Abstract
Removal of nucleoside chain terminator inhibitors mediated by human immunodeficiency virus (HIV) reverse transcriptase (RT) using ATP as an acceptor molecule has been proposed as a novel mechanism of HIV resistance. Recombinant wild-type and mutant HIV type 1 (HIV-1) RT enzymes with thymidine analog resistance mutations D67N, K70R, and T215Y were analyzed for their ability to remove eight nucleoside reverse transcriptase inhibitors in the presence of physiological concentrations of ATP. The order for the rate of removal of the eight inhibitors by the mutant RT enzyme was zidovudine (AZT) > stavudine (d4T) ≫ zalcitabine (ddC) > abacavir > amdoxovir (DAPD) > lamivudine (3TC) > didanosine (ddI) > tenofovir. Thymidine analogs AZT and d4T were the most significantly removed by the mutant enzyme, suggesting that removal of these inhibitors by the ATP-dependent removal mechanism contributes to the AZT and d4T resistance observed in patients with HIV expressing thymidine analog resistance mutations. ATP-dependent removal of tenofovir was 22- to 35-fold less efficient than removal of d4T and AZT, respectively. The addition of ATP and the next complementary deoxynucleoside triphosphate caused a reduction of ATP-mediated removal of d4T, ddC, and DAPD, while AZT and abacavir removal was unaffected. The reduction of d4T, ddC, and DAPD removal in the presence of the deoxynucleoside triphosphate could explain the minor changes in susceptibility to these drugs observed in conventional in vitro phenotypic assays using cells that have higher deoxynucleoside triphosphate pools. The minimal removal of abacavir, ddC, DAPD, 3TC, ddI, and tenofovir is consistent with the minor changes in susceptibility to these drugs observed for HIV mutants with thymidine analog resistance mutations.
A large proportion of the human immunodeficiency virus (HIV)-infected patient population in developed countries have had prior antiretroviral therapy experience and have taken multiple nucleoside reverse transcriptase (RT) inhibitors (NRTIs). HIV in many of these patients has developed mutations conferring resistance to the NRTIs. Mutations in RT could confer resistance either by decreasing incorporation of inhibitors or by increasing the removal of inhibitors after they are incorporated. For a number of NRTI mutations, most notably for thymidine analog resistance mutations including D67N, K70R, and T215Y/F, changes in relative binding affinities or incorporation of inhibitors by mutant RT enzymes do not appear to correlate with drug susceptibility changes in viral replication assays (7, 14, 15, 25). However, a mechanism by which inhibitors can be removed by HIV-1 RT after they are incorporated has recently been described, and this removal may contribute to zidovudine (AZT) resistance of some HIV type 1 (HIV-1) mutants (1, 6, 19, 21). Retroviral RTs lack a 3′ exonuclease proofreading activity (3, 28) but are capable of pyrophosphorolysis, the reverse reaction of nucleotide polymerization. In this process, RT complexed with a dideoxynucleoside monophosphate (ddNMP)-terminated primer/template binds pyrophosphate or ATP and catalyzes the removal of the chain-terminating ddNMP from the primer, resulting in an unblocked DNA chain. It has been shown that an RT mutant with D67N, K70R, T215F, and K219Q mutations removes more AZT from an AZT-terminated DNA primer than wild-type RT by both pyrophosphate- and ATP-dependent removal mechanisms (1, 19). HIV-1 RT can carry out the ATP-dependent chain terminator removal reaction at physiological concentrations of ATP (1 to 4 mM) (21). The most physiologically relevant acceptor molecule for removal of chain terminators as a mechanism of resistance in vivo is currently unresolved. It has been reported that the physiological concentrations of pyrophosphate are below the Kd for the reverse reaction (2, 29). However, the physiological concentrations of ATP are within the range of the Km for ATP (0.7 to 4.3 mM), suggesting that ATP is the most likely acceptor for the chain terminator removal reaction in vivo (21, 32).
In additional experiments, it was demonstrated that the next incoming deoxynucleoside triphosphate (dNTP) can form a dead-end complex and inhibit the ATP-dependent removal of stavudine (d4T) but not AZT by HIV-1 RT (20). Removal of d4T by both wild-type and mutant RT was inhibited by the next incoming complementary dNTP at micromolar to submicromolar concentrations, whereas AZT removal was much less sensitive (>50-fold) to inhibition by the next complementary dNTP (20).
Here we have compared the ATP-dependent removal of eight NRTIs by an RT mutant containing the common thymidine analog mutations D67N, K70R, and T215Y in both the presence and absence of the next complementary dNTP. The NRTIs analyzed include those that are currently approved for HIV treatment, AZT, d4T, didanosine (ddI), abacavir, zalcitabine (ddC), lamivudine (3TC), and tenofovir, and one that is in clinical development, amdoxovir (DAPD). Our results show that an HIV-1 mutant RT with common thymidine analog resistance mutations can increase the removal rate of thymidine analogs in the presence of physiological concentrations of ATP and suggest that this ATP-mediated inhibitor removal has effects on the drug susceptibility of HIV with thymidine analog resistance mutations.
