Abstract
We sequenced and phylogenetically analyzed the reverse transcriptase (RT) region of five human immunodeficiency virus type 1 isolates from treatment-naive Ethiopian émigrés to Israel. Heteroduplex mobility assays were performed to confirm the clade C status of env genomic regions. The RT sequences showed that the strains clustered phylogenetically with clade C viruses, and a KVEQ-specific motif of silent mutations (amino acids 65, 106, 138, and 161, respectively) at resistance sites was present in the polymerase region of all studied Ethiopian isolates and subtype C reference strains. In addition, many other silent mutations were observed in the clade C viruses at various resistance sites. In general, the Ethiopian isolates were more closely related genotypically to a clade C reference strain from Botswana (southern Africa) than to previously sequenced Ethiopian reference strains. Genotypic analysis showed that two Ethiopian isolates naturally harbored the mutations K70R and G190A associated with resistance to ZDV and nonnucleoside reverse transcriptase inhibitors, respectively. Phenotypic assays revealed that the K70R substitution in this context did not reduce susceptibility to ZDV, whereas the G190A substitution resulted in high-level resistance to nevirapine (NVP). Moreover, variants resistant to NVP, delavirdine (DLV), and efavirenz (EFV) were more rapidly selected at lower drug doses culture with clade C than with clade B wild-type isolates. In the case of subtype C, selection with NVP and/or EFV led to the appearance of several previously unseen mutations in RT, i.e., V106M and S98I, as well as other mutations that have been previously reported (e.g., K103N, V106A, V108I, and Y181C). After selection with DLV, a polymorphism, A62A, initially observed in the Ethiopian isolate 4762, mutated to A62V; the latter is a secondary substitution associated with multidrug resistance against nucleoside RT inhibitors. Phenotypic analysis of clade C mutants selected against NVP, DLV, and EFV revealed broad cross-resistance, particularly in regard to NVP and DLV. These findings suggest that RT genotypic diversity may influence the emergence of drug resistance.
Human immunodeficiency virus type 1 (HIV-1) has taken on distinct forms globally. Viruses have been stratified into three major phylogenetic groups, namely, M (major), O (outlier), and N (new) (9, 30). Group M viruses can be subclassified into at least 10 different subtypes, designated clades A to J (9, 22). In North America and Europe, subtype B is predominant and, in other regions of the world, various HIV-1 subtypes are endemic, with the greatest diversity found in Central Africa (22). Global epidemics with the group M (non-B, A through J) and O clades are expanding, with ca. 40 million persons currently living with AIDS worldwide and the majority of new infections occurring in young adults in developing countries (38). Sub-Saharan Africa (clades C, A, D, E, F, G, H, J, and O) and Southeast Asia (clades C and E) represent the epicenter of HIV-1 infection, with 69 and 19%, respectively, of the total of HIV-1-infected persons in the world (22, 30, 38). In densely populated regions of southern Africa and India, clade C virus may be responsible for almost 50% of new HIV-1 infections (4, 7, 22, 32, 38). Clade C virus may become the most commonly transmitted HIV-1 subtype worldwide, given the exponentially growing number of infected persons in India and southern Africa (Botswana, South Africa, Malawi, Zambia, Mozambique, and Namibia) (22, 38).
Although there are overall similarities in genomic arrangement among HIV-1 clades, there is marked interstrain divergence with variations of 30 to 40% in env amino acid sequences, whereas intrastrain heterogeneity ranges from 5 to 20% (4, 7, 30). Characterization of the genotypic divergence of pol sequences among different HIV-1 subtypes is not yet complete, although the reverse transcriptase (RT) and protease (PR) enzymes are the major targets of antiretroviral therapy (3, 8, 10, 11). Both in vitro and in vivo evolution of RT polymorphism and the appearance of resistance mutations have been extensively documented for subtype B viruses (8, 16, 17, 31, 33, 35). Little information is available on the impact of viral subtype diversity on natural susceptibility to antiretroviral drugs. Moreover, it is not known whether preexisting polymorphisms of RT and PR can influence the development of drug resistance patterns through various sequence evolution pathways and have an impact on the outcome of antiretroviral therapy (3, 10-13, 37).
We analyzed here RT sequences from five drug-naive Ethiopian émigrés to Israel infected with clade C HIV-1. These sequences were compared to the RT sequences of subtype B and to subtype C reference strains from various regions of the world, as well as to reference strains of other clades. The phenotypic susceptibility of these strains was compared to clade B clinical isolates. We characterized phenotypic and genotypic drug resistance patterns in these clade C Ethiopian clinical isolates grown in increasing concentrations of three different members of the nonnucleoside reverse transcriptase inhibitor (NNRTI) family of drugs, e.g., nevirapine (NVP), delavirdine (DLV), and efavirenz (EFV).
(This research was conducted mostly by Hugues Loemba in partial fulfillment of the requirements for a Ph.D. degree, Faculty of Graduate Studies and Research, McGill University, Montreal, Quebec, Canada.)
MATERIALS AND METHODS
Study subjects and virus isolates.
Five treatment-naive HIV-1-infected patients from Ethiopia were included in the study. The patients were identified as HIV seropositive in 1994 and 1995, shortly after their emigration to Israel. HIV-1 was isolated from patient peripheral blood mononuclear cell samples that were cocultured with umbilical cord blood mononuclear cells (CBMC) obtained from healthy donors. Prior to coculture, CBMC were prestimulated for 3 days with phytohemagglutinin and cultured in RPMI 1640 supplemented with interleukin-2 (Boehringer Mannheim, Inc., Montreal, Quebec, Canada) as described previously (15, 16). Incubation was carried out at 37°C in 5% CO2 in a volume of 5 ml of RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM glutamine, 200 U of penicillin/ml, and 200 μg of streptomycin/ml. Once a week, culture fluids were evaluated for the presence of p24 antigen (Abbott Laboratories, North Chicago, Ill.) and for RT activity as previously described (16, 17). Fresh donor CBMC were added at 1-week intervals, and cultures were considered positive if >20 pg of p24 antigen/ml was detected in each of two consecutive samples and if the second reading was at least three times higher than the first.
DNA was also extracted from 106 CBMC for viral genotyping. The genotypes of the amplified viral stocks were identical to the original plasma viral RNA with respect to silent mutations, polymorphisms, and resistance mutations.
HMA.
