Abstract
Recent work has raised a question as to the involvement of erythrose-4-phosphate, a product of the pentose phosphate pathway, in the metabolism of the methanogenic archaea (R. H. White, Biochemistry 43:7618-7627, 2004). To address the possible absence of erythrose-4-phosphate in Methanocaldococcus jannaschii, we have assayed cell extracts of this methanogen for the presence of this and other intermediates in the pentose phosphate pathway and have determined and compared the labeling patterns of sugar phosphates derived metabolically from [6,6-2H2]- and [U-13C]-labeled glucose-6-phosphate incubated with cell extracts. The results of this work have established the absence of pentose phosphate pathway intermediates erythrose-4-phosphate, xylose-5-phosphate, and sedoheptulose-7-phosphate in these cells and the presence of d-arabino-3-hexulose-6-phosphate, an intermediate in the ribulose monophosphate pathway. The labeling of the d-ara-bino-3-hexulose-6-phosphate, as well as the other sugar-Ps, indicates that this hexose-6-phosphate was the precursor to ribulose-5-phosphate that in turn was converted into ribose-5-phosphate by ribose-5-phosphate isomerase. Additional work has demonstrated that ribulose-5-phosphate is derived by the loss of formaldehyde from d-arabino-3-hexulose-6-phosphate, catalyzed by the protein product of the MJ1447 gene.
Ribose-5-phosphate is generally assumed to arise via the oxidative and nonoxidative pentose phosphate pathways (16). Early labeling studies with the Archaea have indicated that the oxidative part of the pathway is functioning in a number of methanogenic archaea tested (4, 7-9), whereas other data have indicated that the nonoxidative branch of the pathway may be responsible for the biosynthesis of pentoses in Methanococcus voltae (4). Support for the nonoxidative part of the pathway comes from direct enzymatic analysis of the required enzymes in Methanococcus maripaludis (35) and from analyses of methanogen genomes (5, 21, 23, 28). This information indicates that the pentose phosphate pathway is operational in these cells; however, other research suggests that the all or part of the pentose phosphate pathway is nonfunctional in some archaea. Methanothermobacter thermoautotrophicus data excluded the oxidative pathway (10), while Sprott and coworkers concluded that neither the oxidative nor the reductive pentose phosphate pathways were operational in Methanospirillum hungatei (9). They concluded that the ribose-5-phosphate in M. hungatei is derived from carbons 2 through 6 of glucose and is likely formed by oxidative decarboxylation. Daniels and coworkers questioned the operation of the oxidative portion of the pentose phosphate pathway in archaea after failure to detect any glucose-6-phosphate dehydrogenase activity in the three species tested (20). They did, however, consider an oxidative decarboxylation as a possible route to ribose-5-phosphate as observed in M. hungatei (20).
The recent discovery of an erythrose-4-phosphate independent pathway for the biosynthesis of aromatic amino acids in Methanocaldococcus jannaschii (31) questions the necessity of the pentose phosphate pathway in this methanogen (27). Additionally, genes for the nonoxidative pentose phosphate pathway are apparently absent in most archaeal genomes. The nonoxidative part of the pentose-phosphate pathway had been assumed to function in M. jannaschii (23) because initial analysis of the genome indicated several genes that could be involved in catalyzing the individual steps in the cycle. Early work by Whitman et al. (23) stated two possible pathways: (i) a nonoxidative pathway composed of transketolase MJ0679 or MJ0681, transaldolase MJ0960, ribose-5-phosphate epimerase MJ1603, and ribulose-5-phosphate 3-epimerase MJ0680 or (ii) a second pathway involving carboxylation of erythrose-4-phosphate (4, 35).
Genes for the ribulose monophosphate (RuMP) pathway, however, are present in the archaea (25). The RuMP pathway may be functioning as an alternate route to ribose-5-phosphate in these organisms (25). M. jannaschii and M. maripaludis are of particular interest, as they are the only archaea known to have putative genes for both a complete nonoxidative pentose phosphate pathway and an RuMP pathway. The putative M. jannaschii RuMP pathway genes include MJ1247 (3-hexulose-6-phosphate isomerase, phi), MJ1447 (3-oxohexulose-6-phosphate synthase, hps), and MJ1603 (a ribose-5-phosphate isomerase gene). The first enzyme in this pathway, Phi, has been previously characterized, and the crystal structure of the protein has been solved (15). The MJ1447-encoded enzyme, Hps (molecular weight, 42,989), has two domains, one domain of 158 amino acids that is homologous to formaldehyde-activating enzyme (designated the Fae domain) (30) and the other of 223 amino acids that is homologous to 3-hexulose-6-phosphate synthase (33).
Here, we present analytical, enzymatic analyses and metabolic labeling data that show that in M. jannaschii, ribose-5-phosphate is produced exclusively via the RuMP pathway and that these cells do not appear to operate a pentose phosphate pathway. We propose that the function of the Fae domain of Hps in the methanogenic archaea is to catalyze the condensation formaldehyde with tetrahydromethanopterin (H4MPT) to form methylene H4MPT and that the function of ribulose monophosphate synthase domain is to catalyze the loss of the formaldehyde from 3-hexulose-6-phosphate to form ribulose monophosphate, as Verhees et al. and Soderberg and Alver proposed (25, 28).
MATERIALS AND METHODS
Chemicals.
All chemicals were obtained from Aldrich/Sigma unless otherwise indicated. Labeled glucose-6-phosphates were prepared and purified as previously reported (31). Ribulose-5-phosphate was prepared in a final volume of 1 ml containing 100 mM ribose-5-phosphate, 140 U of phosphoriboisomerase type I from spinach (Sigma), and 25 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] buffer (pH 7.5) incubated for 2 h at 30°C.
Preparation of M. jannaschii cell extracts.