MATERIALS AND METHODS
Recombinant RT construction, purification, and kinetic analyses.
Wild-type RT expression construct pRT66, described previously (10), was a gift from M. Wainberg. The pol sequences were amplified by PCR from the HXB2D molecular clone of HIV-1 and cloned into expression vector pKK223-3 (Pharmacia Biotech, Piscataway, N.J.). An RT mutant with D67N, K70R, and T215Y mutations was generated by oligonucleotide-based site-directed mutagenesis, constructs were completely sequenced, and RT purification was performed as previously described (25). This mutant was analyzed because of reduced susceptibility resulting from the T215Y mutation (25) and prior evidence of increased pyrophosphorolysis resulting from D67N and K70R (1, 19).
Recombinant HIV production and antiviral susceptibility assays.
PCR fragments corresponding to the first 1,000 bp of the coding sequences for wild-type HIV RT and the HIV RT mutant with D67N, K70R, and T215Y mutations were amplified and cotransfected with HIV-1 proviral molecular clone pHXB2-2-261RT, in which the coding sequence for RT is deleted (a gift from C. Boucher) as previously described (4, 23). Replication-competent viruses generated by homologous recombination were harvested after 8 to 18 days when the cultures contained notable syncytia. The genotypes of the recombinant viruses were determined by RT-PCR of viral supernatant followed by sequencing using an ALF Express automated DNA sequencer (Pharmacia Biotech).
Susceptibilities of the recombinant mutant viruses and the wild-type HIV-1 molecular clone HXB2D to tenofovir, AZT, d4T, ddC, abacavir, β-d-dioxolane guanosine (DXG), ddI, and 3TC were evaluated by using a modified XTT (2,3-bis[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide)-based assay of MT-2 cells as previously described (8). All infections were performed with 1.2 × 106 cells at a multiplicity of infection of approximately 0.001 virus particle/cell which resulted in equal levels of cell killing in the absence of the drug over the 5-day assay period. Values for the 50% inhibitory concentration were calculated as averages from three to eight experiments.
Chain terminator removal assay.
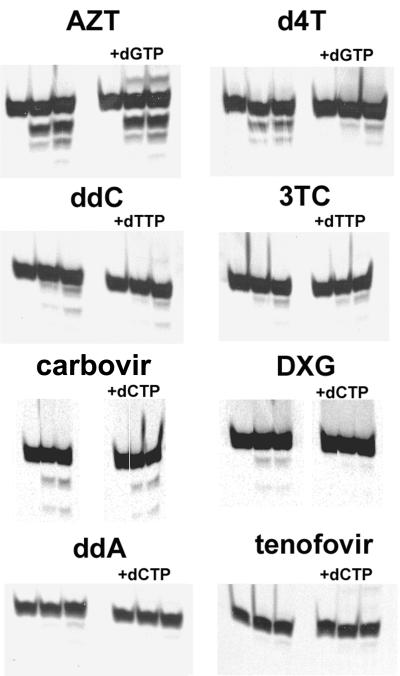
The base for nucleoside analogs AZT and d4T is thymidine, that for ddC and 3TC is cytidine, that for carbovir and DXG is guanosine, and that for ddA and tenofovir is adenosine. Primers corresponding to the natural tRNA primer binding site were 5′ end labeled with [γ-33P]ATP and T4 polynucleotide kinase and terminated essentially as described previously with a 191-bp pHIV-PBS RNA template (1). A 16-nucleotide (nt) prPBS primer, 5′-GTC CCT GTT CGG GCG C-3′, was used for ddA, tenofovir, ddC, and 3TC termination, and 18-nt prPBS primer 5′-GTC CCT GTT CGG GCG CCA-3′ was used for d4T, AZT, DXG, and carbovir termination. The lengths of the chain-terminated primers were 18 nt for ddA and tenofovir termination, 19 nt for ddC and 3TC termination, 20 nt for d4T and AZT termination, and 21 nt for DXG and carbovir termination. Longer 24-nt ddA- and tenofovir-terminated primers with the sequence 5′-GTC CCT GTT CGG GCG CCA CTG CTA-3′ were also tested for removal of ddA and tenofovir. The end-labeled chain-terminated primers were gel purified twice on a 16% polyacrylamide-8 M urea sequencing gel, excised, and eluted in a solution containing 0.5 M ammonium acetate, 10 mM Tris, pH 7.5, 0.1 mM EDTA, 2 mM MgCl2, and 0.01% sodium dodecyl sulfate, followed by desalting through NAP10 columns (Pharmacia Biotech). The purified primers were annealed to a pHIV-PBS RNA template, and a 2.5 