Subtype determination of the different clinical isolates was performed by heteroduplex mobility assays (HMAs) by using the protocol and reagents from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Disease, National Institutes of Health. DNA was extracted from CBMC with the QIAamp DNA purification kit (Qiagen, Inc., Chatsworth, Calif.). A region of the env gene spanning the C2-V5 sequence was amplified by using two rounds of PCR. A γ-32P-labeled 3′ primer was used during the second round of PCR to generate 0.7-kb labeled fragments. A 5-μl sample from the nested PCR product of each uncharacterized HIV-1 isolate was mixed with 5 μl of corresponding DNA fragments, amplified from plasmids containing env genes from reference strains representing the different HIV-1 subtypes A2, B1, B2, B3, C1, C2, C3, and E1. Then, 1.1 μl of HMA annealing buffer (100 mM NaCl, 10 mM Tris [pH 7.2], 2 mM EDTA) was added to each sample mixture. Homo- and heteroduplexes were formed between sample and reference strains by thermal denaturation at 94°C for 2 min and reannealing after rapid cooling on wet ice. The duplexes were mixed with loading dye and then loaded onto a 5% acrylamide gel and separated by electrophoresis on a gel apparatus (160 by 160 by 1.5 mm; Protean II Cell; Bio-Rad, Hercules, Calif.) at a constant voltage of 150 V for 4 h. The mobility of heteroduplexes was visualized by autoradiography.
HIV-1 RT sequencing.
Viral RNA was isolated from culture supernatants of infected cells by using the QIAamp viral extraction kit (Qiagen). The TruGene HIV-1 Assay Gene Kit was used in conjunction with the Open Gene automated DNA sequencing system (Visible Genetics, Inc., Toronto, Ontario, Canada) to sequence the PR and RT regions of HIV-1 cDNA. Testing involved simultaneous clip sequencing of PR and codons 35 to 244 of RT from amplified cDNA in both the 3′ and the 5′ directions. Sequences were aligned and compared to a LAV-1 consensus sequence by using Gene Librarian software (Visible Genetics, Toronto, Ontario, Canada).
Phylogenetic analysis.
A multiple alignment of five drug-naive Ethiopian clade C isolates was performed with four reference clade C strains: ETH2220 (from Ethiopia), 92BR025.8 (from Brazil), IN21068 (from India), and 96BW05.02 (from Botswana). In addition, genotypic variations of the Ethiopian clinical isolates were compared to reference isolates of subtype A (U455 and 92UG037.1 from Uganda and Q2317 from Kenya), subtype D (NDK from the Democratic Republic of Congo), subtype E (CM240 from Thailand, 90CF402.1 from Central African Republic, and 93TH253.3 from Thailand), subtype F (93BR020.1 from Brazil), subtype G (SE61165 from both Sweden and the Democratic Republic of Congo), subtype H (90CF056.1 from Central African Republic), subtype J (SE9280.9 from Sweden), subtype O (MVP5180 from Cameroon), subtype CPZ (CPZGAB from Gabon), and subtype B (LAV from France and JRFL from the United States). The sequences of all subtype reference strains were obtained from the HIV sequence database (http://hiv-web.lanl.gov). All alignments were gap stripped, and a total of 25 sequences, each with 397 bp corresponding to the same polymerase region (mainly the fingers and palm subdomains) of RT, were generated by using Genetool and Peptool software (both from Biotools, Inc., Edmonton, Alberta, Canada). A multiple alignment pairwise matrix, based on the percent identity of the sequences, was performed. Phylogenetic trees based on distances between sequences were constructed by the neighbor-joining method and by using the phylogenetic programs Dnadist, Neighbor, and Drawtree/Drawgram (HIV-WEB Treemaker interface [http//hiv-web.lanl.gov]).
Phenotypic drug susceptibility assay.
Drug susceptibility was measured by determining the extent to which antiretroviral drugs inhibit in vitro HIV replication (15, 16). Clade C and B isolates were amplified and quantified by RT enzyme assays in order to generate defined and titrated clinical isolates with a minimum of interinoculum effects (16, 17). CBMC infected with patient isolates were then plated in 96-well plates both in the absence and in the presence of a variety of ARV concentrations. After 7 days, RT enzyme assays were used to determine the 50% drug inhibitory concentration (IC50) (18, 19; P. Herman, 1st IAS Conf. HIV Pathogen. Treatment, abstr. 396, 2001). The observed IC50 values of patient viral isolates were then compared to the known IC50 values of treatment-naive clade B isolates and clinical isolates known to possess resistance to select ARVs as drug-resistant controls (16, 17).
Selection for NNRTI resistance.
By using procedures previously described in our laboratory, we selected for resistance to the NNRTIs, i.e., NVP, DLV, and EFV, by growing cells in the presence of increasing concentrations of drugs. In these experiments, clade B and C isolates were grown in parallel by repeated passage of wild-type clinical isolates in CBMC in the presence of increasing concentrations of NNRTIs for 8 to 15 weeks for NVP or DLV and for 13 to 30 weeks for EFV. Suboptimal doses of the drugs, i.e., <IC50 values, were used at the beginning of the selection process, i.e., 0.001 to 0.01 μM for NVP and DLV and 0.001 μM for EFV. Final concentrations reached levels of 2.0 to 10.0 μM for NVP and DLV and of 0.01 to 1.0 μM for EFV. RT assays were performed weekly to assess viral replication. At the times of RT peaks, genotyping was performed to identify changes associated with drug resistance. The times to development of resistance and the genotypic profiles were compared in clade B and C isolates.
Nucleotide sequence accession numbers.
The RT sequences of the five Ethiopian isolates—4742, 4743, 4761, 4762, and 4766—described here are available under GenBank accession numbers AF492618, AF492619, AF492620, AF492621, and AF492622, respectively.
RESULTS
Phylogenetic analysis of env regions.
