Abstract
Full characterization of methicillin-resistant Staphylococcus aureus (MRSA) requires definition of not only the bacterial genetic background but also the structure of the complex and heterologous mec element these bacteria carry, which is associated with drug resistance determinant mecA. We report the development, validation, and application of a multiplex PCR strategy that allows quick presumptive characterization of the mec element types based on the structural features that were shown to be typical of mec elements carried by several MRSA clones. The strategy was validated by using a representative collection of pandemic MRSA clones in which the full structure of the associated mec elements was previously determined by hybridization and PCR screenings and also by DNA sequencing. The method was tested together with multilocus sequence typing and other typing methods for the characterization of 18 isolates representative of the MRSA clones recovered during a hospital outbreak in Barcelona, Spain. The multiplex PCR was shown to be rapid, robust, and capable in a single assay of identifying five structural types of the mec element among these strains, three major and two minor variants, each one of which has been already been seen among MRSA characterized earlier. This technique should be a useful addition to the armamentarium of molecular typing tools for the characterization of MRSA clonal types and for the rapid tentative identification of structural variants of the mec element.
Molecular typing techniques have been used with increasing frequency in studies of the epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) and also for a better understanding of the evolutionary relationships among MRSA clones (3, 7, 9, 26). One of the conclusions emerging from these studies was that a complete characterization of MRSA lineages requires not only identification of the genetic background of the bacteria but also identification of the structural types of the large and heterologous mec element, which carries methicillin resistance determinant mecA (11, 26, 27).
Studies by Ito et al. have elucidated the complete structure of three major mec elements, also referred to as the staphylococcal chromosomal cassette (SCCmec). Type I (34 kb) was identified in the first MRSA strain isolated in 1961 in the United Kingdom (strain NCTC10442), type II (52 kb) was identified in an MRSA strain isolated in 1982 in Japan (strain N315), and type III (66 kb) was identified in an MRSA strain isolated in 1985 in New Zealand (strain 82/2082) (12, 13). More recently, a smaller fourth mec element, SCCmec type IV (20 to 24 kb), was independently identified among representatives of the Pediatric clone (26) and in two community-acquired MRSA strains (18).
In a recent report we described the characterization of the structural types of the mec elements carried by 28 MRSA isolates belonging to five widespread epidemic clones, which were all characterized by multilocus sequence typing (MLST) as single-locus or double-locus variants of two completely different genetic backgrounds (26). The full structure of each mec element carried by these MRSA isolates was determined by Southern blot analysis using three restriction enzymes and several key probes specific for each SCCmec type, by PCR detection of several loci covering the entire mec element structures, and by DNA sequencing, based on the information described in references 12 and 13.
However, these methods are laborious and time-consuming. In this paper we describe a multiplex PCR strategy which was designed to detect the structural variations observed in the mec element in our previous study. Such a method should provide a useful tool for the rapid tentative identification of the structural type of the mec elements in MRSA isolates.
MATERIALS AND METHODS
Strain selection.
The 26 strains used for the validation of the multiplex PCR were selected from among MRSA isolates previously characterized for MLST profile and for the structural type of the mec element (26) (Table 1). Strains COL, N315, and ANS46 were included as controls for the three major structural types of mec elements, as previously described (26). Genome data from strain COL (www.tigr.org) indicate that this strain has a mec element identical to that of strain NCTC10442, used by Ito et al. for the definition of SCCmec I (13). Strain N315 was used in the definition of SCCmec II (13). The physical mapping of the mecA region of strain ANS46 showed that it carried a mec element identical to that of SCCmec III (5, 12). After validation, the multiplex PCR was applied to the characterization of 18 representative isolates of MRSA recovered during an outbreak investigation in a hospital in Barcelona, Spain. These strains had been characterized earlier by pulsed-field gel electrophoresis (PFGE) (4).
TABLE 1.
