Abstract
The mechanism of resistance to the streptogramin antibiotics quinupristin and dalfopristin was studied in a Staphylococcus aureus clinical isolate selected under quinupristin-dalfopristin therapy, in four derivatives of S. aureus RN4220 selected in vitro, and in a mutant selected in a model of rabbit aortic endocarditis. For all strains the MICs of erythromycin, quinupristin, and quinupristin-dalfopristin were higher than those for the parental strains but the MICs of dalfopristin and lincomycin were similar. Portions of genes for domains II and V of 23S rRNA and the genes for ribosomal proteins L4 and L22 were amplified and sequenced. All mutants contained insertions or deletions in a protruding β hairpin that is part of the conserved C terminus of the L22 protein and that interacts with 23S rRNA. Susceptible S. aureus RN4220 was transformed with plasmid DNA encoding the L22 alteration, resulting in transformants that were erythromycin and quinupristin resistant. Synergistic ribosomal binding of streptogramins A and B, studied by analyzing the fluorescence kinetics of pristinamycin IA-ribosome complexes, was abolished in the mutant strain, providing an explanation for quinupristin-dalfopristin resistance.
Streptogramin antibiotics are a mixture of two classes of chemically distinct components, designated streptogramins A and B. Quinupristin-dalfopristin is a semisynthetic injectable streptogramin, a mixture of quinupristin and dalfopristin, which are semisynthetic derivatives of pristinamycin IA (PIA; streptogramin B) and pristinamycin IIA (PIIA; streptogramin A), respectively. The binding of these factors to the 50S ribosomal subunit causes inhibition of protein synthesis. Alone, each factor has a moderate bacteriostatic activity, but the combination can display a bactericidal synergistic effect, which has been attributed to the synergistic binding of the factors to their ribosomal target site (11).
Quinupristin-dalfopristin is active against a wide range of gram-positive organisms including methicillin-resistant staphylococci and vancomycin-resistant Enterococcus faecium (33). Since streptogramins A and B are chemically unrelated and have different binding sites, the mechanisms of resistance to these two streptogramins are different. Resistance to each component of the streptogramins in both staphylococci and enterococci has been reported (13, 32). The most common type of resistance to streptogramin B antibiotics is related to the production of ribosomal methylases encoded by erm genes. Resistance results from decreased component B binding to the ribosome. Cross-resistance between streptogramins B and all macrolides and lincosamides occurs because these antimicrobials have overlapping binding sites, yielding the so-called macrolide-lincosamide-streptogramin B (MLSB) phenotype. The synergistic inhibitory activity of the two streptogramin components is conserved even when the 23S rRNA is modified by an Erm methylase although the bactericidal activity of the streptogramin combination is altered. By contrast, resistance to streptogramins A appears to suppress, at least in part, the synergism displayed by the streptogramin combination (1, 13). Resistance to streptogramins A is usually due to genes encoding acetyltransferases (vatA, vatB, and vatC) or putative efflux pumps (vgaA and vgaB) (2 to 7). Streptogramin A resistance is often associated with streptogramin B inactivation by lyases encoded by vgb genes, which results in a higher level of resistance to the streptogramin mixture (1, 13, 24, 34).
The emergence of resistance in Staphylococcus aureus during quinupristin-dalfopristin therapy has rarely been reported (23). In one case of clinical failure after treatment with quinupristin-dalfopristin, resistance was due to the acquisition of a vat gene (23). We have investigated the basis of streptogramin resistance in another clinical isolate as well as in mutants selected in vitro or in an in vivo model of aortic endocarditis in rabbits (44). We found that mutations in the L22 protein were responsible for streptogramin resistance in each resistant strain, indicating the importance of this protein in synergism between the two streptogramin components.
MATERIALS AND METHODS
Bacterial strains.
Two pre- and posttreatment pairs of S. aureus isolates susceptible and resistant, respectively, to erythromycin and quinupristin-dalfopristin were studied (Table 1). One pair, S. aureus 740 and 740-1, were isolated from a patient enrolled in an emergency use program of quinupristin-dalfopristin. The other pair, S. aureus RP13 and RP13-1, were isolated from the cardiac vegetations of a rabbit with experimental aortic endocarditis (44). The members of each pair were considered to be genetically related since the patterns of SmaI-restricted DNA determined by pulsed-field gel electrophoresis were identical (44) (data not shown). Pretreatment strain S. aureus RP13 was susceptible to quinupristin and quinupristin-dalfopristin but resistant to dalfopristin (MIC = 32 μg/ml) and lincomycin by an unknown mechanism (44). Initially, it was reported that the mutant derived from S. aureus RP13 was resistant to erythromycin and quinupristin and that the MIC of dalfopristin for it (>256 μg/ml) was greater than that for RP13. However, in our hands, dalfopristin resistance was lost after serial subcultures in antibiotic-free medium and a stable mutant, RP13-1, was obtained. This mutant was still resistant to erythromycin, quinupristin, and quinupristin-dalfopristin, but the MIC of dalfopristin for it was identical to that for RP13. The identity of S. aureus RP13-1 and RP13 was verified by pulsed-field gel electrophoresis (data not shown).
TABLE 1.