nM template/chain-terminated primer was incubated with 20 nM mutant recombinant RT enzyme for 5 min at 37°C in RT buffer. Then ATP (3.2 mM final concentration) was added. Experiments analyzing the effect of the next complementary nucleotide included the addition of 50 μM (final concentration) dCTP for the tenofovir-, ddA-, DXG-, and carbovir-terminated primers/templates, dTTP for ddC- and 3TC-terminated primers/templates, and dGTP for the AZT- and d4T-terminated primers/templates. The dNTPs were added at the same time as the ATP to start the reaction. Reactions were stopped by the addition of an equivalent amount of sequencing gel loading buffer at 0, 2.5, 5, 10, and 20 min. Samples were heated at 95°C, run on 16% polyacrylamide-8 M urea sequencing gel, and quantitated by PhosphorImager analysis. The percentages removed shown in Fig. 2 and 3 were determined by quantitating the upper chain-terminated primer band, dividing this by the total for all bands in the lane, and subtracting from 100%. No degradation of the primer band was detected when each RT enzyme was incubated with the template/primer for 15 to 30 min at 37°C in the absence of ATP or sodium pyrophosphate, thus ruling out exonuclease activity. The ATP (Pharmacia Biotech) is 98% triphosphate and 99.5% adenosine by fast protein liquid chromatography. For comparison of wild-type RT and the mutant RT, 2.5 nM template/chain-terminated primer was incubated with 20 nM wild-type or mutant recombinant RT enzymes, with equivalent activity determined as previously described (25), for 5 min at 37°C in RT buffer and then ATP (3.2 mM final concentration) was added for 0, 15, 45, and 90 min as shown in Fig. 4 (0, 15, and 45 min for d4T). Catalytic efficiencies for removal of AZT, d4T, and tenofovir from terminated primers by wild-type and mutant RT were determined at ATP concentrations of 0.36, 1.1, 2.2, 3.2, and 4.5 mM at 0.1, 1.5, 3, 10, and 30 min, respectively. The Vmax for ATP-dependent removal and the Km for ATP were determined by using the Michaelis-Menten hyperbolic relationship curve and fitting to the equation y = (Vmaxx)/(Km + x) with SigmaPlot, version 4.01. The second-order rate constant, or catalytic efficiency, was determined by calculating kcat from Vmax and dividing by the Km for ATP.
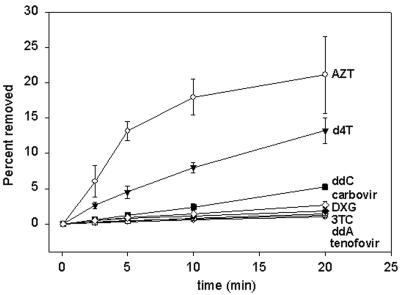
FIG. 2.
Comparison of NRTI removal by mutant RT. Shown is removal of NRTIs AZT (open circle), d4T (filled inverted triangle), ddC (filled square), carbovir (open diamond), DXG (filled diamond), 3TC (open square), ddA (open triangle), and tenofovir (filled circle) averaged from two or three experiments with standard deviations following addition of 3.2 mM ATP in the absence of the next complementary dNTP.
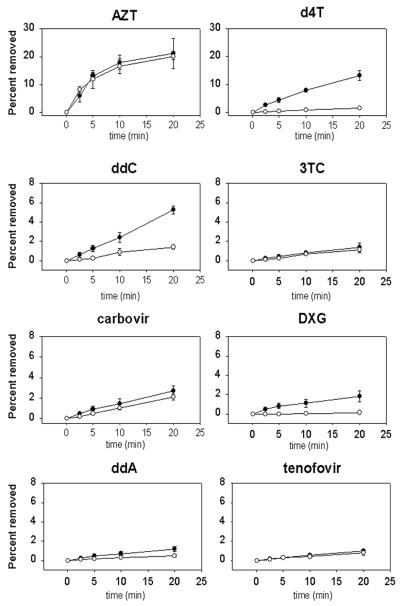
FIG. 3.
Effect of the next complementary dNTP on ATP-dependent removal. Shown is removal of AZT, d4T, ddC, 3TC, carbovir, DXG, ddA, and tenofovir averaged from two or three experiments with standard deviations in the presence (open circles) and absence (filled circles) of 50 μM concentrations of the next complementary dNTP. Note that the y axes for the AZT and d4T graphs extend from 0 to 30% while those for the ddC, 3TC, carbovir, DXG, ddA, and tenofovir graphs extend from 0 to 8%.
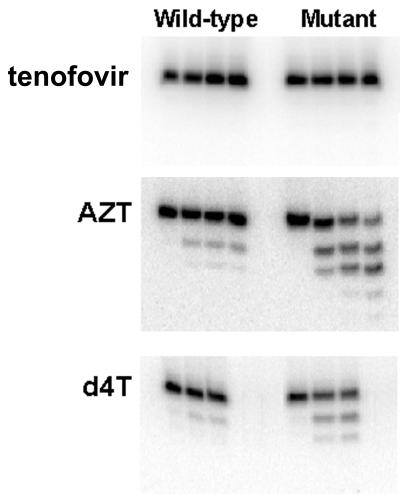
FIG. 4.
Analysis of AZT, d4T, and tenofovir ATP-dependent removal by wild-type and mutant RT. Shown is removal of tenofovir, AZT, and d4T from tenofovir-, AZT-, and d4T-terminated primers/templates by wild-type RT and the RT mutant with D67N, K70R, and T215Y RT mutations following the addition of 3.2 mM ATP.