MRSA strains used for the validation of the multiplex PCR
| Strain | Isolation
|
Clonal typeb | MLST profile | SCCmec type by:
|
||
|---|---|---|---|---|---|---|
| Origina | Yr | Full characterization (26) | Multiplex PCR | |||
| UK13136 | UK | 1960 | Archaic | 3-3-1-1-4-4-16 | I | I |
| BK793 | Egypt | 1961 | Archaic | 3-3-1-1-4-4-16 | I | I |
| COL | UK | 1965 | Archaic | 3-3-1-1-4-4-16 | I | I |
| E2125 | Denmark | 1964 | Archaic | 3-3-1-12-4-4-16 | I | I |
| PER34 | Spain | 1989 | Iberian | 3-3-1-1-4-4-16 | IA | IA |
| HPV107 | Portugal | 1992 | Iberian | 3-3-1-12-4-4-16 | IA | IA |
| BK1953 | USA | 1995 | Iberian | 3-3-1-12-4-4-16 | IA | IA |
| PER184 | Spain | 1991 | Iberian | 3-3-1-12-4-4-16 | IA | I |
| PER88 | Spain | 1992 | Iberian | 3-3-1-12-4-4-16 | IA | IA |
| N315 | Japan | 1982 | NY/Japan | 1-4-1-4-12-1-10 | II | II |
| BK2464 | USA | 1996 | NY/Japan | 1-4-1-4-12-1-10 | II | II |
| JP1 | Japan | 1997 | NY/Japan | 1-4-1-4-12-1-10 | II | II |
| ANS46c | Australia | 1982 | 2-3-1-1-4-4-3 | III | III | |
| R35c | USA | 1987 | 2-3-1-1-4-4-3 | III | III | |
| HU25 | Brazil | 1993 | Brazilian | 2-3-1-1-4-4-3 | IIIA | IIIA |
| HSJ216 | Portugal | 1997 | Brazilian | 2-3-1-1-4-4-3 | IIIA | IIIA |
| HDG2 | Portugal | 1993 | Brazilian | 2-3-1-1-4-4-3 | IIIB | IIIB |
| HUSA304 | Hungary | 1993 | Hungarian | 2-3-1-1-4-4-3 | III | III |
| HU106 | Hungary | 1996 | Hungarian | 2-3-1-1-4-4-3 | III | III |
| BM18 | USA | 1989 | Pediatric | 1-4-1-4-12-1-10 | IV | IV |
| PL72 | Poland | 1991 | Pediatric | 1-4-1-4-12-1-10 | IV | IV |
| POL3 | Poland | 1992 | Pediatric | 1-4-1-4-12-1-10 | IV | IV |
| HDE288 | Portugal | 1996 | Pediatric | 1-4-1-4-12-1-10 | IV | IV |
| COB3 | Colombia | 1996 | Pediatric | 1-4-1-4-12-1-10 | IV | IV |
| BK2529 | USA | 1996 | Clone Vd | 3-3-1-1-4-4-3 | IV | IV |
| BARGII17 | USA | 1996 | Clone Vd | 3-3-1-1-4-4-3 | IV | IV |
UK, United Kingdom; USA, United States.
Clonal types were defined by a combination of three molecular typing techniques: polymorphisms in the vicinity of ClaI::mecA; ClaI::Tn554 insertion patterns, and SmaI PFGE macrorestriction profiles (see Materials and Methods).
DNA isolation.
Chromosomal DNAs from three to five isolated colonies were prepared by using the Wizard genomic DNA preparation kit (Promega, Madison, Wis.), with lysostaphin at 0.5 mg/ml and RNase at 0.3 mg/ml for the lysis step.
MLST and spaA typing.
Molecular typing based on the sequences of seven housekeeping genes (MLST) and the typing of the polymorphic region of protein A (spaA typing) were performed according to previously described procedures (7, 25, 33).
Multiplex PCR for mec element type assignment.
The multiplex PCR includes eight loci (A through H) selected on the basis of previously described mec element sequences (12, 13, 26) (Table 2). Also included in the protocol as an internal positive control was the mecA gene. Locus A is located downstream of the pls gene and is specific for SCCmec type I; locus B is internal to the kdp operon, which is specific for SCCmec type II; locus C is internal to the mecI gene present in SCCmec types II and III; locus D is internal to the dcs region, present in SCCmec types I, II, and IV; locus E is located in the region between integrated plasmid pI258 and transposon Tn554, specific for SCCmec type III; locus F, which is also specific for SCCmec type III, is located in the region between Tn554 and the chromosomal right junction (orfX). Loci G and H were included to distinguish structural variants IA and IIIA, respectively. Locus G is the left junction between IS431 and pUB110, and locus H is the left junction between IS431 and pT181. Primers were designed manually and were commercially obtained (Gibco, Invitrogen Corporation, Carlsbad, Calif.). Primer length and GC contents were kept as uniform as possible in order to minimize differences in the annealing temperature and kinetics, and primer sequences were checked for specificity against available S. aureus genomes and SSCmec sequences with the BLAST utility available through the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov). Amplicon sizes ranged from 162 to 495 bp, differing by at least 40 bp in size from one another (Table 2).