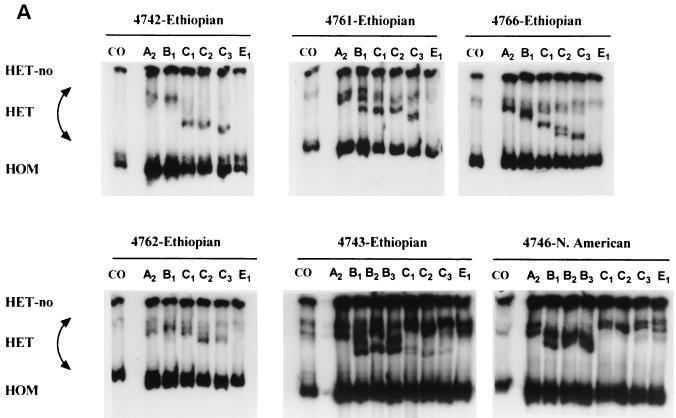
HIV-1 clinical isolates were obtained from treatment-naive Ethiopian émigrés to Israel who entered the country between 1994 and 1995. Previous phylogenetic screening of env regions performed in Israel indicated that this immigrant population, in general, harbored clade C infections. HMAs were performed to confirm the phylogenetic classifications of amplified viral isolates from five individuals. Subtype determination was based on evaluation of the mobility of heteroduplexes formed by DNA fragments amplified from the viral clinical samples and the corresponding PCR product from the env gene of various reference strains. As shown in Fig. 1A, clinical isolates 4742, 4761, 4762, and 4766 showed electrophoretic mobilities consistent with clade C viral reference strains. In contrast, isolate 4743 envelope appeared to be a clade B and C mosaic and displayed a heterogeneous heteroduplex mobility with evolutionary relationships to both subtype B and C reference viruses. The North American clinical isolate, 4246, was used as a control for subtype B viruses.
FIG. 1.
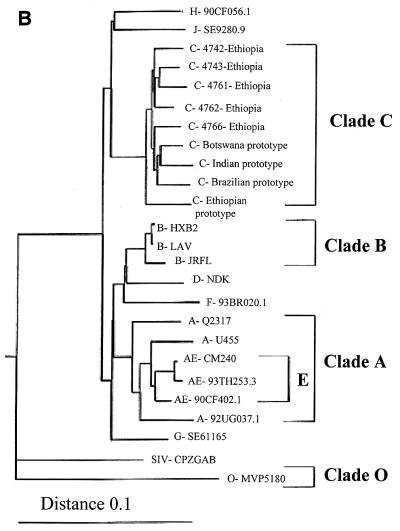
Phylogenetic profiles of clinical isolates from five drug-naive Ethiopian individuals. (A) HMA of isolates from five Ethiopian drug-naive patients. HMAs were performed as described in Materials and Methods. Heterocomplexes were formed by mixing the amplified DNA from the viral isolates with the PCR-amplified env sequences of reference strains. A more rapid migration on acrylamide gels indicates the relative degree of similarity between the unknown isolate and the reference strain sequences. For comparative purposes, the results with a wild-type clade B virus (i.e., 4746 virus) is included. (B) Phylogenetic analysis of reverse transcriptase sequences from HIV-1 Ethiopian isolates. Phylogenetic analysis comparing the RT regions of HIV-1 pol genes from five Ethiopian clinical isolates and 20 different reference strains. Tree topology was inferred from the neighbor-joining method and was based on an alignment of 397 nucleotides from which columns containing gaps were deleted. The subtype O prototype isolate was treated as an outgroup.
Several clones harboring env genes of certain subtype reference strains were used in the HMA. The subtype C reference strains C1, C2, and C3 correspond to the HIV-1 clones pCMA959 (Thailand), HIV-1 pZM18 (Zimbabwe), and HIV-1 pIN868 (India), respectively. For the other subtypes, A2 represents the subtype A reference strain clone IC144 (Ivory Cost), E1 designates subtype E pTH22 (Thailand), whereas B1, B2, and B3 represent subtype B pBR20 (Brazil), pTH14 (Thailand), and pSF162 (USA), respectively. Except for recombinant strain 4743, all of the Ethiopian clade C isolates in this HMA harbored an envelope gene that was phylogenetically closer to subtype C strain C3 from India and C2 from Zimbabwe than to the reference virus C1 from Thailand (Fig. 1A).
RT genotypic analysis.
Direct sequencing of the RT region was performed by using Visible Genetics technology. The RT sequences of five Ethiopian isolates were aligned with a panel of reference strain RT sequences from different geographic regions. The Ethiopian isolate RT sequences had an average divergence of 6.8 to 10% from the different subtype B reference strains, whereas RT sequence variation was just 3.5 to 5.8% between the Ethiopian viruses and the clade C reference strains.
The inter- and intraspecies diversity of RT is depicted on the phylogenetic tree in Fig. 1B. The neighbor-joining tree was constructed by using multiple alignment of 25 RT sequences of the Ethiopian viruses and a broad panel of reference strains. As shown, the RT sequences of the Ethiopian clinical isolates obtained from the Israeli immigrants were more closely related to the clade C reference strain from Botswana, southern Africa, than to the Ethiopian reference strain 2220 (northeastern Africa). The RT regions of the clade C viruses clustered together apart from clade B and from the majority of other non-subtype B viruses.
The amino acid diversity at codons associated with drug resistance for the five Israeli Ethiopian isolates, compared to clade C isolates from four other geographic locations, is summarized in Table 1. As shown, natural polymorphisms were present in some of these subtype C isolates at key codon sites associated with resistance to NNRTIs and ZDV (16, 38). Amino acid substitutions at positions 98, 138, 139, and 190, which have been associated with phenotypic or secondary (low-level) resistance to NNRTIs, were also observed. Additional mutations were detected, including K70R implicated in resistance to ZDV (8, 35) and the common polymorphism L214F that has been previously suggested to play an accessory role in the presence of certain mutations that confer dual resistance to ZDV and lamivudine (37).
TABLE 1.