RESULTS
Removal by the ATP-dependent chain terminator removal mechanism.
We evaluated the ATP-dependent chain terminator removal of NRTIs using a mutant RT enzyme containing mutations D67N, K70R, and T215Y. Removal of the active metabolites of NRTIs AZT, d4T, ddI, abacavir, ddC, 3TC, DAPD, and tenofovir were analyzed. The active metabolites of ddI, abacavir, and DAPD are ddA, carbovir, and DXG, respectively. The thymidine analogs, AZT and d4T, were most efficiently removed by the addition of ATP, with 21% of the AZT removed and 13% of the d4T removed at 20 min (Fig. 1 and 2). There was reduced removal of ddC, carbovir, DXG, 3TC, ddA, and tenofovir compared to removal of AZT and d4T, with less than 5% of these chain terminators removed by 20 min. The order of removal at 20 min was determined from multiple experiments as AZT (21%) > d4T (13%) ≫ ddC (5%) > carbovir (3%) > DXG (2%) > 3TC (1.4%) > ddA (1.2%) > tenofovir (1%) (Fig. 2). Rates of removal were as follows: AZT, 1.8%/min; d4T, 0.64%/min; ddC, 0.26%/min, carbovir, 0.13%/min; DXG, 0.08%/min; 3TC, 0.07%/min; ddA, 0.06%/min; tenofovir, 0.05%/min. No difference in removal of ddA and tenofovir was seen when a longer chain-terminated primer of 24 nt was also used (data not shown). Therefore, the inefficient removal of ddA and tenofovir in these experiments does not appear to be primer length dependent.
FIG. 1.
ATP-dependent removal of chain terminator inhibitors by mutant RT. Removal of AZT, d4T, ddC, 3TC, carbovir, DXG, ddA, and tenofovir at 0, 10, and 20 min following addition of 3.2 mM ATP both in the absence and presence (+dNTP) of a 50 μM concentration of the next complementary dNTP as indicated.
Drug susceptibility in viral replication assays.
The antiviral drug susceptibilities of the mutant virus to the eight NRTIs were determined (Table 1). The mutant showed a 24-fold decrease in susceptibility to AZT compared to wild-type virus but less than fourfold changes in susceptibility to the other drugs (Table 1). The drug susceptibility results with these recombinant RT mutant viruses are similar to the drug susceptibility results obtained from patient viruses containing these mutations in a background of additional RT mutations (23). The efficient ATP-dependent removal of AZT by the mutant RT is consistent with the significant decrease in susceptibility of the mutant virus. The inefficient removal of ddC, carbovir, DXG, ddA, 3TC, and tenofovir by the RT mutant is also consistent with the minor changes in the susceptibility of the mutant virus to these inhibitors.
TABLE 1.
NRTI susceptibilities for recombinant HIV
| NRTI | IC50a (μM) for:
|
Fold change | |
|---|---|---|---|
| HXB2D (wild type) | Mutantb | ||
| Tenofovir | 3.0 ± 0.2 | 7.2 ± 1.1 | 2.4 |
| AZT | 0.18 ± 0.02 | 4.4 ± 1.40 | 24.0 |
| d4T | 8.0 ± 0.4 | 11.8 ± 1.5 | 1.5 |
| 3TC | 2.2 ± 0.4 | 8.4 ± 1.2 | 3.8 |
| ddC | 3.8 ± 0.5 | 6.0 ± 0.5 | 1.6 |
| ddI | 4.0 ± 0.5 | 5.8 ± 0.5 | 1.5 |
| Abacavir | 0.3 ± 0.03 | 0.7 ± 0.08 | 2.4 |
| DXG | 1.9 ± 0.2 | 2.7 ± 0.6 | 1.4 |
Values are averages from three to eight experiments ± standard errors. IC50, 50% inhibitory concentration.
Mutant with D67N, K70R, and T215Y mutations.
Effect of the next complementary nucleotide on chain terminator removal.
The increase in ATP-dependent removal of d4T seen with the mutant RT appears inconsistent with the in vitro susceptibility changes seen for d4T. We have demonstrated efficient removal of d4T with an RT containing thymidine analog resistance mutations, in agreement with the results of others (Fig. 1 and 2) (20). However, susceptibility shifts of less than twofold to d4T for viruses with thymidine analog resistance mutations are observed in phenotypic in vitro susceptibility assays, therefore showing little cross-resistance between d4T and AZT (Table 1) (11, 16, 18, 27). Nevertheless, thymidine analog resistance mutations are selected in d4T-treated patients (9, 17, 18, 26, 30, 31), and patients with thymidine analog resistance mutations show reduced responses to d4T therapy (24, 26). An explanation for this apparent discrepancy between in vitro phenotypic data and clinical results has recently been described: the next incoming complementary dNTP can inhibit removal of some NRTIs by forming a dead-end complex (19, 20).
We analyzed the effect of the next complementary nucleotide on removal of each of the NRTIs (Fig. 1 and 3). The presence of the next nucleotide had little effect on the removal of AZT and carbovir. However, removal of d4T, ddC, and DXG was decreased by more than threefold in the presence of the next dNTP. The minimal removal of tenofovir, 3TC, and ddA made it difficult to assess the effect of the next complementary dNTP. Our results support the data of others showing that removal of d4T by RT was inhibited by the next complementary dNTP, whereas AZT removal was much less sensitive to this inhibition (20).