TABLE 2.
Primers used in the multiplex PCR
| Locus | Primer | Oligonucleotide sequence (5′-3′) | Location | Amplicon size (bp) | Specificitye (SCCmec type) |
|---|---|---|---|---|---|
| A | CIF2 F2 | TTCGAGTTGCTGATGAAGAAGG | 18398-18419a | 495 | I |
| CIF2 R2 | ATTTACCACAAGGACTACCAGC | 18892-18871a | |||
| B | KDP F1 | AATCATCTGCCATTGGTGATGC | 10445-10467b | 284 | II |
| KDP R1 | CGAATGAAGTGAAAGAAAGTGG | 10728-10707b | |||
| C | MECI P2 | ATCAAGACTTGCATTCAGGC | 42428-42447b | 209 | II, III |
| MECI P3 | GCGGTTTCAATTCACTTGTC | 42636-42617b | |||
| D | DCS F2 | CATCCTATGATAGCTTGGTC | 38011-37992a | 342 | I, II, IV |
| DCS R1 | CTAAATCATAGCCATGACCG | 37670-37689a | |||
| E | RIF4 F3 | GTGATTGTTCGAGATATGTGG | 45587-45607c | 243 | III |
| RIF4 R9 | CGCTTTATCTGTATCTATCGC | 45829-45809c | |||
| F | RIF5 F10 | TTCTTAAGTACACGCTGAATCG | 59573-59594c | 414 | III |
| RIF5 R13 | GTCACAGTAATTCCATCAATGC | 59986-59965c | |||
| G | IS431 P4 | CAGGTCTCTTCAGATCTACG | 49963-49982b | 381 | |
| pUB110 R1 | GAGCCATAAACACCAATAGCC | 50343-50323b | |||
| H | IS431 P4 | CAGGTCTCTTCAGATCTACG | 29654-29673c | 303 | |
| pT181 R1 | GAAGAATGGGGAAAGCTTCAC | 29976-29956c | |||
| mecA | MECA P4 | TCCAGATTACAACTTCACCAGG | 1190-1211d | 162 | Internal control |
| MECA P7 | CCACTTCATATCTTGTAACG | 1351-1332d |
Loci G and H were included to distinguish variants IA from I and IIIA from III, respectively.
The optimization of the multiplex PCR followed the general principles described by Henegariu et al. (10). Each pair of primers was first tested for amplification specificity and robustness at annealing temperatures of 55 and 60°C. For the multiplex PCR, we found it necessary to decrease the annealing temperature, increase the extension time, and adjust primer amounts for some loci. These alterations were tested empirically in small steps. The multiplex PCR was performed in a 50-μl volume with the GeneAmp PCR kit (Applied Biosystems, Foster City, Calif.) containing the following: 1× PCR buffer II; 200 μM (each) deoxynucleoside triphosphate; 400 nM concentrations of primers CIF2 F2, CIF2 R2, MECI P2, MECI P3, RIF5 F10, RIF5 R13, pUB110 R1, and pT181 R1; 800 nM concentrations of primers DCS F2, DCS R2, MECA P4, MECA P7, and IS431 P4; 200 nM concentrations of primers KDP F1, KDP R1, RIF4 F3, and RIF4 R9; 1.25 U of AmpliTaq; and approximately 5 ng of template DNA. PCR amplifications were performed in a DNA Thermal Cycler 480 (Applied Biosystems) with the following parameters: predenaturation for 4 min at 94°C; 30 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 1 min; postextension for 4 min at 72°C; and soaking at 4°C. PCR products (10 μl) were resolved in a 2% SeaKem LE (BioWhittaker Molecular Applications, Rockland, Maine) agarose gel in 0.5× Tris-borate-EDTA buffer (Bio-Rad, Hercules, Calif.) at 100 V and visualized with ethidium bromide.
RESULTS AND DISCUSSION
Multiplex PCR was first developed in 1988 by Chamberlain et al. (2). For staphylococci, this technique has been used to specifically detect MRSA (1, 8, 16, 19, 21, 30, 36), to discriminate between S. aureus and coagulase-negative staphylococci together with detection of oxacillin resistance (19, 21, 30), and to detect several resistance determinants (19, 20, 24) and staphylococcal toxin genes simultaneously (22, 23, 31, 32).
Strategy for selecting loci included in the multiplex PCR.