MICs of macrolides, lincosamides, and streptogramins against staphylococcal strains
| S. aureus straina | MIC (μg/ml)b of:
|
FIC index Q-Dc | ||||||
|---|---|---|---|---|---|---|---|---|
| ERY | TEL | LIN | QUI | DAL | Q-D | PRI | ||
| RP13 | 0.25 | 0.06 | 8 | 2 | 32 | 0.5 | 1 | 0.09 |
| RP13-1 | 8 | 4 | 8 | 8 | 32 | 4 | 4 | 0.24 |
| 740 | 0.25 | 0.06 | 0.5 | 2 | 4 | 0.25 | 0.25 | 0.08 |
| 740-1 | 8 | 4 | 0.5 | 8 | 4 | 4 | 4 | 0.85 |
| RN4220 | 0.25 | 0.12 | 1 | 2 | 4 | 0.25 | 0.25 | 0.08 |
| RN4220M1 | 4 | 2 | 0.5 | 8 | 4 | 2 | 2 | 0.43 |
| RN4220M3 | 4 | 2 | 0.5 | 8 | 4 | 2 | 2 | 0.43 |
| RN4220M6 | 8 | 4 | 0.5 | 16 | 4 | 4 | 4 | 0.78 |
| RN4220M8 | 8 | 4 | 0.5 | 16 | 4 | 4 | 4 | 0.78 |
| RN4220(pJIM2246) | 0.25 | 0.12 | 1 | 2 | 4 | 0.25 | 0.25 | 0.08 |
| RN4220(pJIM2246ΩrplV) | 0.25 | 0.12 | 1 | 2 | 4 | 0.25 | 0.25 | 0.08 |
| RN4220(pJIM2246ΩrplVR) | 4 | 2 | 0.25 | 8 | 4 | 2 | 2 | 0.43 |
pJIM2246 is the shuttle vector used to clone the rplV genes (39). rplV was cloned from S. aureus RP13, and rplVR was cloned from S. aureus RP13-1.
Abbreviations: ERY, erythromycin; TEL, telithromycin; LIN, lincomycin; QUI, quinupristin; DAL, dalfopristin; Q-D, quinupristin-dalfopristin; PRI, pristinamycin.
FIC index Q-D, FIC index for the combination of quinupristin and dalfopristin, calculated as explained in Materials and Methods.
No sequences for rRNA methylase genes (ermA, ermB, and ermC), for streptogramin A acetyltransferase genes (vat genes), for the streptogramin A efflux gene (vga), or for the streptogramin B lyase genes (vgb genes) were detected by PCR using primers specific for each class of genes (1) or individual genes (2, 40) for any strain. No inactivation of macrolides or of streptogramins A and B was found by a microbiological assay (13).
S. aureus RN4220, susceptible to macrolides and streptogramins, and Escherichia coli DH10B were used as recipient strains in transformation experiments (30).
In vitro selection of mutants.
Spontaneous mutants were selected by plating an inoculum of approximately 108 to 109 CFU of S. aureus RN4220 onto brain heart infusion (BHI) agar containing 1, 2, or 4 μg of quinupristin-dalfopristin/ml (4 to 16 times the MIC). The plates were incubated at 37°C for 48 h, and all mutants recovered were analyzed further for stability of resistance in antibiotic-free medium and for susceptibility to macrolides, lincosamides, and streptogramins. The mutation frequency relative to the total count of viable organisms plated was determined.
Antibiotic susceptibility.
Antibiotic susceptibility was determined by disk diffusion. Disks of antibiotics were purchased from Bio-Rad (Marnes-la-Coquette, France), except for the disks of quinupristin (10 μg) and dalfopristin (20 μg) and of quinupristin-dalfopristin (10 and 20 μg, respectively), which were made in our laboratory. MICs of antibiotics were determined by agar dilution using Mueller-Hinton medium according to the recommendations of the Comité de l'Antibiogramme de la Société Française de Microbiologie (18). The enhancing interaction between quinupristin and dalfopristin was tested qualitatively by the double-disk synergy test with disks of quinupristin and dalfopristin placed 25 or 30 mm apart center to center (28). The enhancing interaction between the streptogramins A and B was characterized by an enlargement of the inhibition zones. Synergism was expressed quantitatively as fractional inhibitory concentration (FIC) indices calculated from MICs by agar dilution. MICs were determined three times, and identical values from at least two determinations were used for calculation. The FIC for each drug was calculated by dividing the MIC of quinupristin-dalfopristin by the MIC of the streptogramin (dalfopristin or quinupristin) alone. The FIC index was calculated according to the following formula: (A)/MICA + (B)/MICB = FIC index, where (A) and (B) are the concentrations of dalfopristin and quinupristin, respectively, at the MIC of quinupristin-dalfopristin and MICA and MICB are MICs of dalfopristin and quinupristin alone (31). The 30:70 ratio of quinupristin to dalfopristin in the streptogramin mixture was taken into account in the FIC calculation. Synergism is defined as a FIC index ⩽0.5, additivity is defined as a FIC index >0.5 and <2.0, and antagonism is defined as a FIC index ⩾4. Erythromycin was purchased from Sigma-Aldrich (L'isle d'Abeau Chesnes, France). Dalfopristin, quinupristin, quinupristin-dalfopristin, pristinamycin, and telithromycin (HMR 3647) were supplied by their manufacturer (Aventis Pharma, Vitry-sur-Seine, France). Lincomycin was from Pharmacia & Upjohn (Kalamazoo, Mich.).
Molecular techniques.
Portions of genes for domains II and V of 23S rRNA and the genes for ribosomal proteins L4, L10, L11, L16, L22, and L24 were amplified by PCR with the oligonucleotides shown in Table 2. The oligonucleotide primers were designed after analysis of the sequences of S. aureus COL and NCTC8325, obtained from the Institute for Genomic Research and the Oklahoma University websites (http://www.tigr.org and http://www.genome.ou.edu, respectively). Sequencing of DNA was performed on an ABI PRISM 377 (Perkin-Elmer Corp., Norwalk, Conn.) by using the big dye terminator according to the protocol supplied by the manufacturer. Deduced amino acid sequences of wild-type and mutant L22 proteins were used to model the structures with Swiss-Model software (http://www.expasy.ch/swissmod/SWISS-MODEL.html) (38). The Swiss-Pdb viewer software was used for viewing and manipulating proteins (27). For cloning, the rplV genes (for the L22 protein) of S. aureus RP13 and RP13-1 were amplified with primers modified by insertion of restriction sites in the 5′ (BamHI) and 3′ (SalI) ends (Table 2). PCR involved a precycle of 5 min at 94°C and then 30 cycles of 30 s at 94°C, 30 s at 50°C, and 30 s at 72°C, followed by a cycle of 10 min at 72°C. The amplicons were digested with BamHI and SalI restriction enzymes, cloned in pUC18, and introduced by transformation into E. coli DH10B. The inserts were then subcloned in shuttle multicopy plasmid pJIM2246 (chloramphenicol resistance) (39) and introduced into E. coli DH10B and subsequently into S. aureus RN4220 as described previously (13).