Catalytic efficiencies for removal of AZT, d4T, and tenofovir by wild-type and mutant RT.
Removal of AZT and d4T by wild-type RT was detectable but reduced compared to removal by the mutant RT (Table 2 and Fig. 4). Removal of the other NRTIs by wild-type RT was minimal (data not shown). ATP-dependent removal of the thymidine analogs, AZT and d4T, was compared to removal of the adenosine analog, tenofovir, by both wild-type and mutant RT. When ATP was added at a physiological concentration, AZT and d4T were readily removed by both wild-type RT and the RT mutant within 15 min (Fig. 4). Removal of tenofovir by both wild-type RT and mutant RT was not easily detectable even for incubation periods of up to 90 min (Fig. 4). The catalytic efficiencies (kcat/Km) of ATP-dependent removal of AZT, d4T, and tenofovir were determined for both the wild-type and mutant RT enzymes. AZT and d4T were both removed by the ATP-dependent mechanism more efficiently than tenofovir: 15-fold more efficiently by wild-type RT and 35-fold and 22-fold more efficiently, respectively, by the mutant RT (Table 2). The catalytic efficiencies of ATP-dependent removal of these inhibitors in the presence of the next complementary dNTP were also determined (Table 2). Tenofovir was minimally removed by the mutant RT, with a catalytic efficiency of 1.7 × 104 to 1.9 × 104 M−1 s−1 regardless of the presence of the next complementary dNTP. AZT removal by the mutant RT was not significantly decreased by the next complementary dNTP, maintaining a fourfold increase in removal efficiency compared to that for wild-type RT. Consistent with our results above, d4T removal by the ATP-dependent mechanism was decreased in the presence of the next dNTP from 38 × 104 M−1 s−1 to 14 × 104 M−1 s−1, returning the efficiency of removal to the level for the wild-type RT.
TABLE 2.
Catalytic efficiency of ATP-dependent removal
| NRTI |
Kcat/Km (M−1 s−1, 104)c for:
|
||
|---|---|---|---|
| Wild-type RTa | Mutant RTb | Mutant RT + next dNTP | |
| Tenofovir | 0.94 ± 0.17 | 1.7 ± 0.3 | 1.9 ± 0.1 |
| AZT | 13.5 ± 0.7 | 60 ± 11 | 49 ± 4 |
| d4T | 13.5 ± 2.1 | 38 ± 7.1 | 14 ± 2 |
Wild-type RT average Vmax values for removal of tenofovir, AZT, and d4T: 0.037 ± 0.02, 0.45 ± 0.17, and 0.12 ± 0.00 s−1, respectively; average Km values: 3.9 ± 1.9, 3.3 ± 1.5, and 0.9 ± 0.1 μM, respectively.
Mutant RT average Vmax values for removal of tenofovir, AZT, and d4T: 0.020 ± 0.006, 1.7 ± 0.8, and 0.6 ± 0.3 s−1, respectively; average Km values: 2.1 ± 0.4, 4.1 ± 0.9, and 2.2 ± 0.5 μM, respectively.
Values are averages ± standard errors.
ATP-dependent removal and drug susceptibility.
The RT mutant demonstrated increased efficiency of removal of AZT, d4T, and tenofovir compared to wild-type RT (Table 2 and Fig. 4). AZT was removed four- to fivefold more efficiently by the mutant RT than by wild-type RT, further supporting the observations of others that increased AZT removal by RT mutants with thymidine analog resistance mutations may contribute to the significant decrease in the AZT susceptibility of viruses containing these mutations. The mutant RT showed a less-than-twofold increase in tenofovir removal compared to wild-type RT. These results are consistent with the twofold changes in tenofovir susceptibility (Table 1). The mutant RT removes d4T threefold more efficiently by the ATP-dependent removal mechanism than wild-type RT (Table 2). However, we have demonstrated that the presence of the next dNTP can inhibit d4T removal and decrease the efficiency of d4T removal by the RT mutant to levels comparable to those for wild-type RT (Table 2), which is consistent with the less-than-twofold change in d4T susceptibility seen for this mutant (Table 1).
DISCUSSION
The lack of correlation between changes in inhibitor binding for AZT and the significant changes in AZT susceptibility for viruses with thymidine analog resistance mutations has led to research into other resistance mechanisms. Recent work has suggested that an increased rate of AZT excision by pyrophosphorolysis (reverse nucleotide polymerization) or a similar mechanism using ATP instead of pyrophosphate could account for part of the AZT resistance resulting from thymidine analog resistance mutations (1, 6, 19, 21). Our results confirm that HIV-1 RT can remove chain-terminating RT inhibitors in the presence of physiological concentrations of ATP. A comparison of levels of ATP-dependent chain terminator removal of NRTIs by a mutant RT containing common thymidine analog resistance mutations D67N, K70R, and T215Y demonstrated that the thymidine analogs, AZT and d4T, were the most efficiently removed. This mutant RT showed intermediate levels of removal of ddC, carbovir, and DXG, followed by minimal removal of 3TC, ddA, and tenofovir. The inefficient removal of ddC, carbovir, DXG, ddA, 3TC, and tenofovir by the RT mutant is consistent with the minor in vitro changes in the susceptibility of the mutant virus to these inhibitors.
