Abstract
Campylobacter jejuni, a gram-negative organism causing gastroenteritis in humans, is increasingly resistant to antibiotics. However, little is known about the drug efflux mechanisms in this pathogen. Here we characterized an efflux pump encoded by a three-gene operon (designated cmeABC) that contributes to multidrug resistance in C. jejuni 81-176. CmeABC shares significant sequence and structural homology with known tripartite multidrug efflux pumps in other gram-negative bacteria, and it consists of a periplasmic fusion protein (CmeA), an inner membrane efflux transporter belonging to the resistance-nodulation-cell division superfamily (CmeB), and an outer membrane protein (CmeC). Immunoblotting using CmeABC-specific antibodies demonstrated that cmeABC was expressed in wild-type 81-176; however, an isogenic mutant (9B6) with a transposon insertion in the cmeB gene showed impaired production of CmeB and CmeC. Compared to wild-type 81-176, 9B6 showed a 2- to 4,000-fold decrease in resistance to a range of antibiotics, heavy metals, bile salts, and other antimicrobial agents. Accumulation assays demonstrated that significantly more ethidium bromide and ciprofloxacin accumulated in mutant 9B6 than in wild-type 81-176. Addition of carbonyl cyanide m-chlorophenylhydrazone, an efflux pump inhibitor, increased the accumulation of ciprofloxacin in wild-type 81-176 to the level of mutant 9B6. PCR and immunoblotting analysis also showed that cmeABC was broadly distributed in various C. jejuni isolates and constitutively expressed in wild-type strains. Together, these findings formally establish that CmeABC functions as a tripartite multidrug efflux pump that contributes to the intrinsic resistance of C. jejuni to a broad range of structurally unrelated antimicrobial agents.
Bacterial pathogens have evolved multiple mechanisms for resistance to antimicrobial agents, which has greatly compromised the effectiveness of antibiotic treatments and poses a serious threat to public health (13, 17). As one of the general resistance mechanisms, antibiotic efflux systems extrude structurally diverse antimicrobial agents out of bacterial cells (6, 8, 27, 29, 36). One important family of drug transporters that contribute to multidrug resistance (MDR) in gram-negative bacteria are the resistance-nodulation-cell division (RND) efflux systems, which consist of an inner membrane transporter, a periplasmic fusion protein, and an outer membrane protein (39). Genetically, many of the RND-type MDR efflux systems or pumps are encoded by a three-gene operon located on the bacterial chromosome (25, 39). However, some efflux pumps, such as AcrAB from Escherichia coli (19), have an outer membrane component that is encoded by a separate gene physically unattached with the other two members on the bacterial chromosome. Expression of these pumps is controlled by regulatory proteins, and their overexpression is usually mediated by mutations in the regulatory elements resulting in an MDR phenotype (27, 29, 36). Even without overexpression, the MDR efflux pumps work synergistically with other nonefflux resistance mechanisms (such as target mutations) to confer high levels of antimicrobial resistance in bacteria (18, 23, 26, 37).
Campylobacter jejuni is the leading bacterial cause of human enteritis in many industrialized countries (31). This pathogenic organism causes watery diarrhea and/or hemorrhagic colitis and is also associated with Guillain-Barre syndrome, an acute flaccid paralysis that may lead to respiratory muscle compromise and death (21). The increasing resistance of C. jejuni to a broad range of antibiotics including fluoroquinolones (FQs) and macrolides has become a major concern for public health (9, 32, 35). Despite the appreciated magnitude of the problem, little has been known about the MDR mechanisms in Campylobacter. In an early study (5), Charvalos et al. selected MDR C. jejuni isolates by in vitro plating and provided evidence suggesting the association between the MDR phenotype and a possible efflux system. However, the identity of the efflux system was not determined. Recently, the genomic sequence of C. jejuni NCTC 11168 has been completed, and it has revealed several unique features associated with Campylobacter (24). Unlike other gram-negative pathogens, C. jejuni lacks inserted sequence elements, prophages, and transposons (24), which often carry genes encoding for drug resistance. Although the genomic sequence of NCTC 11168 revealed the presence of several genes that share significant homology with known multidrug transporters, none of them have been functionally characterized, nor is anything known about their contributions to drug resistance in C. jejuni.
In a recent study by our laboratory, we successfully used the EZ::TN <KAN-2> transposon for random mutagenesis in C. jejuni 81-176 (L. Michel, J. Lin, and Q. Zhang, Abstract, Int. J. Med. Microbiol. 291[Suppl. 31]:79, 2001). One of the identified mutants, named 9B6, had a transposon inserted in a gene that is homologous to Cj0366c of NCTC 11168 and shares significant homology with typical multidrug efflux transporters belonging to the RND superfamily in gram-negative bacteria. Comparison of Cj0366c and its flanking genes (Cj0365c and Cj0367c) with known bacterial multidrug efflux systems suggested that these three genes likely encode a tripartite efflux system. This finding prompted us to dissect the function of the putative efflux pump by taking advantage of isogenic mutant 9B6 derived from strain 81-176. In this study, we have demonstrated that in 81-176 the three genes that are homologous to Cj0365c, Cj0366c, and Cj0367c encode an energy-dependent efflux system contributing to Campylobacter resistance to structurally unrelated antimicrobial agents. To reflect the function of the three genes, we designated them cmeA (cme for Campylobacter multidrug efflux), cmeB, and cmeC, respectively.