In choosing the main loci (A through F) for the multiplex PCR we followed three criteria. The first two were to include for each SCCmec type one locus located upstream of the mecA gene and a second locus located downstream of the mecA gene. The third was to select one of the loci to be unique for a single SCCmec type.
In addition to those criteria, the genetic organizations of the SCCmec types and the similarities among them conditioned the choice of loci (12, 26). SCCmec types I and II have very similar downstream region organizations, explaining the common downstream locus D and specific upstream loci A and B. Most of the SCCmec type III upstream sequence is similar to that of part of the type II upstream region, whereas the downstream region of type III differs from and is much longer than those of types I and II, explaining the nondifferentiating upstream locus C and the two specific downstream loci, E and F. SCCmec type IV is closely related to type I, having the same downstream region and part of the upstream region. Moreover, the specific upstream region (i.e., the one not present in type I) is variable from strain to strain, so that this SCCmec type is more accurately defined by the absence of the pls region (18 kb) in the mecA upstream region (26). Therefore, strains harboring SCCmec type IV are positive for downstream locus D and negative for upstream locus A, located in the pls region.
To distinguish SCCmec type variants IA (from I) and IIIA (from III), primers to amplify the left junctions between IS431 and integrated plasmids pUB110 (present in variant IA) and pT181 (absent in variant IIIA) were included. The distinction between SCCmec types I and IA is important, since type IA is associated with a widely spread MRSA clone, the Iberian clone. In the same way, type IIIA is associated with another widely spread MRSA clone, the Brazilian clone.
Validation of the multiplex PCR strategy.
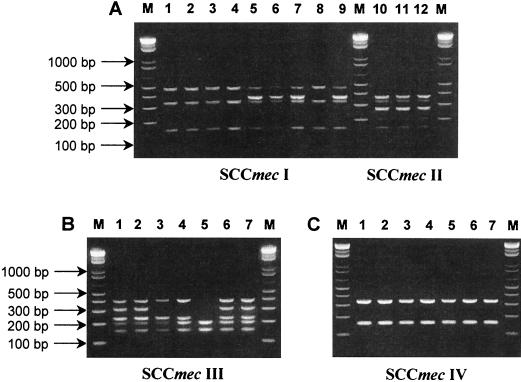
The amplification profiles obtained for the strains included in the validation collection (Table 1) are shown in Fig. 1. All strains were positive for the 162-bp internal fragment of the mecA gene, included in the multiplex PCR as an internal positive control. The four major SCCmec types are easily distinguishable: type I strains display two bands of 495 and 342 bp; type II strains display four bands of 381, 342, 284, and 209 bp; type III strains display four bands of 414, 303, 243, and 209 bp; and type IV strains display a single band of 342 bp. SCCmec variant IA is distinguished from type I by an extra band of 381 bp corresponding to the pUB110 insertion (Fig. 1A, lanes 5 to 7 and 9); SCCmec variant IIIA is distinguished from type III by the absence of the 303-bp band corresponding to the pT181 insertion (Fig. 1B, lanes 3 and 4); and SCCmec variant IIIB is discriminated from type III by the absence of all downstream bands (Fig. 1B, lane 5). Strain PER184 (Fig. 1A, lane 8) shows, by multiplex PCR, a pattern characteristic of type I. In a detailed previous study, this strain was assigned to variant IA because, in spite of not having the integrated pUB110 copy in the mecA downstream vicinity, it has a truncated HVR region (1-kb deletion), which is the specific characteristic of type IA (26), since linearized plasmid pUB110 is also present in SCCmec type II (13). The distinction between SCCmec types I and IA by the multiplex strategy is based on the detection of a pUB110-specific fragment, so the multiplex PCR fails to discriminate these types. However, the absence of pUB110 in SCCmec type IA strains is rare and can be detected on the basis of ClaI::mecA vicinity polymorphisms (28).
FIG. 1.
Validation of the SCCmec multiplex PCR strategy. Major SCCmec types are given at the bottom. (A) SCCmec type I (lanes 1 to 4 and 8), variant IA (lanes 5 to 7 and 9), and SCCmec type II (lanes 10 to 12). (B) SCCmec type III (lanes 1, 2, 6, and 7), variant IIIA (lanes 3 and 4), and variant IIIB (lane 5). (C) SCCmec type IV. Lane numbers are correlated with the order of strains (from top to bottom) in Table 1 (e.g., panel A, lane 1 represents strain UK13136 and panel B, lane 1 represents strain ANS46). M, DNA molecular size marker (1-kb DNA Ladder Plus; Gibco BRL, Invitrogen Corporation).