A cell extract of M. jannaschii was prepared by sonication of 4.67 g of frozen cells suspended in 10 ml of TES buffer (50 mM TES-K+, 10 mM MgCl2, pH 7.5) under argon for 5 min at 3°C. M. jannaschii cells were grown as previously described (18). The resulting mixture was centrifuged under argon (27,000 × g; 10 min) and stored frozen at −20°C until used. The M. jannaschii extract protein concentration was 30 mg/ml. Protein concentration was measured using the BCA Total Protein assay (Pierce) with bovine serum albumin as a standard.
Cloning and expression of the MJ1447-derived protein in Escherichia coli.
The M. jannaschii gene MJ1447 (Swiss-Prot accession number Q58842) was amplified by PCR from genomic DNA using oligonucleotide primers synthesized by Invitrogen: Fwd (5′-GGTCATATGATAAAATTTGGAG-3′) and Rev (5′-GCTGGATCCTCATAATTCCTC-3′). PCR was performed as described previously using a 50°C annealing temperature (12). The primers introduced an NdeI site at the 5′ end and a BamHI site at the 3′ end. The amplified PCR product was purified with a QIAQuick spin column (Invitrogen), digested with restriction enzymes NdeI and BamHI, and then ligated into the compatible sites in plasmid pT 7-7 (USB). T4 DNA ligase (Invitrogen) was used to make the recombinant plasmid pMJ1447. DNA sequences were verified by dye-terminator sequencing at the Virginia Bioinformatics Institute's DNA facility. The resulting plasmid, pMJ1447, was transformed into E. coli BL21-CodonPlus(DE3)-RIL (Stratagene) cells. The transformed cells were grown in Luria-Bertani medium (200 ml; Difco) supplemented with 100-μg/ml ampicillin at 37°C with shaking until they reached an absorbance of 1.0 at 600 nm. Recombinant protein production was induced by the addition of lactose to a final concentration of 28 mM. After an additional culture of 2 h, the cells were harvested by centrifugation (4,000 × g; 5 min) and frozen at −20°C. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% acrylamide) analysis of total cellular proteins, using a Tris-glycine buffer system, confirmed induction of the desired protein. E. coli cells expressing recombinant protein were resuspended in 4 ml extraction buffer (50 mM TES [pH 7.0], 10 mM MgCl2, 20 mM dithiothreitol) and lysed by sonication. MJ1447-derived protein was purified by anion-exchange chromatography on a MonoQ HR column (1 by 8 cm; Amersham Bioscience) with a linear gradient from 0 to 1 M NaCl in 25 mM Tris (pH 7.5) over 55 ml at 1 ml/min. Protein concentration was determined by Bradford analysis (3).
Analysis of the MJ1447 reaction.
The MonoQ-purified MJ1447 gene product (1.9 μg) was incubated for 1 h at 70°C, with 10 mM formaldehyde and 10 mM ribulose-5-phosphate in 1 ml of buffer (pH 7.4) containing 37 mM TES and 4 mM MgCl2. Following incubation, 2 to 3 mg of sodium borohydride (NaBH4) was dissolved in 200 μl of the incubation mixture, and the reduction was allowed to proceed for 30 min at room temperature. Methanol (240 μl) was then added, and the sample was centrifuged (14,000 × g; 10 min) to remove insoluble protein. The methanol layer was separated and acidified by the addition of 10 μl of acetic acid, and the methanol and water were removed by evaporation with a stream of nitrogen gas. The sample was repeatedly dissolved in methanol (three times, 200 μl each) and evaporated to remove borate. The sample was dissolved in 100 μl of water and 50 μl of glycine buffer (0.1 M glycine [pH 10.4], 1 mM zinc acetate, and 1 mM MgCl2). Alkaline phosphatase (0.2 U) was added, and the sample was incubated 1 h at 37°C. The resulting sample was desalted by passing the solution first through a column of Dowex 50W-X8 H+ 200/400 mesh (2.5 by 5 mm) and then through a column of Dowex 1-X8 OH− 200/400 mesh (2.5 by 5 mm). After the samples were evaporated to dryness with a stream of nitrogen gas, the resulting polyols were reacted with 20 μl trimethylsilyl (TMS) reagent prior to gas chromatography-mass spectrometry (GC-MS) analysis as described below.
Analysis of hexose-6-phosphate metabolism in M. jannaschii cell extracts.
The incubation mixture was treated with either borohydride or borodeuteride, and the resulting polyol phosphates were isolated using a DEAE column. The sample was then treated with phosphatase to remove the phosphate; the released polyols were isolated, and their TMS derivatives were formed for GC-MS analysis. With this procedure, erythrose-4-phosphate produced erythritol, ribose-5-phosphate produced ribitol, xylose-5-phosphate produced xylitol, arabinose-5-phosphate produced arabitol, ribulose-5-phosphate produced an equal mixture of arabitol and ribitol, glucose 6-phosphate produced glucitol, fructose phosphates produced an equal mixture of mannitol and glucitol, arabino-3-hexulose phosphate produced an equal mixture of mannitol and d-altritol (d-talitol), and sedoheptulose-7-phosphate gave rise to sedoheptulitol. GC-MS analysis of the TMS derivatives was used to measure the amount of the polyols present and to establish the extent of labeling in each polyol produced. Quantitation was based on comparing the peak heights of the polyols to the peak height of the inositol that was added as a known amount to each sample to serve as an internal standard. With the column used for the GC-MS analyses, the TMS derivatives of arabitol, xylitol, ribitol, mannitol, glucitol, altritol, and sedoheptulitol were all separated in the order listed.
Incubation of cell extracts with precursors.