TABLE 2.
Oligonucleotides used for amplification of portions of 23S rRNA and ribosomal protein genes
| Gene (ribosomal RNA or protein) | Oligonucleotide primer
|
Position | Product size (bp) | |
|---|---|---|---|---|
| Designation | Sequence (5′-3′)a | |||
| rrl (23S rRNA) | ||||
| Domain II | 23SDIIa | + CGGAAGGGGAGTGAAATAGAAC | 487-509b | 858 |
| 23SDIIb | − ACTAACCCAGAGCGGACGAGC | 1324-1344b | ||
| Domain V | 23SDVa | + GAAAATAGGTGCCCGTACCGC | 1585-1606b | 1,054 |
| 23SDVb | − CAAATTTCCTACGCCCACGAC | 2618-2638b | ||
| rplD (L4) | RPL4a | + AATAATAAGAAGTGAAAGGAGG | −31c | 776 |
| RPL4b | − GCCATTTTTACTTGTGTTTTG | +745c | ||
| rplJ (L10) | RPL10a | + TTGACTTGAACGTGATGATCT | −112c | 901 |
| RPL10b | − TAATCCTAAACCAGTTGCTTC | +789c | ||
| rplK (L11) | RPL11a | + CTTTTAGAGCGCCCATTTCGT | −93c | 572 |
| RPL11b | − TTATCTCTGTTATCTGCTTCATCGTT | +479c | ||
| rplP (L16) | RPL16a | + GAAGCTGACACTACTTACGG | −98c | 622 |
| RPL16b | − TAGCTAACTGAAAGCGTAGG | +524c | ||
| rplV (L22) | RPL22a | + CAAAGGACACGTTGCAGACGACAAGAAA | −68c | 456 |
| RPL22b | − ATTTTTTGACCCACAGTATTCCCTCCTT | +388c | ||
| RPL22 BamHI | + CAAAGGATCCGTTGCAGACGACAAGAAA | −68c | 456 | |
| RPL22 SalI | − ATTTGTCGACCCACAGTATTCCCTCCTT | +388c | ||
| rplX (L24) | RPL24a | + AGTACTTTAATTAATAACAACC | −45c | 503 |
| RPL24b | − CATGTTCACAACGATTTTATC | +458c | ||
+, sense primer; −, antisense primer. Modified sequences for insertion of restriction sites are underlined.
E. coli numbering.
Base relative to ATG.
Ribosome binding assay.
For ribosome isolation, the staphylococcal strains were grown overnight at 37°C in BHI agar and the cultures were diluted 1/100 in fresh BHI agar and incubated at 37°C until an optical density at 600 nm of 0.6 was reached. After centrifugation, the cells were washed at 4°C in buffer A (20 mM HEPES [pH 7.6], 6 mM Mg[CH3COO]2, 30 mM NH4Cl, 4 mM β-mercaptoethanol), the pellet was suspended in the same buffer (1 ml/g of pellet), and the cells were lysed with lysostaphin (70 IU/ml) at 37°C for 1 h, sonicated, and centrifuged at 12,000 × g for 1 h. The supernatant was dialyzed three times for 45 min against buffer A at 4°C. The ribosomes were isolated from the extract by ultracentrifugation (22,000 rpm at 4°C for 17 h in a Beckman Ti50 rotor). The crude ribosome pellet was resuspended in buffer A (2 ml) for ultracentrifugation through a sucrose gradient (10 to 40% [wt/vol] in buffer A) at 18,000 rpm for 18 h at 4°C. 70S purified ribosomal particles were stored at −80°C. Ribosome concentrations were determined from the absorbance at 260 nm assuming that 1 A260 unit corresponds to 24 pmol of 70S ribosomes.
Ribosome binding was tested by a fluorescence assay adapted from that described by Beyer et al. (12). The type B component is endowed with intrinsic fluorescence because of its picolinic moiety, and its binding to ribosomes results in a sharp increase of the fluorescence intensity proportional to the number of antibiotic-ribosome complexes formed (22). Erythromycin displaces ribosome-bound type B streptogramin by competition. This competitive effect is abolished by type A streptogramin (16). The interactions of ribosomes and PIA (RP 27404), erythromycin, and PIIA (RP 12536) were measured by a spectrofluorometric procedure (F-2000 fluorescence spectrophotometer; Hitachi Co., Tokyo, Japan). The kinetics of the binding of PIA at different concentrations to ribosomes were studied as follows. A suspension of 30 pmol of 70S was diluted in 800 μl of fluorescence buffer. Increasing concentrations of PIA, from 0.01 to 0.15 μM, were added to 0.037 μM ribosomes, and fluorescence was measured (λexcitation = 350 nm, λemission = 411 nm). At 0.15 μM, the displacement of PIA as a function of time in the presence of 0.08 μM erythromycin added twice every 240 s, followed by two challenges with 0.12 μM of PIIA every 240 s, was studied. Values were expressed as arbitrary units of fluorescence. The same experimental protocol in the absence of ribosomes was done to determine background, and all experimental numbers were corrected.
RESULTS
Antibiotic resistance patterns.