Genotypic diversity in the RT nucleotide sequences of five treatment-naive clade C HIV-1 Ethiopian isolates versus the RT sequences of clade C reference strains from Botswana, Ethiopia, and Indiaa
| Clade B resistance mutation | HIV-1 clade B
|
Drug | Sequence of HIV-1 Ethiopian clade C isolate
|
Sequence of clade C reference strain from:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild-type codon | Resistance codon | 4743 | 4742 | 4761 | 4762 | 4766 | Botswana | Ethiopia | India | ||||
| Resistance mutations
|
|||||||||||||
| K70R | AAA | AGG | ZDV | R(AGG) | |||||||||
| A98G | GCA | GGA | NNRTI | S(TCA) | S(TCA) | ||||||||
| E138K | GAG | AAG | NNRTI | A(GCA) | |||||||||
| T139I | ACA | ATA | NNRTI | A(GCA) | |||||||||
| G190A | GCA | GCA | NVP | A(GCA) | |||||||||
| L214F | CTT | TTT | ZDV | F(TTT) | F(TTC) | F(TTC) | F(TTC) | F(TTC) | F(TTC) | F(TTC) | F(TTC) | ||
| Silent mutations at resistance sites
|
|||||||||||||
| A62V | GCC | GTA | NRTI | A(GCT) | |||||||||
| K65R | AAA | AGA | dd/ddC/Aba | K(AAG) | K(AAG) | K(AAG) | K(AAG) | K(AAG) | K(AAG) | K(AAG) | K(AAG) | ||
| K70R | AAA | AGG | ZDV | K(AAG) | K(AAG) | R(AGG) | K(AAG) | K(AAG) | K(AAG) | K(AAG) | |||
| F77L | TTC | CTA | NRTI | F(TTT) | |||||||||
| V106A | GTA | GCA | NVP | V(GTG) | V(GTG) | V(GTG) | V(GTG) | V(GTG) | V(GTG) | V(GTG) | V(GTG) | ||
| F116Y | TTT | TAT | NRTI | F(TTC) | F(TTC) | F(TTC) | F(TTC) | F(TTC) | F(TTC) | ||||
| E138K | GAG | AAG | NNRTI | E(GAA) | A(GCA) | E(GAA) | E(GAA) | E(GAA) | E(GAA) | E(GAA) | E(GAA) | ||
| Q161L | CAA | CTA | Foscarnet | Q(CAG) | Q(CAG) | Q(CAG) | Q(CAG) | Q(CAG) | Q(CAG) | Q(CAG) | Q(CAG) | ||
| Y181 C/I | TAT | TGT/ATT | NNRTI | Y(TAC) | |||||||||
| K219 Q/E | AAA | CAA/GAA | ZDV | K(AAG) | K(AAG) | K(AAG) | K(AAG) | K(AAG) | |||||
| Silent mutations in the YMDD motif
|
|||||||||||||
| Y183 | TAC | Y(TAT) | Y(TAT) | Y(TAT) | Y(TAT) | Y(TAT) | Y(TAT) | Y(TAT) | Y(TAT) | ||||
| D186N | GAT | AAC | D(GAC) | D(GAC) | D(GAC) | D(GAC) | D(GAC) | D(GAC) | D(GAC) | D(GAC) | |||
RT sequences are the same as the clade B consensus sequence unless otherwise noted. RT regions of drug-naive Ethiopian clade C viruses were compared to clade C reference strains from the HIV Los Alamos database.
Specific silent mutations were observed in all clade C clinical isolates and in all subtype C reference strains at sites encoding resistance to antiretroviral drugs, including the highly conserved YMDD motif of RT located close to the catalytic site. A KVEQ specific motif of silent mutations (amino acids 65, 106, 138, and 161) at resistance sites was observed in the polymerase region of all clade C strains studied, including all subtype C reference strains. An additional silent mutation at amino acid 116, a site encoding cross-resistance to nucleoside analogues, was specifically noted in the Ethiopian clade C isolates and the Ethiopian subtype C reference strain (Table 1).
Phenotypic drug susceptibility.
The sensitivities of three drug-naive Ethiopian isolates to a panel of nucleoside reverse transcriptase inhibitor (NRTI) (i.e., ZDV and 3TC) and NNRTIs (i.e., NVP, DLV, and EFV) were investigated in cell culture as indicated in Material and Methods. The presence of the primary G190A resistance mutation in the clinical isolate 4743 resulted in an approximately 100-fold resistance to NVP (data not shown). This isolate remained relatively sensitive to the other NNRTIs, i.e., DLV and EFV. The overall susceptibility of the Ethiopian clade C isolates, compared to the clade B control, as judged by the IC50 values, was relatively low with respect to NVP and was higher for EFV, DLV, and the NRTIs. The presence of the K70R and L214F polymorphisms did not significantly reduce phenotypic susceptibility to ZDV in our study (data not shown).
Selection of drug-resistant variants.
Selection of resistance to NVP, EFV, and DLV was performed to identify genotypic variations that may arise in clade C versus clade B isolates. As shown in Tables 2, 3, and 4, some of the mutations that arose in clade C viruses selected for resistance against NNRTIs were the same as those seen in subtype B, although a few clade C mutations may have appeared through either synonymous or nonsynonymous codon change. As indicated, the concentrations of NVP, EFV, and DLV that generated primary resistance mutations were 10, 1, and 10 μM, respectively, for the subtype B controls and 2 to 4, 0.01, and 4 μM, respectively, for the Ethiopian subtype C isolates. The numbers of weekly passages needed to generate mutations associated with phenotypic resistance to NVP, EFV, and DLV were 15, 30, and 15, respectively, for the clade B isolates and 9, 13, and 8, respectively, for the Ethiopian subtype C viruses. At the mid-point of the selection period, the clade B viruses still contained a mixture of wild-type and mutated types, whereas the clade C isolates already harbored primary mutations. Several previously unreported amino acid changes were noted in the clade C isolates, e.g., A98I in the 4742 clade C NVP-resistant virus and V106M in the EFV-resistant virus (Tables 2 and 4); the latter mutation is usually detected in clade B viruses as V106A and is associated with resistance to NVP. During DLV selection, a codon change from GCC to GTA was generated at position 62 as previously noted in the Ethiopian clade C isolate 4762 with a silent mutation at codon 62 (Table 1). This, in turn, yielded a secondary mutation, A62V, associated with multinucleoside resistance (Table 3). Another mutation V75E was detected in the Ethiopian isolate 4743 after selection with DLV (Table 3). A similar substitution is also associated with multidrug resistance against NRTIs in clade B strains (V75T).
TABLE 2.
Selection of viruses resistant to NVP by using HIV-1 clade C and B clinical isolatesa
| Wk in culture | 4746 clade B
|
4742 clade C
|
4761 clade C
|
4762 clade C
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| NVP concn (μM) | Genotype | NVP concn (μM) | Genotype | NVP concn (μM) | Genotype | NVP concn (μM) | Genotype | ||
| 0 (drug naive)
|
0 | A98S, E138A, T139A | 0 | K70R | 0 | A98S | |||
| 7 | 4 | V106A/V, Y181Y/C | 4 | S98I, E138A, T139A, Y181C | 2 | K70R, Y181C | 0.1 | A98S, V108I | |
| 9 | 2 | A98S, V108I, K103N | |||||||
| 15 | 10 | V106A/V, Y181C, V108I/V | |||||||
Resistance mutations and wild-type-resistant-strain mixtures are presented. The drug concentrations reached during the selection are noted. S98I is a novel mutation not previously reported for clade B isolates. K103N, V108I, and Y181C confer phenotypic resistance to NVP, whereas S98I, on its own, does not.
TABLE 3.