In a 0.5-ml plastic centrifuge tube, 100-μl portions of cell extracts were mixed with 10 μl of 0.1 M solution of labeled glucose-6-phosphates and 10 μl of the indicated cofactors, and the samples were incubated for 15 or 20 min at 70°C. After incubation, the extracts were cooled to room temperature, and 1 μl of 10 mM inositol and 2 to 3 mg of sodium borohydride or sodium borodeuteride were added with mixing. After 30 min at room temperature, 240 μl of methanol was added, and the precipitated proteins were removed by centrifugation (10 min; 14,000 × g). The clear extract was separated, acidified to pH ∼3 by the addition of HCl, and evaporated to dryness with a stream of nitrogen gas. To remove any borate, the samples were repeatedly suspended in methanol (three suspensions, each 0.5 ml) and the methanol was evaporated to dryness with a stream of nitrogen gas. The resulting sample was dissolved in 200 μl of water and adjusted to pH 8 to 9 by the addition of 1 M NaOH. The sample was applied to a small column of DEAE-Sephadex (2 by 5 mm), which was washed with 1 ml of water; the sugar phosphates were eluted with 1 ml of 2 M NH4HCO3. Evaporation of the sample to dryness with a stream of nitrogen gas while the sample was held at 100°C removed any NH4HCO3. The phosphate was removed from the isolated polyol phosphates as described above. The sample was then passed through a Dowex 50W-X8-pyridinium column (2 by 10 mm) followed by a Dowex 1-X8-200 OH− column (2 by 10 mm) prior to evaporation of water and formation of the TMS derivative.
Formation of TMS derivatives.
Samples to be assayed as TMS derivatives were dried by evaporation with a stream of nitrogen gas while being held at 100°C in a water bath and were then reacted with 20 μl of a mixture of pyridine, hexamethyldisilazane, and chlorotrimethylsilane (9:3:1[vol/vol/vol]) for 2 min at 100°C.
GC-MS analysis.
GC-MS analysis of the sugars in the cell extracts was obtained using a VG-70-70EHF gas chromatography-mass spectrometer operating at 70 eV and equipped with an HP-5 column (0.32 μm by 30 m) programmed from 95°C to 280°C at 10°C per min. Reduction of the sugar phosphates present in the sample with NaBH4 was done so that each sugar phosphate would produce only one GC peak. Under the GC-MS conditions used, the TMS derivatives of the following compounds had the following retention times (in minutes) and mass spectral data (molecular weight, base peak, the most abundant ions with masses of >150 m/z are listed in order of decreasing intensities): erythritol (TMS)4 derivative (9.05) (410, 73, 205, 217, 189, 191, 307), xylitol (TMS)5 derivative (13.65) (512, 73, 217, 205, 319, 307, 422), arabitol (TMS)5 derivative (13.98) (512, 73, 217, 205, 319, 307, 422), ribitol (TMS)5 derivative (14.09) (512, 73, 217, 205, 319, 307, 422), xylitol (TMS)5 derivative (13.65) (512, 73, 217, 205, 319, 307, 422), mannitol (TMS)6 derivative (18.49) (614, 319, 205, 217, 307), glucitol (TMS)6 derivative (18.65) (614, 319, 205, 217, 307), galactitol (TMS)6 derivative (18.73) (614, 319, 205, 217, 307), altritol (TMS)6 derivative (18.73) (614, 319, 205, 217, 307), and perseitol (an isomer of sedohepulitol) (TMS)7 derivative (24.29) (716, 319, 307, 421, 409 [range, 250 to 400]). The distribution of label incorporated into the polyols was calculated by adjusting the observed normalized isotopic cluster intensities of the labeled with the nonlabeled samples as previously described (2).
RESULTS AND DISCUSSION
Levels and identity of sugars present in M. jannaschii cell extracts and incubation mixtures.
Analysis of the TMS derivatives of the polyols generated by borohydride reduction and dephosphorylation of the sugar phosphates present in the incubation mixtures was used to determine the sugars present in the different samples. By using an internal standard of inositol, which is not produced by the cells, the levels of each of the compounds produced could also be determined. The specific compounds assayed by GC-MS were glycerol arising from the dihydroxyacetone, hydroxypyruvaldehyde, glyceraldehyde, and/or their phosphates; erythritol arising from erythrose-3-phosphate; ribitol arising from ribose-5-phosphate; xylitol arising from xylose-5-phosphate; arabitol arising from arabinose-5-phosphate; arabitol and ribitol arising from ribulose-5-phosphate; glucitol arising from glucose-6-phosphate; mannitol and glucitol arising from the fructose phosphates; d-altritol and mannitol arising from arabino-3-hexulose-6-phosphate; and sedoheptitol arising from sedoheptulose-7-phosphate.
By reduction of the sugars with either borohydride or borodeuteride, the number of carbonyl groups in each sugar can be determined by the increase in the nominal mass of the derivative. Changes in the masses of the fragment ions allowed for the determination of the positions where the reduction occurred. By isolating the sugar phosphates after the incubation using DEAE-Sephadex chromatography, it was possible to establish that each sugar was present as the phosphate esters. Analysis of the label distribution of the incorporated stable isotopes allowed the pattern of the label incorporation to be determined. Analysis of cell extracts incubated either in the presence or absence of glucose-6-phosphate by the methods described showed the presence of glycerol, d-arabitol, ribitol, mannitol, d-glucitol, and d-altritol (Table 1). Erythritol, xylitol, and sedoheptitol were never detected; if present, the amounts would have to be 50 times smaller than the glucose-6-phosphate present in the cell extracts. This limit is based on the single ion plots of the m/z 205, 217, 189, 191, 307 ions from erythritol (TMS)4 derivative; the m/z 217, 205, 319, 307, 422 for the xylitol (TMS)5 derivative, and the m/z 319, 307, 421, 409 for the perseitol (TMS)7 derivative. Table 1 shows that incubation of the cell extracts with each of the labeled glucose-6-phosphates increased the amount of each polyols over the control and that in each incubation the polyols contained label derived from the labeled glucose-6-phosphates.
TABLE 1.