The addition of ATP and the next complementary dNTP reduced ATP-mediated removal of d4T, ddC, and DXG, while AZT and carbovir removal was unaffected. Tenofovir, ddA, and 3TC were minimally removed in the presence or absence of the next complementary dNTP. The catalytic efficiency of tenofovir removal by the mutant RT enzyme was 35- and 22-fold less efficient than removal of AZT and d4T, respectively. Furthermore, mutant RT showed a less-than-twofold increase in tenofovir removal compared to wild-type RT, consistent with the twofold changes in tenofovir susceptibility for the mutant virus. AZT removal by the mutant RT remained four- to fivefold more efficient than that by wild-type RT even in the presence of the next dNTP, consistent with susceptibility changes for HIV mutants with thymidine analog resistance mutations and showing AZT resistance. A mutant containing thymidine analog mutations M41L and K219Q in addition to D67N, K70R, and T215Y has been analyzed and has been shown to have a 131-fold decrease in tenofovir removal compared to AZT removal. Tenofovir removal by the mutant RT with M41L, D67N, K70R, T215Y, K219Q mutations is not significantly different from that of wild-type RT, but AZT removal is increased by sevenfold compared to that by wild-type RT. These results are consistent with the changes in the susceptibility of the mutant viruses to these inhibitors (L. K. Naeger, unpublished results).
While d4T is readily removed by the RT mutant with D67N, K70R, T215Y mutations, the next incoming dNTP can inhibit the efficiency of d4T removal by the mutant to levels comparable to those for wild-type RT. These findings with d4T suggest that current in vitro antiviral phenotypic assays may not adequately measure susceptibility to d4T. The higher dNTP concentrations of the activated peripheral blood lymphocytes and transformed human cells used in in vitro phenotypic assays (11-13) may inhibit removal of d4T. Cell culture results for HIV-1 containing thymidine analog resistance mutations commonly demonstrate resistance to AZT and sensitivity to d4T, but the true cross-resistance of d4T in these cells may be masked by high dNTP concentrations. Under conditions of low dNTP levels (e.g., quiescent primary T cells or macrophages), d4T removal would be expected to occur at a higher efficiency, thus possibly explaining the observed in vivo cross-resistance to d4T of HIV-1 mutants containing thymidine analog resistance mutations. The inhibition of ddC and DXG removal by the next complementary dNTP also suggests that current in vitro phenotypic assays showing less-than-twofold changes in ddC and DXG susceptibility for viruses producing RT mutants with thymidine analog resistance mutations may not adequately reflect their in vivo susceptibilities.
The modeling of an AZT-terminated primer/template with HIV-1 RT has shown that translocation of the terminating AZT to the P site (primer site) from the N site (incoming dNTP site) would result in the azide group of AZT having unfavorable interactions (steric clash) with amino acid D185, suggesting that the AZT-terminated end of the primer would preferentially occupy the N site and block the incoming dNTP (5). Furthermore, if the terminated AZT primer end translocated to the P site, steric hindrance from the azide group would interfere with the binding of the incoming dNTP. Therefore, AZT is more likely to occupy the N position, even when dNTPs are present, compared to other dideoxynucleoside triphosphates, resulting in AZT being in a favorable position for removal by ATP. Conversely, dideoxynucleotides, such as d4T, where there is no steric hindrance, would likely favor the P position rather than N position so that the incoming dNTP could interfere with their removal (5). These results support the biochemical data of our group and others showing that removal of AZT is not inhibited by the next incoming dNTP, whereas removal of d4T is inhibited (5). Furthermore, the modeling performed by Boyer et al. proposes that mutations M41L, D67N, K70R, T215Y, and K219Q enhance ATP binding to HIV-1 RT so that, at physiological concentrations of ATP, removal would be expected to be more efficient regardless of the nucleotide (5). Consequently, the specificity of the removal reaction is not dictated by the mutations conferring resistance but rather depends on the structure of the region around the active site of HIV-1 RT and the structure of the nucleoside analog.
The reduced removal of tenofovir might result from the unique phosphonate bond of tenofovir or the unique acyclic structure of tenofovir. The incorporation of tenofovir into the primer may favor the P site rather than the N site, resulting in tenofovir being less accessible to removal by ATP, opposite to what was proposed for AZT. Analyses of crystal structures of tenofovir-terminated primers/templates with HIV-1 RT to address this possibility are under way. Similarly, inefficient removal of 3TC and ddA may also occur because these nucleoside analogs favor the P site rather than the N site. It is also interesting that both tenofovir and ddA are inefficiently removed by ATP, suggesting that adenosine analogs are less likely to be removed by this mechanism than other nucleoside analogs. Our in vitro results reported here showing reduced tenofovir removal by a mutant RT with thymidine analog resistance mutations are consistent with clinical results that have demonstrated significant anti-HIV activity of tenofovir disoproxil fumarate in antiretroviral-experienced patients with extensive resistance mutations, including thymidine analog resistance mutations, in their HIV (22; R. Schooley, R. Myers, P. Ruane, G. Beall, H. Lampiris, M. Miller, R. Mills, and I. McGowan, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 692, p. 293, 2000).