MICs of erythromycin, quinupristin, quinupristin-dalfopristin, and pristinamycin determined by the agar dilution technique increased from 4- to 32-fold for S. aureus RP13-1 and 740-1 strains, compared to those for the wild-type strains (Table 1). For telithromycin, the increase was 66-fold. By contrast, dalfopristin and lincomycin MICs remained unchanged. S. aureus RN4220 resistant mutants could be selected in vitro (MIC of quinupristin-dalfopristin = 0.25 μg/ml) on agar plates containing 1 or 2 μg of quinupristin-dalfopristin/ml but not on plates containing 4 μg/ml. The mutation frequencies were 8 × 10−7 and 1.25 × 10−8 at four and eight times the MIC, respectively. The eight mutants recovered were tested for stability of resistance in the absence of an antibiotic. Only four, S. aureus RN4220M1, RN4220M3, RN4220M6, and RN4220M8, were stable. The increases in MICs of erythromycin, telithromycin, quinupristin, quinupristin-dalfopristin, and pristinamycin for the mutants were similar to those observed for the in vivo-resistant isolates, and no change in dalfopristin and lincomycin activity was observed. The agar diffusion test showed that all resistant strains exhibited cross-resistance to other macrolides, i.e., clarithromycin, azithromycin (15-member ring macrolide), and spiramycin (16-member ring macrolide).
The double disk diffusion test showed that the positive interaction strongly displayed by the combination of quinupristin and dalfopristin against S. aureus RN4220, 740, and RP13 was not present or was strongly diminished when the combination was used against the resistant mutants (Fig. 1). On the basis of agar dilution MICs, FIC indices were equal to 0.08 for S. aureus RN4220 and 740, compared to 0.43 for S. aureus RN4220M1 and RN4220M3, 0.78 for S. aureus RN4220M6 and RN4220M8, and 0.85 for S. aureus 740-1. Although no enhanced interaction between streptogramins A and B could be detected by the double disk diffusion test (data not shown), the FIC index for RP13-1 was still equal to 0.24 versus 0.09 for S. aureus RP13.
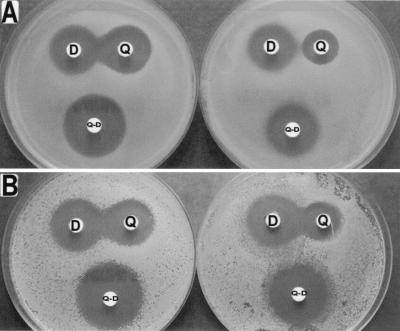
FIG. 1.
Double-disk synergy test. Disks of quinupristin (Q; 10 μg), dalfopristin (D; 20 μg), and quinupristin-dalfopristin (Q-D; 10 and 20 μg, respectively) were placed on Mueller-Hinton agar spread with S. aureus 740 (wild-type L22 protein) (A, left), S. aureus 740-1 (mutated L22 protein) (A, right), S. aureus RN4220(pJIM2246) (wild-type L22 protein) (B, left), and S. aureus RN4220(pJIM2246ΩrplVR) (mutated L22 protein) (B, right). Enlargement of the inhibition zones facing the quinupristin and dalfopristin disks indicated enhanced interaction between the two streptogramins for the two wild-type strains. No interaction and a weak interaction were observed for S. aureus 740-1 and S. aureus RN4220(pJIM2246ΩrplVR), respectively.
Mutations in the L22 protein confer resistance to quinupristin-dalfopristin.
Sequences of domains II and V of the rrl (23S rRNA) genes and of rplD (L4) genes of susceptible and resistant strains selected in vivo were identical. Since S. aureus harbors five or six copies of the rrl gene, we could not exclude the possibility that only a minority of the copies were mutated and therefore not detected by sequencing the genomic DNA as a bulk. To explore this possibility, we used PCR-single-strand conformation polymorphism as described previously (14). After heat denaturation, the single-stranded PCR products were separated by nondenaturant polyacrylamide gel electrophoresis. In Streptococcus pneumoniae, which contains four copies of the rrl genes (43), this technique allowed a single wild copy to be distinguished from three mutated copies on the basis of heterogeneous electrophoretic profiles. Migration profiles of PCR products from S. aureus were homogeneous and identical for parents and mutants. In addition, genes for proteins L10, L11, L16, and L24, which have been thought to be involved in the synergism between streptogramins A and B, in S. aureus RP13 and RP13-1 (9) were sequenced and were found to be identical.
By contrast, sequencing the entire rplV gene (L22) showed that all the resistant strains harbored mutations which were clustered in the conserved C terminus of the L22 protein (Table 3). S. aureus RP13-1 had a 6-bp deletion which removed a glycine and a proline located at positions 79 and 80, respectively. S. aureus 740-1 had a 21-bp duplication resulting in an in-frame insertion of seven amino acids, SAINKRT, at position 101. This duplication was also found in S. aureus RN4220 M8 selected in vitro. The other in vitro mutants had various insertions in the same conserved L22 region. S. aureus RN4220M1 had a 15-bp insertion resulting in the duplication of five amino acids, GPTLK, at position 84. S. aureus RN4220M3 and RN4220M6 had complex mutations resulting probably from a deletion and an insertion at positions located between amino acids 97 and 101.
TABLE 3.
Mutations in the C terminus of the L22 protein
| Strain or consensus | Sequence (amino acids 61-107)b |
|---|---|
| Consensusa | NYDMNTDELVVKEAYANEGPTLK-----RFRPRAQGRASAINKRT-------SHITIVVS |
| RP13-1 | NYDMNTDELVVKEAYANE∗∗TLK-----RFRPRAQGRASAINKRT-------SHITIVVS |
| 740-1 | NYDMNTDELVVKEAYANEGPTLK-----RFRPRAQGRASAINKRTSAINKRTSHITIVVS |
| RN4220M1 | NYDMNTDELVVKEAYANEGPTLKGPTLKRFRPRAQGRASAINKRT-------SHITIVVS |
| RN4220M3 | NYDMNTDELVVKEAYANEGPTLK-----RFRPRAQGRASAIN∗∗TSAINKRTSHITIVVS |
| RN4220M6 | NYDMNTDELVVKEAYANEGPTLK-----RFRPRAQGRASAINK∗T---IHITIHITIVVS |
| RN4220M8 | NYDMNTDELVVKEAYANEGPTLK-----RFRPRAQGRASAINKRTSAINKRTSHITIVVS |
Consensus, sequence identical in S. aureus RP13, 740, and RN4220.
Insertions are in boldface. ∗, deletion.