Levels of the sugar-derived polyols and distribution of their incorporated label generated in M. jannaschii cell extracts incubated with [6,6-2H2]glucose-6-phosphate and [U-13C]glucose-6-phosphate
| Amt of products detected and measured distribution of label with:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controla (μM)d | [6,6-2H2]glucose-6-phosphateb
|
[U-13C]glucose-6-phosphatec
|
||||||||||||||
| μMd | 0e | 1e | 2e | 3e | 4e | μMd | 0e | 1e | 2e | 3e | 4e | 5e | 6e | 7e | ||
| Glycerol | 28f | 135 | 40.8 | 7.1 | 46.4 | 3.8 | 1.8g | 250 | 57.1 | 64.5 | 1.4 | 12.2 | 22g | |||
| d-Arabitolh | 2 | 44 | 73.5 | 4.1 | 22.4i | 11 | 44.2 | 10.5 | 15.2 | 12.2 | 10.0 | 7.9j | ||||
| Ribitolk | 6 | 120 | 82.2 | 3.9 | 13.8i | 16 | 53.9 | 11.6 | 13.2 | 10.2 | 7.0 | 4.0i | ||||
| Mannitoll (0.3) | 49 | 55.6 | 12.5 | 26.0 | 3.0 | 3.1m | 50 | 5.4 | 29.3 | 3.9 | 12.5 | 14.4 | 4.2 | 10.6 | 20m | |
| 55.8 | 10.4 | 34.6n | ||||||||||||||
| 54.8 | 10.4 | 34.6o | ||||||||||||||
| d-Glucitol | 0.3 | 180 | 55.6 | 12.5 | 26.0 | 3.0 | 3.1m | 180 | 5.9 | 36.6 | 3.8 | 11.4 | 22.3 | 1.6 | 8.3 | 10m |
| 54.8 | 10.4 | 34.6n | ||||||||||||||
| 54.8 | 10.4 | 34.6o | ||||||||||||||
| d-Altritol | ND | 7 | 48.3 | 12.9 | 21.0 | 8.8 | 9m | 7 | 5.1 | 27.8 | 4.2 | 14.6 | 13.0 | 5.1 | 10.5 | 20m |
The control sample is the analysis of a 100-μl M. jannaschii cell extract that was not incubated. All sugar assayed, except glycerol, contained one deuterium as a result of the sodium borodeuteride reduction.
In this experiment, 100 μl of M. jannaschii cell extract was incubated for 20 min at 70°C in the presence of 12 mM [6,6-2H2]glucose-6-phosphate and 4 mM (each) NADH, NADPH, ATP, GTP, UTP, TTP, and CTP prior to the analysis of sugars.
In this experiment, M. jannaschii cell extract (100 μl) was incubated for 30 min at 70°C in the presence of 7.7 mM [U-13C]glucose-6-phosphate, 3.8 mM NADH, and 3.8 mM NADPH.
Calculated from peak heights using inositol as an internal standard. ND, not detected.
The distribution of label incorporated into the polyols was calculated by adjusting the observed normalized isotopic cluster intensities of the labeled with the nonlabeled samples as previously described (2). The numbers refer to the percentage of the molecules containing the indicated number of 2H and/or 13C.
Glycerol (TMS)3 M+-15 ion at m/z 293 had 2H0 = 59%, 2H1 = 12%, and 2H2 = 29%; M+-90 ion at m/z 218 had 2H0 = 70.5%, 2H1 = 18%, and 2H2 = 4.8%; and M+-203 ion at m/z 205 had 2H0 = 80%, 2H1 = 7.2%, and 2H2 = 13%.
Measured from the M+-15 ion at m/z 293 from the (TMS)3 derivative of glycerol.
Arabitol was derived from ribulose-5-phosphate during the borohydride reduction.
Measured from the M+-90 ion at m/z 422 from the (TMS)5 derivative of ribitol and l-arabitol.
The m/z 422 ion of the arabitol (TMS)5 derivative was too weak for accurate measurement of the ion intensities. The reported values were calculated from the m/z 307 and the m/z 320 ions that showed the same labeling pattern as seen in the ribitol but with a higher abundance of 13C.
Ribitol is derived both from d-ribose-5-phosphate and d-ribulose-5-phosphate present in the cell extracts.
Mannitol is consider to arise from the borohydride reduction of the fructose-6-phosphate, fructose-1-phosphate, fructose-1,6-phosphate, and/or fructose-1,6-bisP present or generated in the cell extracts.
Measured from the M+-90-90-89 ion at m/z 345 from the (TMS)6 derivative of mannitol, glucitol, and altritol.
Measured from the m/z 307 ion from the (TMS)3 derivative of mannitol and glucitol.
Measured from the m/z 319 ion from the (TMS)6 derivative of mannitol and glucitol.
Sugar metabolism as revealed by labeling studies.