Selection of resistance to EFV by using HIV-1 clade C and B clinical isolatesa
| Wk in culture | 5346 clade B
|
4742 clade C
|
||
|---|---|---|---|---|
| EFV concn (μM) | Genotype | EFV concn (μM) | Genotype | |
| 0 (drug naive) | No mutations | A98S, E138A, T139A | ||
| 6 | 0.004 | L100L/I, K101K/E | 0.01 | A98S, K103E, E138A, T139A |
| 20 | 0.04 | L100L/I | 0.2 | A98S, E138A, T139A, Y188Y/C, G190G/A |
| 30 | 1 | K103N | 1 | A98S, V106M, E138A, T139A, Y188Y/C, G190G/A |
Resistance mutations and wild-type-resistant-strain mixtures are presented. The drug concentration reached during the selection is noted. V106M is a novel mutation found in clade C but not in clade B isolates and confers phenotypic resistance to EFV.
TABLE 4.
Selection of resistance to DLV by using HIV-1 clade C and B isolatesa
| Wk in culture | 4746 clade B
|
4742 clade C
|
4743 clade C
|
4762 clade C
|
||||
|---|---|---|---|---|---|---|---|---|
| DLV concn (μM) | Genotype | DLV concn (μM) | Genotype | DLV concn (μM) | Genotype | DLV concn (μM) | Genotype | |
| 0 (drug naive) | No mutation | A98S, E138A, T139A | G190A | A98S | ||||
| 8 | 4 | Y181Y/C, P236L | 4 | A98S, E138A, T139A, P236L | 2 | V75E, K103T, G190A | 1 | A98S, V108I, Y181C |
| 10 | 10 | V75E, K103T, G190A | 2 | A98S, V108I, K103N | ||||
| 15-16 | 10 | Y181C, P236L | 10 | A62A/V, V108I, Y181C | ||||
Resistance mutations and wild-type-resistant-strain mixtures are noted. The drug concentrations reached during the selection are indicated. P236L, a mutation causing phenotypic resistance to DLV, is commonly selected in vitro but is rarely reported in clade B clinical isolates. The V75E and A62V mutations are associated with the multi-NRTI resistance pathway in clade B isolates.
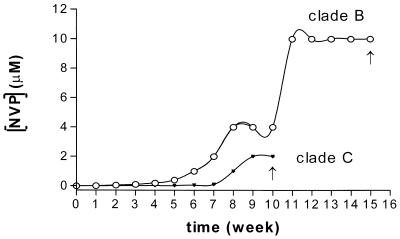
Figure 2 illustrates the main tendencies of progression to resistance against NVP for the clade B and Ethiopian clade C isolates as described in Table 2. Similar trends were observed for EFV and DLV. Clade C Ethiopian isolates generally developed more rapid primary resistance against NNRTIs at lower concentrations of NNRTIs than observed with clade B isolates. Susceptibilities to NRTIs and NNRTIs among the Ethiopian clade C resistant mutants, as well as cross-resistance profiles, were investigated (Table 5). NVP- and DLV-resistant isolates were broadly cross-resistant, and the majority of EFV-resistant mutants did not show significant cross-resistance with the isolates selected against NVP and DLV (data not shown).
FIG. 2.
Progression to resistance against NVP in cell culture. Ethiopian clade C and clade B control viruses were selected for resistance against NVP by growing cells in the presence of increasing concentrations of drugs. Concentrations of NVP selecting for primary resistance mutations and the amount of time required (in weeks) are shown.
TABLE 5.
Phenotypic drug susceptibility values for NNRTIs with clade C isolates selected for resistance to designated NNRTI drugsa
| NNRTI used in selection | Phenotypic drug susceptibility (IC50 [μM]) withb:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 4742 (NVP) | 4742 (DLV) | 4743 (DLV) | 4761 (NVP) | 4761 (DLV) | 4762 (NVP) | 4762 (DLV) | 4762 (WT) | |
| Preselection | 0.060 | 0.010 | 0.005 | 0.021 | 0.014 | 0.400 | 0.064 | |
| NVP | >10 | >2 | >10 | >10 | >2 | >10 | >2 | 0.400 |
| DLV | >10 | >10 | >10 | >10 | >10 | >10 | >10 | 0.064 |
| EFV | 0.00092 | 0.00060 | 0.00074 | 0.00017 | 0.00055 | 0.00019 | >0.4 | 0.07 |
| ZDV | 0.0006 | 0.0014 | 0.021 | >2 | 0.0024 | |||
Clade C viral isolates were selected with the designated NNRTI. The phenotypic susceptibilities of resistant isolates were monitored against all NNRTIs (NVP, DLV and EFV). Viral isolates harboring the K70R or A62V mutations, conferring resistance to ZDV or multiple NRTIs, respectively, were also evaluated for susceptibility to ZDV.
The isolate number is specified with the NNRTI given in parentheses. Genotypes: 4742 with NVP (S98I, E138A, T139A, L214F, Y181C), 4742 with DLV (A98S, E138A, T139A, L214F, P236L), 4743 with DLV (V75E, K103T, G190A, L214F), 4761 with NVP (K70R, Y181C, L214F), 4761 with DLV (L100I, K70R, L214F), 4762 with NVP (A98S, K103N, V108I, L214F), 4762 with DLV (A62V, A98S, V108I, Y181C, L214F), and 4762 wild type (WT; polymorphisms at A98S, L214F).
DISCUSSION
Although numerous studies have phylogenetically classified viral isolates based on variability in env regions, few studies have charted the effects of genetic diversity of RTs among different clades (3, 10, 11, 15, 19, 34). However, diversity in this region may have enormous ramifications and impact on viral replication, drug susceptibility, and the evolution of drug resistance. In this study, we evaluated the genotypic diversity of Ethiopian clade C strains in conjunction with phenotypic drug susceptibility and the emergence of RT mutations conferring drug resistance.
Our findings show that five Ethiopian clinical isolates studied were of clade C origin. Of interest, these isolates were more closely related to Botswana clade C variants than to the Ethiopian strains previously described (1, 23, 27). Although the predominant subtype in Ethiopia is clade C, phylogenetic divergence of some Ethiopian isolates from other HIV-1 strains has previously been observed (1, 14, 27). Similarly, HIV-1 strains from India were reported to be highly divergent from prototypic African and U.S.-European strains but linked to the South African reference strain (14, 26).