The primary criteria originally used for the classification of the mec element into three structural types were the structure of the mecA complex (mecA gene with the upstream regulatory region) and the nature of the ccrAB allele (12), the function of which is associated with the precise insertion and excision mechanisms of the mec element at the specific chromosomal site of orfX (15). The multiplex PCR method described here was designed to provide maximum resolution for the various structural variants of the mec element that we have identified in association with various clones of MRSA (26). This strategy did not include identification of the ccrAB alleles. However, PCR primers to confirm the presence of the specific ccrAB alleles associated with SCCmec types I, II, and III, identified by the multiplex PCR strategy, are available (12). Identification of the recently described type IV mec element is more problematic. This new mec element has a structure identical to that of mec element I in its mecA complex and in the downstream region but showed variability, from one strain to another, further upstream of the mecA complex (18, 26). The ccrAB allele of type IV strains differed from the alleles associated with mec element types I through III. Moreover, different strains carrying the type IV mec element showed variations in ccrAB. In the multiplex PCR described here the type IV mec element is defined through the absence of the pls region (characteristic of SCCmec type I) and a positive reaction for the downstream dcs region.
Overall, the multiplex PCR strategy was able to discriminate the four major mec element types and some of its previously detected variants, such as SCCmec IA and IIIA, associated with widespread MRSA clones (Iberian and Brazilian MRSA clones, respectively). There was a correct assignment of SCCmec types to the strains for which the mec element had been previously extensively characterized except for strain PER184 (see above) (Table 1).
Application of the multiplex PCR strategy.
We tested the multiplex PCR method as part of a protocol for the complete genetic characterization of a group of MRSA isolates recovered in a hospital outbreak in Barcelona, Spain (4). A preliminary, relatively rapid characterization of the genetic background of strains was done by spaA typing, which was shown to be predictive of MLST results in many of the cases (26). Confirmation of the genetic background was done by MLST, and tentative identification of the mec elements was done by multiplex PCR. The strains had already been characterized by PFGE (5). The results of this study are summarized in Table 3.
TABLE 3.
Application of the multiplex PCR to an outbreak collection
| Strain | PFGE patterna | No. of band differences in relation to pattern A1 | SpaA typing | MLST profile | SCCmec type | Clonal type |
|---|---|---|---|---|---|---|
| PER88 | B7 | 6 | YHFGFMBQBLO | 3-3-1-12-4-4-16 | IA | Iberian |
| PER4 | A1 | 0 | YHFGFMBQBLO | NDb | IA | Iberian |
| PER90 | A2 | 2 | YHFGFMBQBLO | ND | IA | Iberian |
| PER110 | B1 | 4 | YHFGFMBQBLO | ND | IA | Iberian |
| PER135 | B2 | 6 | YHFGFMBQBLO | ND | IA | Iberian |
| PER87 | B3 | 4 | YHFGFMBQBLO | ND | IA | Iberian |
| PER170 | B4 | 6 | YHFGFMBQBLO | ND | IA | Iberian |
| PER138 | B5 | 6 | YHFGFMBQBLO | ND | IA | Iberian |
| PER128 | B6 | 4 | YHFGFMBQBLO | ND | IA | Iberian |
| PER46 | B8 | 8 | YHFGFMBQBLO | ND | IA | Iberian |
| PER184 | D | 6 | YHFMBQBLO | 3-3-1-12-4-4-16 | I | Iberian |
| PER92 | E | 6 | YHFGFMBQBLO | 3-3-1-12-4-4-16 | IA | Iberian |
| PER222 | H | 8 | YHFGFMBQBLO | 3-3-1-12-4-4-16 | IA | Iberian |
| PER178 | F | 10 | WGKAOMQ | 2-3-1-1-4-4-3 | IIIA | Brazilian |
| PER127 | G | >10 | WGQQL2LO | 2-2-2-2-6-3-2 | IV | ST30/UK |
| PER2 | I | >10 | WGKAKAOMQQ | 2-2-2-2-6-3-2 | IVA | ST30/UK |
| PER220 | J | >10 | WGKAKAOMQQ | 2-2-2-2-6-3-2 | IV | ST30/UK |
| PER205 | C | 8 | UJGFGMDMGGML2 | 1-4-1-8-4-4-3 | IVA | New |
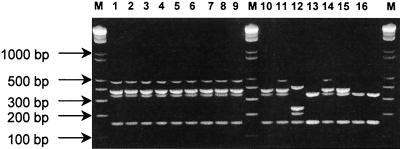
The application of the multiplex PCR to this collection of strains allowed rapid presumptive assignment of the SCCmec types (Fig. 2). The outbreak clone (Iberian clone), classified as having PFGE patterns A and B, carried SCCmec type IA (Fig. 2, lanes 1 to 9). Representative isolates of what appeared to be sporadic lineages, found by MLST and spaA typing to be related to the Iberian clone, also carried SCCmec type IA (Fig. 2, lanes 11 and 14). The single isolate belonging to the epidemic Brazilian clone was shown to have the characteristic SCCmec type IIIA (Fig. 2, lane 12). The isolates representing clone ST30/UK, a single-locus variant of clone E-MRSA 16 (epidemic MRSA clone 16) (7), were assigned to SCCmec type IV or IVA, which differs from type IV by the presence of a 381-bp band due to the integration of pUB110 (Fig. 2, lanes 13, 15, and 16). The new clone, classified as having PFGE pattern C, also carried SCCmec type IVA (Fig. 2, lane 10).