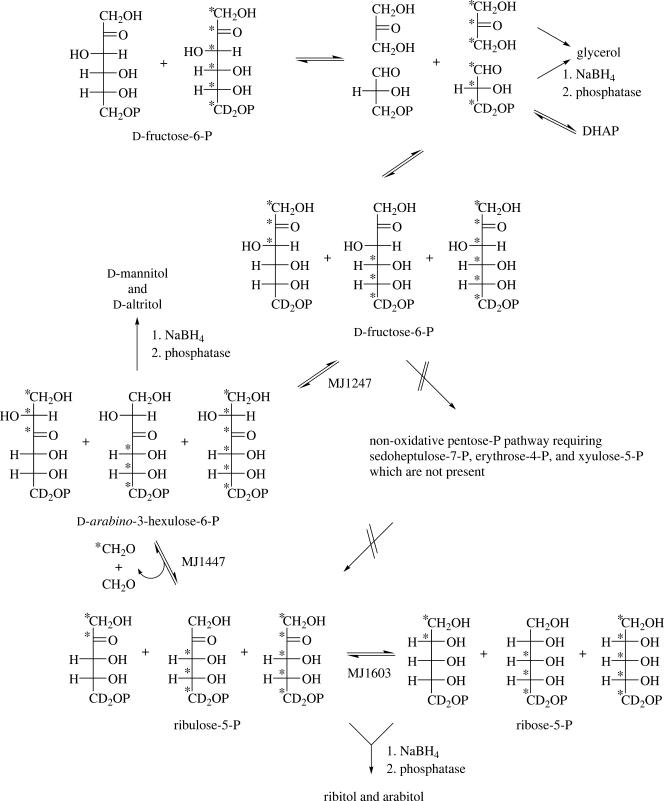
Since M. jannaschii does not rely on the pentose phosphate pathway to produce ribose-5-phosphate, we needed to establish the route for the conversion of glucose-6-phosphate to ribose-5-phosphate. The incorporation of labeled glucose-6-phosphates into dihydroxyacetone-phosphate, glyceraldehyde-3-phosphate, ribose-5-phosphate, ribulose-5-phosphate, d-arabino-3-hexulose-6-phosphate, fructose-6-phosphate, and glucose-6-phosphate was measured. This was accomplished by measuring the incorporation of labeled glucose-6-phosphate into each of the specific polyols. Cell extracts incubated with [6,6-2H2]glucose-6-phosphate produced a fourfold increase in the amount of glycerol isolated, indicating that the amount of triose phosphate and/or glycerol phosphate had increased (Table 1). Since this sample was not reduced with borodeuteride, these different trioses could not be distinguished. Also, since the samples were isolated on DEAE, each sugar would have been present as phosphates. The important measurement is that around 46% of the isolated glycerol contained two deuteriums. This confirmed cleavage of fructose-6-phosphate, derived from glucose-6-phosphate, into triose phosphates labeled with two deuteriums (Fig. 1). This cleavage occurred in the absence of added ATP, which further indicates that fructose-6-phosphate and not fructose-1,6-bisphosphate was the molecule cleaved. This is consistent with the presence of dihydroxyacetone in the cell extract (31). Such cleavage is not consistent with fructose-1,6-diphosphate aldolase, which, despite relaxed acceptor aldehyde specificity, requires a conserved “dihydroxyacetone phosphate” unit in the sugar substrate (1, 24). This same pattern of labeling was also seen in the isolated arabitol and ribitol, where arabitol reflected the labeling present in the ribulose-5-phosphate and ribitol reflected the ribose-5-phosphate in the cells. The higher levels of labeling seen in arabitol over ribitol indicates that these sugars were not completely in equilibrium and that the ribulose was biosynthetically closer to the glucose-6-phosphate than the ribose-5-phosphate. Considering that mannitol, which measures the label in both fructose-6-phosphate and arabino-3-hexulose-6-phosphate, had a label distribution similar to that seen in the glucitol, then all these sugars have been labeled from glucose-6-phosphate. The operation of both glucose-6-phosphate isomerase (MJ1605) and fructose 6-phosphate isomerase (MJ1247) is consistent with the detection of altritol in the cell extracts incubated with glucose-6-phosphate.
FIG. 1.
Labeling of sugar phosphates in M. jannaschii cell extracts incubated with labeled hexose phosphate. *, 13C.
About 3% of these hexitols, but not the pentitols, were found to contain 2H4. This distribution would arise by the breaking down of [6,6-2H2]fructose-6-phosphate, derived from [6,6-2H2]glucose-6-phosphate, to dihydroxyacetone and [6,6-2H2]glyceraldehyde-3-phosphate. After [6,6-2H2]glyceraldehyde-3-phosphate equilibrates to [6,6-2H2]dihydroxyacetone phosphate, the two recombine to form [1,1,6,6-2H4]fructose-1,6-phosphate. The fact that arabitol had only two deuteriums indicates that no transketolase reaction from [6,6-2H2]fructose-6-phosphate was involved in the formation of [5,5-2H2]ribulose-5-phosphate.
The glycerol and the mannitol and glucitol together contain 46% and 35%, respectively, of the molecules with two deuteriums. The presence of unlabeled glucose-6-phosphate and fructose-6-phosphate present in the cell extract diluted the label. Based on the fragmentation of the isolated polyols, these two deuteriums were present at the C-5 position of the ribose-5-phosphate and ribulose-5-phosphate and the C-6 position of the hexose-6-phosphates. Dilution of the label, which began with the unlabeled glucose-6-phosphate in the cell extract, continued as the label moved to the other metabolites and was increased by other unlabeled compounds present in the cell extract. In each case, some of the molecules were found with 2H2. These data clearly established that the ribose-5-phosphate is generated from [6,6-2H2]glucose-6-phosphate when incubated with cell extract, but does not indicate the pathway(s) involved.
Repeating the experiment with [U-13C]glucose-6-phosphate allowed for analysis of the breakdown and reassembly of the carbon chains. Extensive scrambling of the carbon skeleton was seen in all of the polyols from the [U-13C]glucose-6-phosphate experiment (Table 1). This was measured by the generation of hexose-phosphates containing from one to six 13C's. This scrambling most significantly resulted from the breakdown of labeled and unlabeled fructose-6-phosphate to labeled and unlabeled glyceraldehyde-3-phosphate and dihydroxyacetone, followed by random recombining of labeled and unlabeled components to re-form fructose-6-phosphate (Fig. 1). This produced the largest abundance of molecules with 13C0, 13C3, and 13C6. Isolated mannitol and glucitol also have a significant amount of 13C1, 13C2, 13C1 plus 13C3, and 13C3 plus 13C2 units. This resulted, in part, from the equilibration of fructose-6-phosphate and ribulose-phosphate via arabino-3-hexulose-6-phosphate with subsequent incorporation of labeled and unlabeled formaldehyde (Fig. 1). This would not occur with a pentose phosphate pathway, but it is consistent with the RuMP pathway. In addition, altitol, derived from d-arabino-3-hexulose-6-phosphate, was found with about the same distribution of 13C as the mannitol, confirming its direct formation from the fructose-6-phosphate.