To establish the role for the L22 protein in resistance, the L22 gene of S. aureus RP13-1, rplVR, and the corresponding allele of susceptible S. aureus RP13, rplV, were cloned in multicopy shuttle vector pJIM2246. Introduction of the recombinant plasmid bearing the rplVR gene into S. aureus RN4220 resulted in increases in the MICs of erythromycin, telithromycin, quinupristin, quinupristin-dalfopristin, and pristinamycin similar to those for the in vivo and in vitro mutants (Table 1). The MICs of dalfopristin and lincomycin remained unchanged, showing that mutation of rplV did not explain dalfopristin and lincomycin resistance in S. aureus RP13. The positive interaction between the two streptogramin components was weak by the double disk diffusion test for the strain containing the rplVR gene, S. aureus RN4220(pJIM2246ΩrplVR) (Fig. 1), and the FIC index value (0.43) showed that synergy was markedly altered.
Ribosome binding.
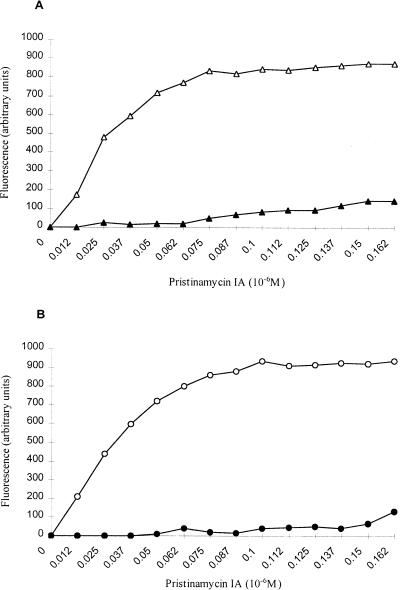
S. aureus 740 and 740-1 and S. aureus RN4220 and RN4220M1 were analyzed for the binding of streptogramins to the ribosomes. Like erythromycin, streptogramin B binds stoichiometrically to the bacterial ribosome (17, 37). When PIA (one of the group B molecules) binds the 50S subunits, its fluorescence increases in proportion to the concentration of the antibiotic-ribosome complex formed (22). Figure 2 shows fluorescence produced by PIA-ribosome complexes. In a first step, 70S ribosomes were saturated with increasing concentrations of PIA. For the susceptible strains, fluorescence increased proportionally to the concentration of PIA until a plateau was reached at 0.1 μM PIA. By contrast, a barely detectable increase in fluorescence was observed for the mutants under the same experimental conditions, indicating dramatic changes in the binding of PIA to the ribosomes. At saturating concentrations of PIA, the percentage of fluorescence expressed by resistant ribosomes relative to that expressed by wild-type ribosomes saturated with PIA was 20%.
FIG. 2.
Binding of PIA to wild-type and mutant ribosomes. (A) S. aureus RN4220 (open triangles) and RN4220M1 (solid triangles); (B) S. aureus 740 (open circles) and 740-1 (solid circles). Increasing concentrations of PIA were added to a ribosome suspension (0.037 μM). Spectrofluorometric measurements were made after each addition of PIA.
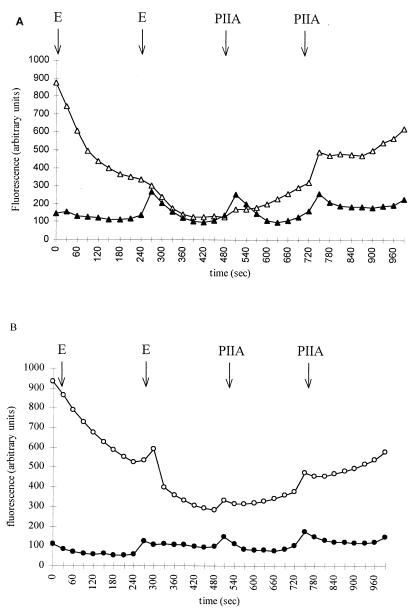
In a second step, the enhanced binding observed with the combination was analyzed by competition and displacement experiments. Erythromycin was used to displace the ribosomally bound type B streptogramin, while suppression of this competition occurred upon introduction of a type A streptogramin (16, 36). Fluorescence decreased after addition of erythromycin as a function of time by 20 and 30% for streptogramin B-susceptible S. aureus RN4220 and S. aureus 740, respectively (Fig. 3). Addition of the type A component resulted in a reemergence of fluorescence due to a higher affinity of ribosomes for the type B compound (21) (Fig. 3). For resistant strains, the addition of the A component was not followed by emission of fluorescence, indicating the inability of ribosomes to bind PIA, even in the presence of PIIA.
FIG. 3.
Displacement of ribosome-bound PIA to wild-type and mutant ribosomes by erythomycin (E) and PIIA. (A) S. aureus RN4220 (open triangles) and RN4220M1 (solid triangles); (B) S. aureus 740 (open circles) and 740-1 (solid circles). Erythromycin (0.08 μM) was added at time zero and at 240 s to the mixture of ribosomes (0.037 μM) and PIA (0.16 μM). PIIA (0.12 μM) was then added at 480 and 720 s.
DISCUSSION
Resistance of clinical isolates of staphylococci to quinupristin-dalfopristin is rare and is usually due to the acquisition of extrinsic genes responsible for inactivation or efflux of streptogramin components (24, 32). Interestingly, in the posttreatment strains and in the in vitro mutants that we studied, resistance was always due to a mutation in the rplV gene. Selection of laboratory mutants resistant to quinupristin-dalfopristin has been previously reported (29, 44). The frequencies at which the staphylococci developed spontaneous resistance to quinupristin-dalfopristin varied from 2 × 10−7 to 7 × 10−8 on agar containing twice the MIC and from 9.5 × 10−10 to less than 10−10 on agar containing four times the MIC. In our study, S. aureus RN4220 developed spontaneous resistance at four times the MIC and mutants could still be obtained at eight times the MIC at a frequency of 1.25 × 10−8. The clinical relevance of this low frequency of mutation was demonstrated in the model of aortic endocarditis in rabbits (44). In this model, cardiac vegetations of one rabbit were infected with S. aureus RP13 and treatment with quinupristin-dalfopristin selected for resistant mutants. Since the bacterial concentration reached approximately 108 CFU per aortic vegetation, the development of mutants in the cardiac vegetations under the selective pressure of quinupristin-dalfopristin could be anticipated (44). Interestingly, the mutants obtained in the laboratory and in the animal model were predictive of mutations observed in the clinical strain.