HMAs confirmed that the Ethiopian isolates were of C subtype. Interestingly, the envelope of isolate 4743 appeared to be a clade B and C mosaic, but RT region sequencing revealed homology to other Ethiopian isolates and to the different subtype C reference strain RTs. The circulation of mosaic viruses that contain resistance mutations argues for extensive RT screening and drug resistance surveillance for non-subtype B viruses. Recombination has been reported to be a common feature among retroviruses and particularly among various HIV-1 strains (19, 28). In addition, mutation and recombination may both contribute to rescuing high-fitness HIV-1 variants that harbor phenotypically relevant genetic alterations. The recent identification of individuals infected with HIV-1 isolates of two subtypes and intersubtype recombinants suggests that this phenomenon may be common among viruses cocirculating in specific regions such as parts of Africa or Asia (19, 21, 28). Recently, a study in Ivory Coast showed that almost all HIV-1 patients were infected with non-B subtypes, with a predominance of recombinant A/G viruses (2). In addition, a high prevalence of 57.4% HIV-1 drug-resistant strains were reported among 68 patients who were treated with NNRTIs, NRTIs, and PR inhibitors between 1998 and 1999 (2). Since various non-subtype B strains are currently being reported to carry resistance mutations, intersubtype mosaics may pose problems for the application of antiviral therapies in populations where the predominant HIV-1 subtypes are non-clade B.
The analysis of Ethiopian isolate RT sequences showed that these strains clustered phylogenetically with clade C reference strain RTs; a KVEQ specific motif of silent mutations (amino acids 65, 106, 138, and 161) at resistance sites has been found in the RT polymerase region. In addition, several silent mutations at codons 183 and 186 were detected in the highly conserved YMDD motif of all studied Ethiopian isolates and previously reported subtype C prototype strains (Table 1). However, the emergence of a mutation at position 186 in RT is unlikely, since the encoded aspartic residue D186 plays a key role in catalysis and in coordinating the presence of the required metal ion (36). To date, the only mutations associated with drug resistance in this region of RT have occurred at residue M184 (39, 40).
Our evaluation of Ethiopian isolate drug susceptibility showed that most viruses displayed similar drug sensitivities, confirming observations reported for clade C strains from Zimbabwe (34). However, numerous resistance mutations, polymorphisms, and silent mutations in RT have been linked to resistance to NNRTIs and NRTIs. Phenotypic drug testing revealed resistance to NVP in isolate 4743, which carried the G190A primary mutation. As confirmed in our studies, this mutation is not associated with primary resistance to DLV, and the IC50 of EFV was only slightly higher than that of the clade B control (5, 6). It is interesting that the Ethiopian clade C isolates 4742 and 4762 initially harbored an A98S secondary mutation associated with resistance to NVP. After cell culture selection with NVP, a novel S98I mutation appeared in isolate 4742. In subtype B HIV-1 strains, the mutation at this position has been reported to be A98G and has been observed in vivo (5, 6). This is the first report of the S98I mutation in RT and suggests that S and not A is the naturally occurring residue at position 98 in the RT of subtype C viruses.
The final drug concentration that selected for primary resistance mutations was significantly higher for the clade B than clade C viruses for each of NVP (10 versus 2 μM), EFV (1 versus 0.01 μM), and Del (10 versus 1 μM), respectively. Furthermore, resistant variants were fully selected more rapidly in the clade C isolates (8 or 9 weeks with NVP or DLV and 13 weeks with EFV) compared to the clade B control (at least 15 weeks with NVP or DLV and 30 weeks with EFV).
In the middle interval of the selection period, the subtype B virus harbored a mixture of both wild-type and mutated forms in regard to all of the NNRTIs (Tables 2 to and 4). These findings suggest that clade C viruses can be rapidly selected for resistance to NNRTIs. Non-B viruses may be more prone to development of resistance after highly active antiretroviral therapy and may show different mutational patterns than B isolates (Herman, abstr. 396). Furthermore, several polymorphisms have been reported at high frequency at resistance sites in clade C RT and PR from individuals who received treatment in Israel (19). These in vivo reports correlate with our cell culture observations on the development of NNRTI resistance mutations not previously observed in vitro for HIV-1 subtype C.
Other polymorphisms and silent mutations in RT may also be linked to the emergence of resistance to NNRTIs and NRTIs. After selection with DLV, a silent mutation, A62A, initially observed in Ethiopian isolate 4762, became A62V, a secondary mutation previously associated with multidrug resistance to NRTIs (24). At late stages of selection with EFV, the novel V106M mutation was detected in the clade C isolate 4742. Another substitution, i.e., V106A, has been reported in clade B viruses after treatment failure with NNRTIs, and its emergence is associated with ∼120-fold resistance to NVP, intermediate levels of resistance to DLV, and low-level resistance to EFV (6). V106M has emerged at a site at which a silent mutation (GTA→GTG) was detected in clade C isolate 4742 (Table 1). The V106M codon change may be more facilitated in clade C than clade B viruses, because only a single nucleotide change is required in this instance (GTG→ATG), as opposed to the situation in clade B viruses, which requires two such events (GTA→ATG). It is possible, therefore, that silent mutations at sites related to drug resistance in clade C RT may facilitate the emergence of resistance.
A similar impact of clade C genetic background on the development of resistance to NNRTIs has been reported in patients failing therapy (29; D. Pillay, K. Sinka, P. Rice, B. Peters, J. Clarke, J. Workman, B. Evans, and P. A. Cane, Antiviral Ther. 5[Suppl. 3]:128, abstr. 163, 2000). Moreover, a different secondary mutation at a site associated with cross-resistance among multiple NRTIs, i.e., V75E, was generated in an Ethiopian clade C isolate during selection with DLV (25). This suggests that a divergent genotypic resistance profile of clade C RT may exist, which may result in enhanced development of resistance to NNRTIs on the part of clade C viruses. Prospective studies need to be conducted in order to assess the incidence of drug resistance-related mutations in populations infected with subtype C strains and undergoing drug therapy.
As stated, the emergence of some NVP resistance mutations may be more accelerated in certain non-B subtypes and facilitated by preexisting polymorphisms. In a recent clinical trial in which NVP was used in Uganda for prevention of mother-to-child transmission of HIV-1, the K103N mutation was generated in 20% of treated women by 6 weeks after a single dose of NVP at the onset of the labor (20).
In our study, the presence of certain secondary mutations associated with resistance to NNRTIs and to ZDV did not significantly decrease the susceptibility of Ethiopian clade C strains to RT inhibitors, except for strain 4743 which harbored a NVP resistance primary mutation. The natural genotypic diversity of HIV RT among different subtypes, variations in drug susceptibility, and the development of resistance to certain drugs all indicate a need for global genotypic and phenotypic surveillance of non-subtype B strains. The emergence of recombinant viruses in areas where various HIV-1 subtypes are endemic may accelerate the selection of highly resistant mosaics and represent another challenge for the treatment of HIV disease.