FIG. 2.
Application of the SCCmec multiplex PCR strategy to an outbreak collection including the sporadic clones. Lanes 1 to 9, PFGE patterns A1, A2, B1 to B6, and B8, respectively; lane 10, pattern C; lanes 11 to 16, patterns E through J, respectively. M, DNA molecular size marker (1-kb DNA Ladder Plus).
We applied the SCCmec multiplex PCR to MRSA isolates collected in the Barcelona outbreak as a kind of field test of the multiplex PCR method. Beyond proving its rapidity and fidelity, the study also yielded some interesting new information. First, some of the “sporadic isolates” were related to the major clone responsible for the outbreak, and others were related to highly epidemic MRSA clones, such as the Brazilian and E-MRSA 16 clones (14, 35). Second, the previously detected presence of SCCmec type IV in different genetic backgrounds was observed, which is compatible with the suggested enhanced mobility of this mec element (18, 27), perhaps because it is smaller than the other SCCmec types. In previous studies, this SCCmec type was found to be associated with MRSA of several different genetic backgrounds, each one of which was also found in association with other SCCmec types (26). Recent data indicate the presence of SCCmec type IV also in community-acquired MRSA (18) and in coagulase-negative staphylococci (11). In this study, SCCmec type IV was found in two distinct MRSA clones: the new clone classified as having PFGE pattern C and clone ST30, which is related to clone E-MRSA 16. A representative strain of clone E-MRSA 16 is being sequenced, and analysis of its preliminary genome sequence indicates that it harbors SCCmec type II, which was confirmed by the multiplex PCR strategy described in this study (data not shown). Another important MRSA clone circulating in British hospitals and also present in Germany, E-MRSA clone 15, was also found to carry SCCmec type IV by SCCmec multiplex PCR (data not shown). It seems that SCCmec type IV is or was a mobile version of the mec element, and SCCmec multiplex PCR has provided more evidence for this mobility (Table 4).
TABLE 4.
Presence of SCCmec type IV in different genetic backgroundsb
| Strain | Origin | MLST profilea | Clonal type | SCCmec type | Reference(s), source |
|---|---|---|---|---|---|
| PER34 | Spain | 3-3-1-1-4-4-16 | Iberian | IA | 4 |
| BK2529 | USA | 3-3-1-1-4-4-3 | Clone V | IV | 26 |
| BARGII17 | USA | 3-3-1-1-4-4-3 | Clone V | IV | 26 |
| N315 | Japan | 1-4-1-4-12-1-10 | NY/Japan | II | 13, 26 |
| BM18 | USA | 1-4-1-4-12-1-10 | Pediatric | IV | 26 |
| PL72 | Poland | 1-4-1-4-12-1-10 | Pediatric | IV | 26 |
| POL3 | Poland | 1-4-1-4-12-1-10 | Pediatric | IV | 26 |
| HDE288 | Portugal | 1-4-1-4-12-1-10 | Pediatric | IV | 26 |
| COB3 | Colombia | 1-4-1-4-12-1-10 | Pediatric | IV | 26 |
| E-MRSA 16 | UK | 2-2-2-2-3-3-2 | E-MRSA 16 | II | 7, this study |
| PER127 | Spain | 2-2-2-2-6-3-2 | ST30/UK | IV | 4, this study |
| PER2 | Spain | 2-2-2-2-6-3-2 | ST30/UK | IVA | 4, this study |
| PER220 | Spain | 2-2-2-2-6-3-2 | ST30/UK | IV | 4, this study |
| PER205 | Spain | 1-4-1-8-4-4-3 | PFGE pattern C | IVA | 4, this study |
| E-MRSA 15 | UK | 7-6-1-5-8-8-6 | E-MRSA 15 | IV | 4, this study |
| USA | ND | CA-MRSA | IV | 18 | |
| Japan | ND | CNS | IV | 11 |
Single-locus variants are underlined.