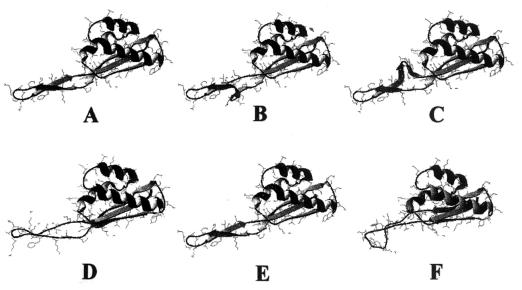
Resistance was related to mutation of the rplV gene encoding the L22 ribosomal protein, which has not, to the best of our knowledge, been reported to confer resistance to the streptogramin mixture. Insertions or deletions in the C terminus of the protein conferred cross-resistance to 14-, 15-, 16-member ring macrolides, telithromycin, and streptogramins B. The first reported description of erythromycin resistance due to an rplV mutation was of an E. coli in vitro mutant (8). This mutant, E. coli N281, was resistant to erythromycin but not to lincosamides (26). The streptogramin B susceptibility was not tested. This mutant had an 82MLN84 deletion (15). Mutations that we observed were clustered in the same region, which was reported as highly conserved in a variety of bacterial species (41). In two recent studies, S. pneumoniae laboratory mutants derived by serial passage with various macrolides were also found to harbor mutations in the C terminus of L22 (14; J. Sutcliffe, A. Tait-Kamradt, A. Walker, and J. Petitpas, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1925, 2000). L22 is a small, 117-amino-acid protein located predominantly on the surface of the 50S ribosomal subunit (35, 41). The molecule consists of a single domain with three α helices packed against a β sheet. Two of the three strands of the β structure form a β hairpin protruding from the core of the protein. All insertions or deletions characterized in our study were located in the β hairpin. This β hairpin has a twisted conformation due to residues G (Gly)-79 and P (Pro)-80. It is interesting that, in two S. aureus mutants, an insertion or a deletion occurred at this position, possibly leading to changes in twisting. Computerized modeling of S. aureus wild-type and mutant L22 proteins based on the crystal structure of Thermus thermophilus L22 determined by Unge et al. (41) shows an apparent disruption of the conformation of the extended β hairpin either by changes in the twist or distortion of the β hairpin structure in mutants (Fig. 4). This hairpin contributes, along with ribosomal protein L4 and rRNA, to the formation of part of the tunnel wall (35). In addition, L22 is an early protein which plays, directly or indirectly, important roles in the assembly of the subunit (41) and is the only protein that interacts with rRNA sequences belonging to all six domains of the 23S rRNA (10). The L22 mutation might change the surface properties or perturb the three-dimensional structure of 23S rRNA at multiple sites, as proposed for E. coli by Gregory and Dahlberg, and therefore might impact antibiotic binding (26). However, Chittum and Champney found that ribosomes of E. coli N281 (L22 mutant) were still able to bind erythromycin very well (15); the binding of PIA to mutated ribosomes was not studied. Our ribosome binding assays showed that PIA bound poorly to mutated ribosomes of S. aureus or in a way which did not produce fluorescence. Recent studies by three-dimensional cryoelectron microscopy of erythromycin-resistant ribosomes of E. coli N281 have shown that L22 mutants had substantial changes in the polypeptide tunnel. The L22 mutant had an enlargement of the entrance and could bind erythromycin but in an ineffective way (25). The reasons for the dissociation between resistance to macrolides and streptogramins B and susceptibility to lincosamides remain to be elucidated. Since the binding site of lincosamides is distinct from that of streptogramins B, these antimicrobials could still bind to mutated ribosomes (21).
FIG. 4.
Ribbon diagrams showing the secondary structures of ribosomal protein L22 of S. aureusRN4220 (A) and streptogramin-resistant mutants. Structures were predicted from the Thermus aquaticus L22 crystal structure (41). Shown are S. aureus RN4220M1 (B), -M3 (C), -M6 (D), -M8, 740-1 (E), and RP13-1 (F). All mutations were located in the β hairpin of the protein. Figures were produced with Swiss-Pdb viewer software (27).
When a plasmid bearing a wild-type rplV gene on a multicopy plasmid was introduced into E. coli N281, the strain became relatively susceptible to erythromycin, suggesting dominance of erythromycin sensitivity over resistance (15). In contrast, introduction of the rplVR gene from S. aureus RP13-1 borne by a multicopy plasmid into erythromycin-susceptible S. aureus RN4220 yielded resistance to erythromycin. Taken together, these observations are consistent with the hypothesis that susceptibility or resistance to erythromycin depends on the proportions of wild-type and mutated ribosomes.