Acknowledgments
This study was supported by the Canadian Institutes of Health Research and the Canadian Foundation for AIDS Research.
REFERENCES
- 1.Abebe, A., V. V. Lukashov, Pollakis, A. Kliphuis, A. L. Fontanet, J. Goudsmit, and T. F. Rinke de Wit. 2001. Timing of the HIV-1 subtype C epidemic in Ethiopia based on early virus strains and subsequent virus diversification. AIDS 15:1555-1561. [DOI] [PubMed] [Google Scholar]
- 2.Adje, C., R. Cheingsong, T. H. Roels, C. Maurice, G. Djomand, W. Verbiest, K. Hertogs, B. Larder, B. Monga, M. Peeters, S. Eholie, E. Bissagene, M. Coulibaly, R. Respess, S. Z. Wiktor, T. Chorba, and J. N. Nkengasong. 2001. High prevalence of genotypic and phenotypic HIV-1 drug-resistant strains among patients receiving antiretroviral therapy in Abidjan, Cote d'Ivoire. J. Acquir. Immune Defic. Syndr. 26:501-506. [DOI] [PubMed] [Google Scholar]
- 3.Apetrei, C., D. Descamps, G. Collin, I. Loussert-Ajaka, F. Damond, M. Duca, F. Simon, and F. Brun-Vezinet. 1998. Human immunodeficiency virus type 1 subtype F reverse transcriptase sequence and drug susceptibility. J. Virol. 72:3534-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayenie, S., B. O. Johansson, M. O. Salminen, P. Leinikki, A. Sonnerborg, D. Work-Zewdie, S. Britton, and O. Strannegard. 1991. HIV-1 in Ethiopia: phylogenetic divergence from other HIV-1 strains. Virus Genes 5:359-366. [DOI] [PubMed] [Google Scholar]
- 5.Bacheler, L., E. D. Anton, P. Kudish, D. Baker, J. Bunville, K. Krakowski, L. Bolling, M. Aujay, X. V. Wang, D. Ellis, M. F. Becker, A. L. Lasut, H. J. George, D. R. Spalding, G. Hollis, and K. Abremski. 2000. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 44:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacheler, L., S. Jeffrey, G. Hanna, R. D'Aquila, L. Wallace, K. Logue, B. Cordova, K. Hertogs, B. Larder, R. Buckery, D. Baker, K. Gallagher, H. Scarnati, R. Tritch, and C. Rizzo. 2001. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J. Virol. 75:4999-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker, M. L. B., G. de Jager, and W. B. Becker. 1995. Analysis of partial gag and env gene sequences of HIV type I strains from Southern Africa. AIDS Res. Hum. Retrovir. 11:1265-1267. [DOI] [PubMed] [Google Scholar]
- 8.Birk, B., and A. Sonnerborg. 1998. Variations in HIV-1 pol gene associated with reduced sensitivity to anti-retroviral drugs in treatment-naive patients. AIDS 12:2369-2375. [DOI] [PubMed] [Google Scholar]
- 9.Burke, D. S., and F. E. McCutchan. 1997. Global distribution of human immunodeficiency virus-1 clades, p. 119-126. In T. Vincent, J. De Vita, S. Hellman, and S. A. Rosenberg (ed.), AIDS: biology, diagnosis, treatment, and prevention. Lippincott-Raven, Philadelphia, Pa.
- 10.Caride, E., K. Hertogs, B. Larder, P. Dehertogh, R. Brindeiro, E. Machado, C. A. de Sa, W. A. Eyer-Silva, F. S. Sion, L. F. Passioni, J. A. Menezes, A. R. Calazans, and A. Tanuri. 2000. Genotyping and phenotyping analysis of B and non-B HIV-1 subtypes from Brazilian patients under HAART. Antiviral Ther. 5(Suppl. 3):128. [DOI] [PubMed] [Google Scholar]
- 11.Cornelissen, M., R. Van Den Burg, F. Zorgdrager, V. Lukashov, and J. Goodsmit. 1997. pol gene diversity of five human immunodeficiency virus type 1 subtypes: evidence for naturally occurring mutations that contribute to drug resistance, limited recombination patterns, and common ancestry for subtypes B and D. J. Virol. 71:6348-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Descamps, D., G. Collin, F. Letourneur, C. Apetrei, F. Damond, I. Loussert-Ajaka, F. Simon, S. Saragosti, and F. Brun-Vezinet. 1997. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analysis. J. Virol. 71:8893-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Descamps, D., G. Collin, I. Loussert-Ajaka, S. Saragosti, F. Simon, and F. Brun-Vezinet. 1995. HIV-1 group O sensitivity to antiviral drugs. AIDS 9:977-978. [PubMed] [Google Scholar]
- 14.Dietrich, U., M. Grez, H. von Briesen, B. Panhans, M. Geissendorfer, H. Kuhnel, J. Maniar, G. Mahambre, M. L. B. Becker, and H. Rubsamen-Waigmann. 1993. HIV-1 strains from India are highly divergent from prototypic African and US/European strains but are linked to a South African isolate. AIDS 7:23-27. [DOI] [PubMed] [Google Scholar]
- 15.Frater, A. J., A. Beardall, K. Ariyoshi, D. Churchill, S. Galpin, J. R. Clarke, J. N. Weber, and M. O. McClure. 2001. Impact of baseline polymorphisms in RT and protease on outcome of highly active antiretroviral therapy in HIV-1-infected African patients. AIDS 15:1493-1502. [DOI] [PubMed] [Google Scholar]
- 16.Gao, Q., Z. Gu, H. Salomon, K. Nagai, M. A. Parniak, and M. A. Wainberg. 1994. Generation of multiple drug resistance by sequential in vitro passage of the human immunodeficiency virus type 1. Arch. Virol. 36:111-122. [DOI] [PubMed] [Google Scholar]
- 17.Gao, Q., Z. Gu, M. A. Parniak, X. Li, and M. A. Wainberg. 1992. In vitro selection of variants of human immunodeficiency virus type 1 resistant to 3′-azido-3′-deoxythymidine and 2′,3′-dideoxyinosine. J. Virol. 66:12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu, Z., Q. Gao, E. A. Faust, and M. A. Wainberg. 1995. Possible involvement of cell fusion and viral recombination in generation of human immunodeficiency virus variants that display dual resistance to AZT and 3TC. J. Gen. Virol. 76:2601-2605. [DOI] [PubMed] [Google Scholar]
