishibori, N., A. Nishii, and H. Takayama. 2001. Detection of free polyamine in coastal seawater using ion exchange chromatography. ICES J. Mar. Sci. 58:1201-1207. [Google Scholar]
- 55.Okujo, N., M. Saito, S. Yamamoto, T. Yoshida, S. Miyoshi, and S. Shinoda. 1994. Structure of vulnibactin, a new polyamine-containing siderophore from Vibrio vulnificus. Biometals 7:109-116. [DOI] [PubMed] [Google Scholar]
- 56.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 57.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rashid, M. H., C. Rajanna, A. Ali, and D. K. Karaolis. 2003. Identification of genes involved in the switch between the smooth and rugose phenotypes of Vibrio cholerae. FEMS Microbiol. Lett. 227:113-119. [DOI] [PubMed] [Google Scholar]
- 59.Shukla, B. N., D. V. Singh, and S. C. Sanyal. 1995. Attachment of non-culturable toxigenic Vibrio cholerae O1 and non-O1 and Aeromonas spp. to the aquatic arthropod Gerris spinolae and plants in the River Ganga, Varanasi. FEMS Immunol. Med. Microbiol. 12:113-120. [DOI] [PubMed] [Google Scholar]
- 60.Stanley, N. R., and B. A. Lazazzera. 2004. Environmental signals and regulatory pathways that influence biofilm formation. Mol. Microbiol. 52: 917-924. [DOI] [PubMed] [Google Scholar]
- 61.Sturgill, G., and P. N. Rather. 2004. Evidence that putrescine acts as an extracellular signal required for swarming in Proteus mirabilis. Mol. Microbiol. 51:437-446. [DOI] [PubMed] [Google Scholar]
- 62.Sugiyama, S., D. G. Vassylyev, M. Matsushima, K. Kashiwagi, K. Igarashi, and K. Morikawa. 1996. Crystal structure of PotD, the primary receptor of the polyamine transport system in Escherichia coli. J. Biol. Chem. 271: 9519-9525. [DOI] [PubMed] [Google Scholar]
- 63.Tabor, C. W., and H. Tabor. 1984. Polyamines. Annu. Rev. Biochem. 53:749-790. [DOI] [PubMed] [Google Scholar]
- 64.Tal, R., H. C. Wong, R. Calhoon, D. Gelfand, A. L. Fear, G. Volman, R. Mayer, P. Ross, D. Amikam, H. Weinhouse, A. Cohen, S. Sapir, P. Ohana, and M. Benziman. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tamplin, M. L., A. L. Gauzens, A. Huq, D. A. Sack, and R. R. Colwell. 1990. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl. Environ. Microbiol. 56:1977-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Throup, J. P., F. Zappacosta, R. D. Lunsford, R. S. Annan, S. A. Carr, J. T. Lonsdale, A. P. Bryant, D. McDevitt, M. Rosenberg, and M. K. Burnham. 2001. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 40:10392-10401. [DOI] [PubMed] [Google Scholar]
- 67.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ulijasz, A. T., A. Grenader, and B. Weisblum. 1996. A vancomycin-inducible LacZ reporter system in Bacillus subtilis: induction by antibiotics that inhibit cell wall synthesis and by lysozyme. J. Bacteriol. 178:6305-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Waldor, M. K., and J. J. Mekalanos. 1994. Emergence of a new cholera pandemic: molecular analysis of virulence determinants in Vibrio cholerae O139 and development of a live vaccine prototype. J. Infect. Dis. 170:278-283. [DOI] [PubMed] [Google Scholar]
- 71.Watnick, P. I., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. Absence of a flagellum leads to altered colony morphology, biofilm development, and virulence in V. cholerae O139. Mol. Microbiol. 39:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamamoto, S., M. A. R. Chowdhury, M. Kuroda, T. Nakano, Y. Kuoumoto, and S. Shinoda. 1990. Further study on polyamine compositions in Vibrionaceae. Can. J. Microbiol. 37:148-153. [DOI] [PubMed] [Google Scholar]
- 75.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPSETr-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497-515. [DOI] [PubMed] [Google Scholar]
- 77.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoshida, M., K. Kashiwagi, G. Kawai, A. Ishihama, and K. Igarashi. 2002. Polyamines enhance synthesis of the RNA polymerase sigma 38 subunit by suppression of an amber termination codon in the open reading frame. J. Biol. Chem. 277:37139-37146. [DOI] [PubMed] [Google Scholar]
- 79.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]