Quinupristin resistance due to the L22 mutation was associated with resistance to quinupristin-dalfopristin. Studies carried out with the isogenic pairs composed of S. aureus RN4220 and mutants or a transformant showed that resistance to the streptogramin combination was due to a partial (mutants M1 and M3 and the rplVR transformant) or total (mutants M6 and M8) loss of synergism between the two streptogramin factors. A similar conclusion can be drawn for clinical pair S. aureus 740 and 740-1. Surprisingly, the FIC index for S. aureus RP13-1 remained low although it was threefold higher than that for S. aureus RP13. However, S. aureus RP13-1 expressed resistance to dalfopristin and lincomycin by an unknown mechanism and expressed resistance to quinupristin by an L22 mutation. This makes it difficult to assess the contribution of quinupristin resistance to the FIC index shift. Fluorescence assays showed that PIA bound efficiently susceptible ribosomes of S. aureus RN4220 and 740 and was displaced by erythromycin. The addition of PIIA restored fluorescence, showing that PIA binds the ribosome with higher affinity than erythromycin in the presence of the other streptogramin. Hypothetically, the ribosome is assumed to have two conformations: in the absence of dalfopristin, a conformation with low affinity for quinupristin and, in the presence of dalfopristin, a conformation with high affinity for quinupristin. The ability of streptogramins A to enhance the affinity of streptogramins B for the ribosome explains the synergism between the two components (12, 19). The observation that streptogramin B resistance can alter synergy apparently contradicts the belief that in staphylococci, resistance to the streptogramin complex requires resistance to streptogramin A (1, 13). This notion, which was based on the observation that all clinical isolates of staphylococci resistant to the streptogramin complex are resistant to the A component but not necessarily to the B component (24), probably holds true for strains harboring only acquired resistance genes. Although the binding of intact dalfopristin to the ribosomes is required for synergy, our study shows that this condition is not sufficient. Other mutations in the binding sites of streptogramins B which have no effect or less-marked effects on synergy have been described. Vanuffel et al. have studied the effect on the synergy between streptogramins A and B of the A2058U mutation of 23S rRNA cloned on a multicopy plasmid and introduced into E. coli (42). The A2058 nucleotide has a key role in the binding of macrolides, lincosamides, and streptogramins B. These authors confirmed that the mutant ribosomes did not bind streptogramins B. However, this mutation did not impact synergy. Indeed, the binding of streptogramin A to mutant ribosomes unmasked a high-affinity site for streptogramin B, explaining why synergy was maintained. Another nucleotide mutation of 23S rRNA at position 2062 was reported to confer resistance to the streptogramin mixture in a clinical isolate of Streptococcus pneumoniae (MICs of quinupristin-dalfopristin and pristinamycin equal to 2 μg/ml) (20). The MIC of each component (MIC of PIA = 32 μg/ml and MIC of PIIA = 16 μg/ml) was high, and the pneumococcal strain was categorized as having intermediate resistance to streptogramins despite the conservation of the synergy between the two components.
The contribution of the L22 mutation to the resistance to quinupristin-dalfopristin of clinical isolates including those containing genes for resistance to streptogramins A and B should be assessed. Finally, this report of L22 mutations conferring resistance to streptogramins should contribute to a better understanding of the mechanism of synergy between the streptogramin components.
Acknowledgments
This work was supported in part by a grant from Fondation de la Recherche Médicale. Bülent Bozdogan was the recipient of a grant from Aventis Pharma.
We thank Dieter Beyer and Jean-François Desnottes for helpful discussions and Stéphane Allouche for help in ribosomal experiments.
REFERENCES
- 1.Allignet, J., S. Aubert, A. Morvan, and N. El Solh. 1996. Distribution of genes encoding resistance to streptogramin A and related compounds among staphylococci resistant to these antibiotics. Antimicrob. Agents Chemother. 40:2523-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allignet, J., and N. El Solh. 1995. Diversity among the gram-positive acetyltransferases inactivating streptogramin A and structurally related compounds and characterization of a new staphylococcal determinant, vatB. Antimicrob. Agents Chemother. 39:2027-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allignet, J., and N. El Solh. 1997. Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transporter conferring resistance to streptogramin A and related compounds. Gene 202:133-138. [DOI] [PubMed] [Google Scholar]
- 4.Allignet, J., N. Liassine, and N. El Solh. 1998. Characterization of a staphylococcal plasmid related to pUB110 and carrying two novel genes, vatC and vgbB, encoding resistance to streptogramins A and B and similar antibiotics. Antimicrob. Agents Chemother. 42:1794-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allignet, J., V. Loncle, and N. El Solh. 1992. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamycin A-like antibiotics. Gene 117:45-51. [DOI] [PubMed] [Google Scholar]
- 6.Allignet, J., V. Loncle, P. Mazodier, and N. El Solh. 1988. Nucleotide sequence of a staphylococcal plasmid gene, vgb, encoding a hydrolase inactivating the B components of virginiamycin-like antibiotics. Plasmid 20:271-275. [DOI] [PubMed] [Google Scholar]
- 7.Allignet, J., V. Loncle, C. Simenel, M. Delepierre, and N. El Solh. 1993. Sequence of a staphylococcal gene, vat, encoding an acetyltransferase inactivating the A-type compounds of virginiamycin-like antibiotics. Gene 130:91-98. [DOI] [PubMed] [Google Scholar]
- 8.Apirion, D. 1967. Three genes that affect Escherichia coli ribosomes. J. Mol. Biol. 30:255-275. [PubMed] [Google Scholar]
- 9.Aumercier, M., S. Bouhallab, M. L. Capmau, and F. Le Goffic. 1992. RP 59500: a proposed mechanism for its bactericidal activity. J. Antimicrob. Chemother. 30(Suppl. A):9-14. [DOI] [PubMed] [Google Scholar]