Acknowledgments
We thank Craig Gibbs, Mick Hitchcock, and Kirsten Lofgren White for review of the manuscript and Melanie Lehwalder for administrative support.
REFERENCES
- 1.Arion, D., N. Kaushik, S. McCormick, G. Borkow, and M. A. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908-15917. [DOI] [PubMed] [Google Scholar]
- 2.Barshop, B. A., D. T. Adamson, D. C. Vellom, F. Rosen, B. L. Epstein, and J. E. Seegmiller. 1991. Luminescent immobilized enzyme test systems for inorganic pyrophosphate: assays using firefly luciferase and nicotinamide-mononucleotide adenylyl transferase or adenosine-5′-triphosphate sulfurylase. Anal. Biochem. 197:266-272. [DOI] [PubMed] [Google Scholar]
- 3.Battula, N., and L. A. Loeb. 1976. On the fidelity of DNA replication. Lack of exodeoxyribonuclease activity and error-correcting function in avian myeloblastosis virus DNA polymerase. J. Biol. Chem. 251:982-986. [PubMed] [Google Scholar]
- 4.Boucher, C. A. B., W. Keulen, T. van Bommel, M. Nijhuis, D. de Jong, M. D. de Jong, P. Schipper, and N. K. T. Back. 1996. HIV-1 drug susceptibility determination by using recombinant viruses generated from patient sera tested in a cell-killing assay. Antimicrob. Agents Chemother. 40:2404-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, P., S. Sarafianos, E. Arnold, and S. Hughes. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canard, B., S. R. Sarfati, and C. C. Richardson. 1998. Enhanced binding of azidothymidine-resistant human immunodeficiency virus 1 reverse transcriptase to the 3′-azido-3′-deoxythymidine 5′-monophosphate-terminated primer. J. Biol. Chem. 273:14596-14604. [DOI] [PubMed] [Google Scholar]
- 7.Carroll, S. S., J. Geib, D. B. Olsen, M. Stahlhut, J. A. Shafer, and L. C. Kuo. 1994. Sensitivity of HIV-1 reverse transcriptase and its mutants to inhibition by azidothymidine triphosphate. Biochemistry 33:2113-2120. [DOI] [PubMed] [Google Scholar]
- 8.Cherrington, J. M., M. D. Fuller, A. S. Mulato, S. J. W. Allen, S. C. Kunder, M. A. Ussery, Z. Lesnikowski, R. F. Schinazi, J.-P. Sommadossi, and M. S. Chen. 1996. Comparative kinetic analysis of interaction of inhibitors with Rauscher murine leukemia virus and human immunodeficiency virus reverse transcriptases. Antimicrob. Agents Chemother. 40:1270-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Mendoza, C., V. Soriano, C. Briones, O. Gallego, P. Barreiro, A. Alvarez, and J. Gonzalez-Lahoz. 2000. Emergence of zidovudine resistance in HIV-infected patients receiving stavudine. J. Acquir. Immune Defic. Syndr. 23:279-281. [DOI] [PubMed] [Google Scholar]
- 10.Gu, Z., Q. Gao, H. Fang, H. Salomon, M. A. Parniak, E. Goldberg, J. Cameron, and M. A. Wainberg. 1994. Identification of a mutation of codon 65 in the IKKK motif of reverse transcriptase that encodes human immunodeficiency virus resistance to 2′,3′-dideoxycytidine and 2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 38:275-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hertogs, K., M. P. de Bethune, V. Miller, T. Ivens, P. Schel, A. Van Cauwenberge, C. Van Den Eynde, V. Van Gerwen, H. Azijn, M. Van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Japour, A. J., D. L. Mayers, V. A. Johnson, D. R. Kuritzkes, L. A. Beckett, J. M. Arduino, J. Lane, R. J. Black, P. S. Reichelderfer, R. T. D'Aquila, et al. 1993. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob. Agents Chemother. 37:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellam, P., and B. A. Larder. 1994. Recombinant virus assay: a rapid, phenotypic assay for assessment of drug susceptibility of human immunodeficiency virus type 1 isolates. Antimicrob. Agents Chemother. 38:23-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerr, S. G., and K. S. Anderson. 1997. Pre-steady-state kinetic characterization of wild type and 3′-azido-3′-deoxythymidine (AZT) resistant human immunodeficiency virus type 1 reverse transcriptase: implication of RNA directed DNA polymerization in the mechanism of AZT resistance. Biochemistry 36:14064-14070. [DOI] [PubMed] [Google Scholar]
- 15.Krebs, R., U. Immendorfer, S. H. Thrall, B. M. Wohrl, and R. S. Goody. 1997. Single-step kinetics of HIV-1 reverse transcriptase mutants responsible for virus resistance to nucleoside inhibitors zidovudine and 3-TC. Biochemistry 36:10292-10300. [DOI] [PubMed] [Google Scholar]