Another route to ribose-5-phosphate.
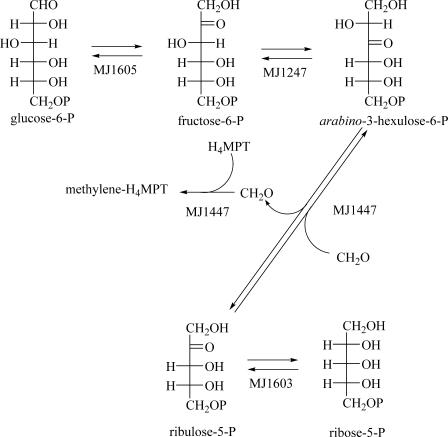
These labeling data indicate that M. jannaschii must have another route to ribose-P that does not utilize the nonoxidative pentose phosphate pathway. Based on genomic analysis, this could proceed through the RuMP pathway (Fig. 2). The RuMP pathway is used by some methylotrophic bacteria for formaldehyde fixation, and several gene clusters have been cloned (13, 33, 34, 36). Soderberg and Alver proposed this pathway as an alternate route to ribose biosynthesis in archaea (25). This alternate route to ribose-5-phosphate would involve the protein products of the MJ1603, MJ1247, and MJ1447 genes. MJ1247 has been previously characterized, and its crystal structure has been solved (15).
FIG. 2.
The RuMP pathway and its associated genes in M. jannaschii. MJ1605 is the gene encoding glucose-6-phosphate isomerase, MJ1247 is the gene encoding fructose-6-phosphate isomerase, MJ1447 is the gene encoding 3-oxohexulose-6-phosphate synthase and the formaldehyde-activating enzyme, and MJ1603 is the gene encoding ribose-6-phosphate isomerase.
We have confirmed the function of the MJ1447 gene by recombinantly expressing its protein product and determining that it does catalyze the addition of formaldehyde to ribulose-5-P. M. jannaschii arabino-3-hexulose-6-phosphate synthase incubated with ribulose-5-phosphate and formaldehyde resulted in the production of arabino-3-hexulose-6-phosphate that was detected as mannitol and talitol by GC-MS of the reduced, dephosphorylated sugar. The reaction was further confirmed by the incorporation of two deuteriums at C-1 when the reaction was conducted with [2H2]formaldehyde. The Fae domain of the molecule is expected to transfer formaldehyde to H4MPT for subsequent metabolism via the methanogenesis pathway. While the manuscript was in preparation, the facB-hpsB gene product from Methanosarcina barkeri, which is a fusion of the Fae and Hps domains as seen in MJ1447, was shown to catalyze both of the reactions discussed above (11). Since there is no clear mechanistic connection between the release of formaldehyde from the ribulose-5-P and the combination of the formaldehyde with H4MPT, this enzyme may represent an example of intraprotein channeling.
Implications of these results.
Our results show that M. jannaschii does not produce any intermediate expected in the pentose phosphate pathway and that the RuMP pathway is functioning in ribose-5-phosphate biosynthesis. An important question to be asked is why this pathway is functioning in M. jannaschii, which contains putative genes for each of the nonoxidative pentose phosphate pathway enzymes. Sodenberg and Alver's genomic analysis of archaeal genomes indicated that genes for the RuMP pathway enzymes are present in the majority of archaea, while a complete nonoxidative pentose phosphate pathway appears to be absent (25). Xavier et al. suggested a novel glycolytic pathway in Thermococcus zilligii, based on formate formation from glucose in this organism (32). Verhees et al. later reinterpreted their results to suggest that the RuMP pathway may be operating in conjunction with a formaldehyde oxidoreductase to produce the observed formate (28). The proposed importance of the RuMP pathway in archaeal metabolism has been supported by Orita et al.'s recent work, which showed that in Pyrococcus horikoshii the two central enzymes of the RuMP pathway, Hps and Phi, are fused into a single polypeptide that is constitutively expressed (19). Several archaea (M. jannaschii, Methanococcus maripaludis, Thermoplasma acidophilum, and Thermoplasma volcanium), however, are predicted to have the complete nonoxidative pentose phosphate pathway (14, 25). Two of these species, T. acidophilum and T. volcanium, do not have homologs of the RuMP enzymes or a homolog of 2-amino-3,7-dideoxy-d-threo-hept-6-ulosonic acid synthase (ADTH synthase) (MJ0400), an enzyme that is involved in the erythrose-4-phosphate-independent pathway to aromatic amino acids (31). It is likely that the nonoxidative pentose phosphate pathway, as well as the typical erythrose-4-phosphate pathway for aromatic amino acid biosynthesis, functions in these organisms. M. jannaschii and M. maripaludis are unique because they contain a complete set of putative genes for both pathways (25). It is interesting that while M. maripaludis has been shown to have a functioning nonoxidative pentose phosphate pathway (14, 27, 35), it does not utilize erythrose-4-phosphate in the formation of aromatic amino acids and instead utilizes the alternate pathway found in M. jannaschii (unpublished results).
The results presented here indicate that the RuMP pathway may be the only pathway for ribose-5-phosphate biosynthesis in M. jannaschii. This raises several evolutionary questions concerning the origins of sugar and aromatic amino acid biosynthesis. In M. jannaschii, condensation of formaldehyde and ribulose to form hexoses, as well as the reverse reaction, may have developed from a prebiotic formaldehyde condensing reaction, the so-called formose reaction, as Decker et al. proposed (6). Alternately, this pathway may have evolved since M. jannaschii no longer required erythrose-4-phosphate for aromatic amino acid biosynthesis; as a result, the cells had no use for erythrose-4-phosphate. This would, in turn, make the nonoxidative pentose phosphate pathway no longer necessary because an alternate route to ribose existed. Although M. jannaschii is predicted to contain a full complement of nonoxidative pentose phosphate pathway enzymes, it apparently does not utilize this pathway for either ribose-5-phosphate or aromatic amino acid biosynthesis. M. maripaludis may represent an evolutionary intermediate between organisms that utilize either the nonoxidative pentose phosphate pathway or the RuMP pathway for ribose biosynthesis. Although M. maripaludis contains both pathways, it utilizes the nonoxidative pentose phosphate pathway for ribose biosynthesis. However, it does not use the erythrose-4-phosphate intermediate for aromatic amino acid biosynthesis but rather uses the archaeal pathway involving aspartate semialdehyde and 6-deoxy-5-ketofructose-1-phosphate.