- 10.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 11.Beyer, D., and K. Pepper. 1998. The streptogramin antibiotics: update on their mechanism of action. Expert Opin. Investig. Drugs 7:591-599. [DOI] [PubMed] [Google Scholar]
- 12.Beyer, D., P. Vannuffel, and K. Pepper. 1998. Quinupristin (RP 57669): a new tool to investigate ribosome-group B streptogramin interactions. Biol. Chem. 379:841-846. [DOI] [PubMed] [Google Scholar]
- 13.Bozdogan, B., and R. Leclercq. 1999. Effects of genes encoding resistance to streptogramins A and B on the activity of quinupristin-dalfopristin against Enterococcus faecium. Antimicrob. Agents Chemother. 43:2720-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chittum, H. S., and W. S. Champney. 1994. Ribosomal protein gene sequence changes in erythromycin-resistant mutants of Escherichia coli. J. Bacteriol. 176:6192-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cocito, C., and G. Chinali. 1985. Molecular mechanism of action of virginiamycin-like antibiotics (synergimycins) on protein synthesis in bacterial cell-free systems. J. Antimicrob. Chemother. 16(Suppl. A):35-52. [DOI] [PubMed] [Google Scholar]
- 17.Cocito, C., and M. Di Giambattista. 1978. The in vitro binding of virginiamycin M to bacteria ribosomes and ribosomal subunits. Mol. Gen. Genet. 166:53-59. [DOI] [PubMed] [Google Scholar]
- 18.Comité de l'Antibiogramme de la Société Française de Microbiologie. 1996. 1996 report of the Comité de l'Antibiogramme de la Société Française de Microbiologie. Technical recommendations for in vitro susceptibility testing. Clin. Microbiol. Infect. 2(Suppl. 1):11-25. [Google Scholar]
- 19.Contreras, A., and D. Vazquez. 1977. Synergistic interaction of the streptogramins with the ribosome. Eur. J. Biochem. 74:549-551. [DOI] [PubMed] [Google Scholar]
- 20.Depardieu, F., and P. Courvalin. 2001. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Giambattista, M., Y. Engelborghs, E. Nyssen, and C. Cocito. 1987. Kinetics of binding of macrolides, lincosamides, and synergimycins to ribosomes. J. Biol. Chem. 262:8591-8597. [PubMed] [Google Scholar]
- 22.Di Giambattista, M., E. Nyssen, A. Pecher, and C. Cocito. 1990. Affinity labeling of the virginiamycin S binding site on bacterial ribosome. Biochemistry 29:9203-9211. [DOI] [PubMed] [Google Scholar]
- 23.Dowzicky, M., G. H. Talbot, C. Feger, P. Prokocimer, J. Etienne, and R. Leclercq. 2000. Characterization of isolates associated with emerging resistance to quinupristin/dalfopristin (Synercid) during a worldwide clinical program. Diagn. Microbiol. Infect. Dis. 37:57-62. [DOI] [PubMed] [Google Scholar]
- 24.El Solh, N., and J. Allignet. 1998. Staphylococcal resistance to streptogramins and related antibiotics. Drug Res. Updates 1:169-175. [DOI] [PubMed] [Google Scholar]
- 25.Gabashvili, I. S., S. T. Gregory, M. Valle, R. Grassucci, M. Worbs, M. C. Wahl, A. E. Dahlberg, and J. Frank. 2001. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol. Cell 8:181-188. [DOI] [PubMed] [Google Scholar]
- 26.Gregory, S. T., and A. E. Dahlberg. 1999. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23 S ribosomal RNA. J. Mol. Biol. 289:827-834. [DOI] [PubMed] [Google Scholar]
- 27.Guex, N., and M. C. Peitsch. 1997. Swiss-Model and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 28.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 29.Kang, S. L., and M. J. Rybak. 1995. Pharmacodynamics of RP 59500 alone and in combination with vancomycin against Staphylococcus aureus in an in vitro-infected fibrin clot model. Antimicrob. Agents Chemother. 39:1505-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 31.Krogstad, D. J., and R. C. Moellering, Jr. 1986. Antimicrobial combinations, p. 537-595. In V. Lorian (ed.), Antibiotics in laboratory medicine, 2nd ed. Williams & Wilkins, Baltimore, Md.
- 32.Lina, G., A. Quaglia, M. E. Reverdy, R. Leclercq, F. Vandenesch, and J. Etienne. 1999. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob. Agents Chemother. 43:1062-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Low, D. E. 1995. Quinupristin/dalfopristin: spectrum of activity, pharmacokinetics, and initial clinical experience. Microb. Drug Resist. 1:223-234. [DOI] [PubMed] [Google Scholar]
- 34.Mukhtar, T. A., K. P. Koteva, D. W. Hughes, and G. D. Wright. 2001. Vgb from Staphylococcus aureus inactivates streptogramin B antibiotics by an elimination mechanism not hydrolysis. Biochemistry 40:8877-8886. [DOI] [PubMed] [Google Scholar]
- 35.Nissen, P., J. Hansen, N. Ban, P. B. Moore, and T. A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920-930. [DOI] [PubMed] [Google Scholar]
- 36.Nyssen, E., M. Di Giambattista, and C. Cocito. 1989. Analysis of the reversible binding of virginiamycin M to ribosome and particle functions after removal of the antibiotic. Biochim. Biophys. Acta 1009:39-46. [DOI] [PubMed] [Google Scholar]
- 37.Parfait, R., M. Di Giambattista, and C. Cocito. 1981. Competition between erythromycin and virginiamycin for in vitro binding to the large ribosomal subunit. Biochim. Biophys. Acta 654:236-241. [DOI] [PubMed] [Google Scholar]
- 38.Peitsch, M. C. 1996. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24:274-279. [DOI] [PubMed] [Google Scholar]
- 39.Renault, P., G. Corthier, N. Goupil, C. Delorme, and S. D. Ehrlich. 1996. Plasmid vectors for gram-positive bacteria switching from high and to low copy number. Gene 183:175-182. [DOI] [PubMed] [Google Scholar]
- 40.Sutcliffe, J., E. T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Unge, J., A. Aberg, S. Al-Kharadaghi, A. Nikulin, S. Nikonov, N. Davydova, N. Nevskaya, M. Garber, and A. Liljas. 1998. The crystal structure of ribosomal protein L22 from Thermus thermophilus: insights into the mechanism of erythromycin resistance. Structure 6:1577-1586. [DOI] [PubMed] [Google Scholar]
- 42.Vannuffel, P., M. Di Giambattista, and C. Cocito. 1992. The role of rRNA bases in the interaction of peptidyltransferase inhibitors with bacterial ribosomes. J. Biol. Chem. 267:16114-16120. [PubMed] [Google Scholar]
- 43.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zarrouk, V., B. Bozdogan, R. Leclercq, L. Garry, C. Carbon, and B. Fantin. 2000. Influence of resistance to streptogramin A type antibiotics on the activity of quinupristin-dalfopristin in vitro and in experimental endocarditis due to Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1168-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]