- 16.Larder, B. A., B. Chesebro, and D. D. Richman. 1990. Susceptibilities of zidovudine-susceptible and -resistant human immunodeficiency virus isolates to antiviral agents determined by using a quantitative plaque reduction assay. Antimicrob. Agents Chemother. 34:436-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, P. F., C. J. Gonzalez, B. Griffith, G. Friedland, V. Calvez, F. Ferchal, R. F. Schinazi, D. H. Shepp, A. B. Ashraf, M. A. Wainberg, V. Soriano, J. W. Mellors, and R. J. Colonno. 1999. Stavudine resistance: an update on susceptibility following prolonged therapy. Antivir. Ther. 4:21-28. [PubMed] [Google Scholar]
- 18.Lin, P.-F., H. Samanta, R. E. Rose, A. K. Patrick, J. Trimble, C. M. Bechtold, D. R. Revie, N. C. Khan, M. E. Federici, H. Li, A. Lee, R. E. Anderson, and R. J. Colonno. 1994. Genotypic and phenotypic analysis of human immunodeficiency virus type 1 isolates from patients on prolonged stavudine therapy. J. Infect. Dis. 170:1157-1164. [DOI] [PubMed] [Google Scholar]
- 19.Meyer, P. R., S. E. Matsuura, A. M. Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35-43. [DOI] [PubMed] [Google Scholar]
- 20.Meyer, P. R., S. E. Matsuura, R. F. Schinazi, A. G. So, and W. A. Scott. 2000. Differential removal of thymidine nucleotide analogues from blocked DNA chains by human immunodeficiency virus reverse transcriptase in the presence of physiological concentrations of 2′-deoxynucleoside triphosphates. Antimicrob. Agents Chemother. 44:3465-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer, P. R., S. E. Matsuura, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 95:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, M., N. Margot, R. Schooley, R. Mills, and I. McGowan. 2000. Anti-HIV responses and development of RT mutations in antiretroviral-experienced patients adding tenofovir DF therapy: 48 week genotypic analysis of study 902. AIDS 14:S111. [Google Scholar]
- 23.Miller, M. D., K. E. Anton, A. S. Mulato, P. D. Lamy, and J. M. Cherrington. 1999. Human immunodeficiency virus type 1 expressing the lamivudine-associated M184V mutation in reverse transcriptase shows increased susceptibility to adefovir and decreased replication capability in vitro. J. Infect. Dis. 179:92-100. [DOI] [PubMed] [Google Scholar]
- 24.Montaner, J. S., T. Mo, J. M. Raboud, S. Rae, C. S. Alexander, C. Zala, D. Rouleau, and P. R. Harrigan. 2000. Human immunodeficiency virus-infected persons with mutations conferring resistance to zidovudine show reduced virologic responses to hydroxyurea and stavudine-lamivudine. J. Infect. Dis. 181:729-732. [DOI] [PubMed] [Google Scholar]
- 25.Naeger, L. K., N. A. Margot, and M. D. Miller. 2001. Altered drug susceptibility of HIV-1 RT mutants containing M184V and zidovudine-resistance mutations: analysis of RT processivity, viral replication, and chain-terminator removal. Antivir. Ther. 6:111-123. [PubMed] [Google Scholar]
- 26.Pellegrin, I., J. Izopet, J. Reynes, M. Denayrolles, B. Montes, J. L. Pellegrin, P. Massip, J. Puel, H. Fleury, M. Segondy, et al. 1999. Emergence of zidovudine and multidrug-resistance mutations in the HIV-1 reverse transcriptase gene in therapy-naive patients receiving stavudine plus didanosine combination therapy. AIDS 13:1705-1709. [DOI] [PubMed] [Google Scholar]
- 27.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts, J. D., K. Bebenek, and T. A. Kunkel. 1988. The accuracy of reverse transcriptase from HIV-1. Science 242:1171-1173. [DOI] [PubMed] [Google Scholar]
- 29.Russell, R. G. 1976. Metabolism of inorganic pyrophosphate (PPi). Arthritis Rheum. 19:465-478. [DOI] [PubMed] [Google Scholar]
- 30.Schinazi, R. F., B. A. Larder, and J. W. Mellors. 2000. Mutations in retroviral genes associated with drug resistance: 2000-2001 update. Int. Antivir. News 8:65-91. [Google Scholar]
- 31.Soriano, V., U. Dietrich, N. Villalba, A. Immelmann, A. Gil-Aguado, S. Echevarria, B. Clotet, I. Ocana, J. M. Santamaria, E. Bouza, V. Barona, J. M. Gatell, and J. Gonzalez-Lahoz. 1997. Lack of emergence of genotypic resistance to stavudine after 2 years of monotherapy. AIDS 11:696-697. [PubMed] [Google Scholar]
- 32.Traut, T. 1994. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140:1-22. [DOI] [PubMed] [Google Scholar]