- 19.Grossman, Z., N. Vardinon, D. Chemtob, M. L. Alkan, Z. Bentwich, M. Burke, G. Gottesman, V. Istomin, I. Levi, S. Maayan, E. Shahar, and J. M. Schapiro. 2001. Genotypic variation of HIV-1 reverse transcriptase and protease: comparative analysis of clade C and clade B. AIDS 15:1453-1460. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, J. B., G. Becker-Pergola, L. A. Guay, P. Musoke, M. Mracna, M. G. Fowler, L. M. Mofenson, M. Mirochnick, F. Mmiro, and S. H. Eshleman. 2001. Identification of the K103N resistance mutation in Ugandan women receiving nevirapine to prevent HIV-1 vertical transmission. AIDS 14:F111-F115. [DOI] [PubMed] [Google Scholar]
- 21.Janini, L., A. Tanuri, M. Schechter, J. M. Peralta, A. C. P. Vicente, N. D. Torre, N. J. Pieniazek, C. C. Luo, A. Ramos, V. Soriano, G. Schochetman, M. A. Rayfield, and D. Pieniazek. 1998. Horizontal and vertical transmission of human immunodeficiency virus type 1 dual infections caused by viruses of subtypes B and C. J. Infect. Dis. 177:227-231. [DOI] [PubMed] [Google Scholar]
- 22.Janssens, W., A. Buve, and J. N. Nkengasong. 1997. The puzzle of HIV-1 subtypes in Africa. AIDS 11:705-712. [DOI] [PubMed] [Google Scholar]
- 23.Kefenie, H., S. Butto, B. Desta, P. Virani, F. Titti, L. Sernicola, M. Rapicetta, P. Pasquini, and G. B. Rossi. 1989. Serological survey of human immunodeficiency virus in Ethiopia. J. Med. Virol. 28:21-24. [DOI] [PubMed] [Google Scholar]
- 24.Kuritzkes, D. 1998. Progress in drug resistance testing in the management of anti-retroviral therapy, p. 261-274. In R. Trip Gulick and F. Hecht (ed.), The PRN notebook. Mosby, New York, N.Y.
- 25.Lacey, S. F., and B. A. Larder. 1994. Novel mutation (V75) in human immunodeficiency virus type 1 reverse transcriptase confers resistance to 2′,3′-didehydro-2′,3′-dideoxythymidine in cell culture. Antimicrob. Agents Chemother. 38:1428-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Scheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louwagie, J., W. Janssens, J. Mascola, L. Heyndrickx, P. Hegerich, G. van der Groen, F. E. McCutchan, and D. S. Burke. 1995. Genetic diversity of the envelope glycoprotein from human immunodeficiency virus type 1 isolates of African origin. J. Virol. 69:263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miguel, E. Q., and E. J. Arts. 1999. Recombination in HIV-1: update and implications. AIDS Rev. 1:89-100. [Google Scholar]
- 29.Mital, D., and D. Pillay. 2001. The impact of HIV-1 subtype on drug resistance. J. HIV Ther. 6:56-60. [PubMed] [Google Scholar]
- 30.Myers, G., B. Korber, and B. Hahn. 1995. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 31.Najera, I., A. Holguin, M. E. Quinones-Mateu, M. A. Munoz-Fernandez, R. Najera, C. Lopez-Galindez, and E. Domingo. 1995. pol gene quasispecies of human immunodeficiency virus from patients undergoing no drug therapy. J. Virol. 69:23-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novitsky, V. A., M. A. Montano, M. F. McLane, B. Renjifo, F. Vannberg, B. T. Foley, T. P. NDung'u, M. Ravaman, M. J. Makhema, R. Marlink, and M. Essex. 1999. Molecular cloning and phylogenetic analysis of Human immunodeficiency virus type 1 subtype C: a set of 23 full-length clones from Botswana. J. Virol. 73:4427-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richman, D. D., D. Havlir, J. Corbeil, D. Looney, C. Ignacio, S. A. Spector, J. Sullivan, S. Cheeseman, K. Barringer, D. Pauletti, C. K. Shich, M. Myers, and J. Griffin. 1994. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J. Virol. 68:1660-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shafer, R. W., J. A. Eisen, T. C. Merigan, and D. A. Katzenstein. 1997. Sequence and drug susceptibility of subtype C reverse transcriptase from human immunodeficiency virus type 1 seroconverters in Zimbabwe. J. Virol. 71:5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shafer, R. W., M. A. Winters, S. Palmer, and T. C. Merigan. 1998. Multiple concurrent reverse transcriptase and protease mutations and multi-drug resistance of HIV-1 isolates from heavily treated patients. Ann. Intern. Med. 128:906-911. [DOI] [PubMed] [Google Scholar]
- 36.Tantillo, C., A. Ding, and R. Jacobs-Molina. 1994. Locations of anti-AIDS drug binding sites and resistance mutations in three-dimensional structure of HIV-1 reverse transcriptase: implications for mechanisms of drug inhibition and resistance. J. Mol. Biol. 243:369. [DOI] [PubMed] [Google Scholar]
- 37.Torti, C., Y. Gilleece, K. Hertogs, B. G. Gazzard, and A. L. Pozniak. 2001. R211K and L214F do not invariably confer high-level phenotypic resistance to thymidine analogs in zidovudine-naive patients with M184V. J. Acquir. Immune Defic. Syndr. 26:514-515. [DOI] [PubMed] [Google Scholar]
- 38.UNAIDS. 2001. AIDS epidemic update-December. UNAIDS, Geneva, Switzerland.
- 39.Wainberg, M. A. 1999. HIV resistance to antagonists of viral reverse transcriptase, p. 223-249. In A. Dalgleish and R. Weiss (ed.), AIDS and the new viruses, 2nd ed. Academic Press, London, England.
- 40.Wainberg, M. A., Z. Gu, Q. Gao, E. Arts, R. Geleziumas, S. Bour, R. Beaulieu, C. Tsoukas, J. Singer, and J. Montaner. 1993. Clinical correlates and molecular basis of HIV drug resistance. J. Acquir. Immune Defic. Syndr. 6:S36-S46. [PubMed] [Google Scholar]
- 41.Xin, K., X. Ma, K. Crandall, H. Bukawa, Y. Ishigatsubo, S. Kawamoto, and K. Okuda. 1995. Dual infection with HIV-1 Thai subtypes B and E. Lancet 346:1372-1373. [DOI] [PubMed] [Google Scholar]