Abbreviations: ND, not determined; CA-MRSA, community-acquired MRSA; CNS, coagulase-negative staphylococci; UK, United Kingdom; USA, United States.
The protocol for the multiplex PCR developed in this study was kept as simple as possible in order to allow rapid presumptive assignment of SCCmec types to MRSA strains. This strategy allows the presumptive classification of the SCCmec type resident in each MRSA isolate in a single PCR, a technique nowadays commonly available in clinical microbiology laboratories. The routine assignment MRSA isolates to SCCmec types, now feasible with the SCCmec multiplex PCR typing tool, may provide researchers with important information concerning the origin of MRSA clones and the evolutionary relationships among them, since it probes the genetic organization of the mec element, which harbors the central genetic component of methicillin resistance.
Acknowledgments
We thank Alexander Tomasz for suggestions, support, and critical reading and revision of the manuscript. Some observations were made possible by the information available at the MLST database (www.mlst.net) and the COL and E-MSRA 16 genome-sequencing projects available at www.tigr.org and www.sanger.ac.uk, respectively.
Partial support for this study was provided by project POCTI/1999/ESP/34872 from Fundação para a Ciência e Tecnologia, Lisbon, Portugal, awarded to H. de Lencastre, by a grant from Fundação Calouste Gulbenkian, Lisbon, Portugal, awarded to H. de Lencastre, and by a grant from the U.S. Public Health Service awarded to A. Tomasz, project RO1 AI37275. D.C. Oliveira was supported by a doctoral grant from Fundação Calouste Gulbenkian.
REFERENCES
- 1.Barski, P., L. Piechowicz, J. Galinski, and J. Kur. 1996. Rapid assay for detection of methicillin-resistant Staphylococcus aureus using multiplex PCR. Mol. Cell. Probes 10:471-475. [DOI] [PubMed] [Google Scholar]
- 2.Chamberlain, J. S., R. A. Gibbs, J. E. Ranier, P. N. Nguyen, and C. T. Caskey. 1988. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res 16:11141-11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin susceptible and resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominguez, M. A., H. de Lencastre, J. Linares, and A. Tomasz. 1994. Spread and maintenance of a dominant methicillin-resistant Staphylococcus aureus (MRSA) clone during an outbreak of MRSA disease in a Spanish hospital. J. Clin. Microbiol. 32:2081-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubin, D. T., S. G. Chikramane, B. Inglis, P. R. Matthews, and P. R. Stewart. 1992. Physical mapping of the mec region of an Australian methicillin-resistant Staphylococcus aureus lineage and a closely related American strain. J. Gen. Microbiol. 138:169-180. [DOI] [PubMed] [Google Scholar]
- 6.Dubin, D. T., P. R. Matthews, S. G. Chikramane, and P. R. Stewart. 1991. Physical mapping of the mec region of an American methicillin-resistant Staphylococcus aureus strain. Antimicrob. Agents Chemother. 35:1661-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geha, D. J., J. R. Uhl, C. A. Gustaferro, and D. H. Persing. 1994. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J. Clin. Microbiol. 32:1768-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goering, R. V. 1993. Molecular epidemiology of nosocomial infection: analysis of chromosomal restriction fragment patterns by pulsed-field gel electrophoresis. Infect. Control Hosp. Epidemiol. 14:595-600. [DOI] [PubMed] [Google Scholar]
- 10.Henegariu, O., N. A. Heerema, S. R. Dlouhy, G. H. Vance, and P. H. Vogt. 1997. Multiplex PCR: critical parameters and step-by-step protocol. BioTechniques 23:504-511. [DOI] [PubMed] [Google Scholar]
- 11.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9:486-493. [DOI] [PubMed] [Google Scholar]
- 12.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, A. P., H. M. Aucken, S. Cavendish, M. Ganner, M. C. Wale, M. Warner, D. M. Livermore, and B. D. Cookson. 2001. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS). J. Antimicrob. Chemother. 48:143-144. [DOI] [PubMed] [Google Scholar]
- 15.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kearns, A. M., P. R. Seiders, J. Wheeler, R. Freeman, and M. Steward. 1999. Rapid detection of methicillin-resistant staphylococci by multiplex PCR. J. Hosp. Infect. 43:33-37. [DOI] [PubMed] [Google Scholar]