An important secondary question that remains to be addressed is the function of the annotated gene products that were considered to be involved in the nonoxidative pentose phosphate pathway in M. jannaschii. Soderberg recently showed that MJ0960 encodes a transaldolase that catalyzed the reaction of fructose-6-phosphate with erythrose-4-phosphate to form sedoheptulose-7-phosphate (25). Based on sequence homology this enzyme was expected to be like the transaldolase enzyme characterized from E. coli (22, 26). This result is in conflict, since our results show no erythrose-4-phosphate in the cell extracts of M. jannaschii and may simply represent an example of a promiscuous enzyme capable of utilizing a wide number of other sugars. We suggest that each of the proposed pentose phosphate pathway gene products carry out a function(s) not directly related to the operation of the pentose phosphate cycle. These genes may have become nonfunctional or may function in ribose-5-phosphate biosynthesis under certain physiological conditions. Another alterative is that they are used for the biosynthesis of presently unknown sugars involved in the production of S-layer glycoproteins (29) or flagellum glycans (17).
Acknowledgments
This research was supported by U.S. National Science Foundation grant MCB 0231319 to R.H.W.
We thank Kim Harich for assistance with GC-MS analysis and Walter Niehaus for assistance with editing the manuscript.
REFERENCES
- 1.Bednarski, M. D., E. S. Simon, N. Bischofberger, W. Fessner, M. Kim, W. Lees, T. Saito, H. Waldmann, and G. M. Whitesides. 1989. Rabbit muscle aldolase as a catalyst in organic synthesis. J. Am. Chem. Soc. 111:627-635. [Google Scholar]
- 2.Biemann, K. 1962. Mass spectrometry: organic chemical applications. McGraw-Hill, New York, N.Y.
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Choquet, C. G., J. C. Richards, G. B. Patel, and G. D. Sprott. 1994. Ribose biosynthesis in methanogenic bacteria. Arch. Microbiol. 161:481-488. [Google Scholar]
- 5.Dandekar, T., S. Schuster, B. Snel, M. Huynen, and P. Bork. 1999. Pathway alignment: application to the comparative analysis of glycolytic enzymes. Biochem. J. 343:115-124. [PMC free article] [PubMed] [Google Scholar]
- 6.Decker, P., H. Schweer, and R. Pohlmann. 1982. Identification of formose sugars, presumable prebiotic metabolites, using capillary gas chromatography/gas chromatography-mass spectrometry of n-butoximine trifluoroacetates on OV-225. J. Chromatogr. 244:281-291. [Google Scholar]
- 7.Eisenreich, W., and A. Bacher. 1991. Biosynthesis of 5-hydroxybenzimidazolylcobamid (factor III) in Methanobacterium thermoautotrophicum. J. Biol. Chem. 266:23840-23849. [PubMed] [Google Scholar]
- 8.Eisenreich, W., B. Schwarzkopf, and A. Bacher. 1991. Biosynthesis of nucleotides, flavins, and deazaflavins in Methanobacterium thermoautotrophicum. J. Biol. Chem. 266:9622-9631. [PubMed] [Google Scholar]
- 9.Ekiel, I., I. C. Smith, and G. D. Sprott. 1983. Biosynthetic pathways in Methanospirillum hungatei as determined by 13C nuclear magnetic resonance. J. Bacteriol. 156:316-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs, G., and E. Stupperich. 1980. Acetyl CoA, a central intermediate of autotrophic CO2 fixation in Methanobacterium thermoautotrophicum. Arch. Microbiol. 127:267-272. [Google Scholar]
- 11.Goenrich, M., R. K. Thauer, H. Yurimoto, and N. Kato. 2005. Formaldehyde activating enzyme (Fae) and hexulose-6-phosphate synthase (Hps) in Methanosarcina barkeri: a possible function in ribose-5-phosphate biosynthesis. Arch. Microbiol. 2005:1-8. [Online.] [DOI] [PubMed] [Google Scholar]
- 12.Graham, D. E., H. Xu, and R. H. White. 2002. Identification of coenzyme M biosynthetic phosphosulfolactate synthase: a new family of sulfonate-biosynthesizing enzymes. J. Biol. Chem. 277:13421-13429. [DOI] [PubMed] [Google Scholar]
- 13.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrickson, E. L., R. Kaul, Y. Zhou, D. Bovee, P. Chapman, J. Chung, E. Conway de Macario, J. A. Dodsworth, W. Gillett, D. E. Graham, M. Hackett, A. K. Haydock, A. Kang, M. L. Land, R. Levy, T. J. Lie, T. A. Major, B. C. Moore, I. Porat, A. Palmeiri, G. Rouse, C. Saenphimmachak, D. Soll, S. Van Dien, T. Wang, W. B. Whitman, Q. Xia, Y. Zhang, F. W. Larimer, M. V. Olson, and J. A. Leigh. 2004. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 186:6956-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Cruz, L. A., M. K. Dreyer, D. C. Boisvert, H. Yokota, M. L. Martinez-Chantar, R. Kim, and S. H. Kim. 2002. Crystal structure of MJ1247 protein from M. jannaschii at 2.0 A resolution infers a molecular function of 3-hexulose-6-phosphate isomerase. Structure (Cambridge) 10:195-204. [DOI] [PubMed] [Google Scholar]
- 16.Melendez-Hevia, E., T. G. Waddell, R. Heinrich, and F. Montero. 1997. Theoretical approaches to the evolutionary optimization of glycolysis-chemical analysis. Eur. J. Biochem. 244:527-543. [DOI] [PubMed] [Google Scholar]