- 17.Kreiswirth, B., J. Kornblum, R. D. Arbeit, W. Eisner, J. N. Maslow, A. McGeer, D. E. Low, and R. P. Novick. 1993. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science 259:227-230. [DOI] [PubMed] [Google Scholar]
- 18.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martineau, F., F. J. Picard, L. Grenier, P. H. Roy, M. Ouellette, and M. G. Bergeron. 2000. Multiplex PCR assays for the detection of clinically relevant antibiotic resistance genes in staphylococci isolated from patients infected after cardiac surgery. The ESPRIT Trial. J. Antimicrob. Chemother. 46:527-534. [DOI] [PubMed] [Google Scholar]
- 20.Martineau, F., F. J. Picard, N. Lansac, C. Menard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 2000. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mason, W. J., J. S. Blevins, K. Beenken, N. Wibowo, N. Ojha, and M. S. Smeltzer. 2001. Multiplex PCR protocol for the diagnosis of staphylococcal infection. J. Clin. Microbiol. 39:3332-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehrotra, M., G. Wang, and W. M. Johnson. 2000. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 38:1032-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monday, S. R., and G. A. Bohach. 1999. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J. Clin. Microbiol. 37:3411-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nunes, E. L., K. R. dos Santos, P. J. Mondino, M. Bastos, and M. Giambiagi-de Marval. 1999. Detection of ileS-2 gene encoding mupirocin resistance in methicillin-resistant Staphylococcus aureus by multiplex PCR. Diagn. Microbiol. Infect. Dis. 34:77-81. [DOI] [PubMed] [Google Scholar]
- 25.Oliveira, D. C., I. Crisostomo, I. Santos-Sanches, P. Major, C. R. Alves, M. Aires-de-Sousa, M. K. Thege, and H. de Lencastre. 2001. Comparison of DNA sequencing of the protein A gene polymorphic region with other molecular typing techniques for typing two epidemiologically diverse collections of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 39:574-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 27.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 28.Oliveira, D. C., S. W. Wu, and H. de Lencastre. 2000. Genetic organization of the downstream region of the mecA element in methicillin-resistant Staphylococcus aureus isolates carrying different polymorphisms of this region. Antimicrob. Agents Chemother. 44:1906-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts, R. B., A. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, and A. Tomasz. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. MRSA Collaborative Study Group. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 30.Schmitz, F. J., C. R. Mackenzie, B. Hofmann, J. Verhoef, M. Finken-Eigen, H. P. Heinz, and K. Kohrer. 1997. Specific information concerning taxonomy, pathogenicity and methicillin resistance of staphylococci obtained by a multiplex PCR. J. Med. Microbiol. 46:773-778. [DOI] [PubMed] [Google Scholar]
- 31.Schmitz, F. J., M. Steiert, B. Hofmann, J. Verhoef, U. Hadding, H. P. Heinz, and K. Kohrer. 1998. Development of a multiplex-PCR for direct detection of the genes for enterotoxin B and C, and toxic shock syndrome toxin-1 in Staphylococcus aureus isolates. J. Med. Microbiol. 47:335-340. [DOI] [PubMed] [Google Scholar]
- 32.Sharma, N. K., C. E. Rees, and C. E. Dodd. 2000. Development of a single-reaction multiplex PCR toxin typing assay for Staphylococcus aureus strains. Appl. Environ. Microbiol. 66:1347-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song, M. D., M. Wachi, M. Doi, F. Ishino, and M. Matsuhashi. 1987. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 221:167-171. [DOI] [PubMed] [Google Scholar]
- 35.Teixeira, L. A., C. A. Resende, L. R. Ormonde, R. Rosenbaum, A. M. Figueiredo, H. de Lencastre, and A. Tomasz. 1995. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J. Clin. Microbiol. 33:2400-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vannuffel, P., J. Gigi, H. Ezzedine, B. Vandercam, M. Delmee, G. Wauters, and J. L. Gala. 1995. Specific detection of methicillin-resistant Staphylococcus species by multiplex PCR. J. Clin. Microbiol. 33:2864-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]