- 17.Mukhopadhyay, B., E. F. Johnson, and R. S. Wolfe. 2000. A novel PH2 control on the expression of flagella in the hyperthermophilic strictly hydrogenotrophic methanarchaeaon Methanococcus jannaschii. Proc. Natl. Acad. Sci. USA 97:11522-11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukhopadhyay, B., E. F. Johnson, and R. S. Wolfe. 1999. Reactor-scale cultivation of the hyperthermophilic methanarchaeon Methanococcus jannaschii to high cell densities. Appl. Environ. Microbiol. 65:5059-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orita, I., H. Yurimoto, R. Hirai, Y. Kawarabayasi, Y. Sakai, and N. Kato. 2005. The archaeon Pyrococcus horikoshii possesses a bifunctional enzyme for formaldehyde fixation via the ribulose monophosphate pathway. J. Bacteriol. 187:3636-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purwantini, E., T. P. Gillis, and L. Daniels. 1997. Presence of F420-dependent glucose-6-phosphate dehydrogenase in Mycobacterium and Nocardia species, but absence from Streptomyces and Corynebacterium species and methanogenic Archaea. FEMS Microbiol. Lett. 146:129-134. [DOI] [PubMed] [Google Scholar]
- 21.Ronimus, R. S., and H. W. Morgan. 2003. Distribution and phylogenies of enzymes of the Embden-Meyerhof-Parnas pathway from archaea and hyperthermophilic bacteria support a gluconeogenic origin of metabolism. Archaea 1:199-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schurmann, M., and G. A. Sprenger. 2001. Fructose-6-phosphate aldolase is a novel class I aldolase from Escherichia coli and is related to a novel group of bacterial transaldolases. J. Biol. Chem. 276:11055-11061. [DOI] [PubMed] [Google Scholar]
- 23.Selkov, E., N. Maltsev, G. J. Olsen, R. Overbeek, and W. B. Whitman. 1997. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene 197:GC11-GC26. [DOI] [PubMed] [Google Scholar]
- 24.Siebers, B., H. Brinkmann, C. Dorr, B. Tjaden, H. Lilie, J. van der Oost, and C. H. Verhees. 2001. Archaeal fructose-1,6-bisphosphate aldolases constitute a new family of archaeal type class I aldolase. J. Biol. Chem. 276:28710-28718. [DOI] [PubMed] [Google Scholar]
- 25.Soderberg, T., and R. C. Alver. 2004. Transaldolase of Methanocaldococcus jannaschii. Archaea. 1:255-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sprenger, G. A. 1995. Genetics of pentose-phosphate pathway enzymes of Escherichia coli K-12. Arch. Microbiol. 164:324-330. [DOI] [PubMed] [Google Scholar]
- 27.Tumbula, D. L., Q. Teng, M. G. Bartlett, and W. B. Whitman. 1997. Ribose biosynthesis and evidence for an alternative first step in the common aromatic amino acid pathway in Methanococcus maripaludis. J. Bacteriol. 179:6010-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verhees, C. H., S. W. Kengen, J. E. Tuininga, G. J. Schut, M. W. Adams, W. M. De Vos, and J. Van Der Oost. 2003. The unique features of glycolytic pathways in Archaea. Biochem. J. 375:231-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voisin, S., R. S. Houliston, J. Kelly, J. R. Brisson, D. Watson, S. L. Bardy, K. F. Jarrell, and S. M. Logan. 2005. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J. Biol. Chem. 280:16586-16593. [DOI] [PubMed] [Google Scholar]
- 30.Vorholt, J. A., C. J. Marx, M. E. Lidstrom, and R. K. Thauer. 2000. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J. Bacteriol. 182:6645-6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White, R. H. 2004. l-Aspartate semialdehyde and a 6-deoxy-5-ketohexose 1-phosphate are the precursors to the aromatic amino acids in Methanocaldococcus jannaschii. Biochemistry 43:7618-7627. [DOI] [PubMed] [Google Scholar]
- 32.Xavier, K. B., M. S. da Costa, and H. Santos. 2000. Demonstration of a novel glycolytic pathway in the hyperthermophilic archaeon Thermococcus zilligii by 13C-labeling experiments and nuclear magnetic resonance analysis. J. Bacteriol. 182:4632-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanase, H., K. Ikeyama, R. Mitsui, S. Ra, K. Kita, Y. Sakai, and N. Kato. 1996. Cloning and sequence analysis of the gene encoding 3-hexulose-6-phosphate synthase from the methylotrophic bacterium, Methylomonas aminofaciens 77a, and its expression in Escherichia coli. FEMS Microbiol. Lett. 135:201-205. [DOI] [PubMed] [Google Scholar]
- 34.Yasueda, H., Y. Kawahara, and S.-i. Sugimoto. 1999. Bacillus subtilis yckG and yckF encode two key enzymes of the ribulose monophosphate pathway used by methylotrophs, and yckH is required for their expression. J. Bacteriol. 181:7154-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, J. P., J. Ladapo, and W. B. Whitman. 1994. Pathway of glycogen metabolism in Methanococcus maripaludis. J. Bacteriol. 176:325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yurimoto, H., R. Hirai, H. Yasueda, R. Mitsui, Y. Sakai, and N. Kato. 2002. The ribulose monophosphate pathway operon encoding formaldehyde fixation in a thermotolerant methylotroph, Bacillus brevis S1. FEMS Microbiol. Lett. 214:189-193. [DOI] [PubMed] [Google Scholar]