(This study was presented in part at the 11th International Workshop on Campylobacter, Helicobacter and Related Organisms, 1-5 September 2001, Freiburg, Germany.)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. jejuni strain 81-176 is a human isolate and has been described in a previous study (3). C. jejuni ATCC 33291 and Campylobacter coli ATCC 33559 were obtained from the American Type Culture Collection, Rockville, Md. T48768, W52546, and F26747 were human C. jejuni isolates obtained from the Microbiology Laboratory of the Ohio State University hospital. Other C. jejuni strains, including five human isolates (M36292, H49024, H30769, E46972, and X77136), two chicken isolates (S2B and 21190), two ovine isolates (19084571 and 19094451), and one bovine isolate (15046764) were described in a previous publication (40). These isolates were routinely grown in Mueller-Hinton (MH) broth (Difco) or agar at 42°C under microaerophilic conditions, which were generated using a Campypak Plus (Becton Dickinson) gas pack in an enclosed jar.
Construction of cmeB mutants.
Mutant 9B6 of strain 81-176 was originally isolated in a mutagenesis study (Michel et al., abstract, 2001) using EZ::TN <KAN-2> Tnp Transposome (Epicentre). The insertion site of the transposon in mutant 9B6 was in a gene that is homologous to Cj0366c of NCTC 11168, which is named cmeB in this study (Fig. 1). The insertional mutation in 9B6 was back-crossed to wild-type 81-176, and the isogenic mutant 9B6 from the back-crossing experiment was used in this study for characterizing the function of cmeABC. In addition, a cmeB-specific insertional mutation was generated in strain 21190, which is genetically divergent from 81-176 as determined by pulsed-field gel electrophoresis and major outer membrane protein-based sequence polymorphism (40). For creating the cmeB mutant in strain 21190, genomic DNA of 9B6 was purified using a Wizard Genomic Purification Kit (Promega). The purified DNA was used to transform strain 21190 using the standard biphasic method for natural transformation (38). Transformants were plated on MH plates with 30 μg of kanamycin/ml. A single Kanr colony was selected, and the mutation in this isolate was confirmed to be in cmeB by PCR using a transposon-specific primer (Epicentre) and a cmeB-specific primer (data not shown). This cmeB mutant of 21190 was also used for drug susceptibility testing.
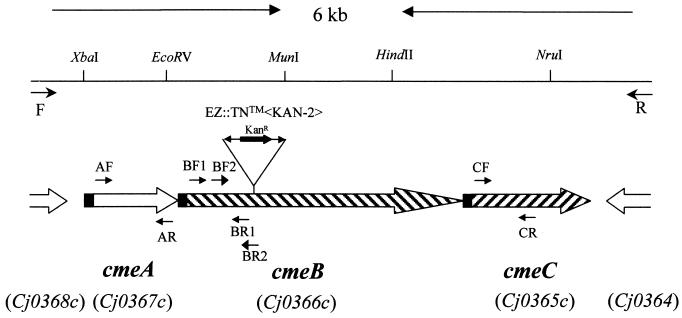
FIG. 1.
Genomic organization and features of the cmeABC operon in C. jejuni 81-176. The line at the top depicts the 6-kb fragment amplified by primers F and R. Selected restriction sites are labeled on top of the line. The identified ORFs are indicated by boxed arrows below the line. The solid box in each ORF represents a predicted signal peptide. The open boxes at both ends indicate ORFs flanking the cmeABC operon. The corresponding gene loci in C. jejuni NCTC 11168 are listed in parentheses. The locations of various primers used in this study are indicated by arrows. The location and orientation of the transposon insertion in 9B6 is indicated in cmeB.
PCR.
PCR was performed in a volume of 100 μl containing 200 μM concentrations of each of the deoxynucleoside triphosphates, 200 nM concentrations of primers, 2.5 mM MgSO4, 50 ng of Campylobacter genomic DNA, and 5 U of Taq DNA polymerase (Promega) or Pfu Turbo DNA polymerase (Stratagene). Cycling conditions varied according to the estimated annealing temperatures of primers and the expected sizes of products. To amplify a 6-kb fragment from C. jejuni 81-176 which contains the entire operon encoding the CmeABC efflux system, primers F (5′-TCATCTTAATGTTTTAATTAACGCTCC-3′) and R (5′-GGCTTATGAAATTACAGATGCAGA-3′) were designed from the genomic sequence of C. jejuni NCTC 11168 (24) and used in PCR with genomic DNA of 81176 and Pfu Turbo DNA polymerase. PCR products were purified using the QIAquick PCR Purification Kit (Qiagen) and subsequently sequenced. To determine the distribution of the cmeABC operon, cmeB-specific primers BF2(5′-GGTACAGATCCTGATCAAGCC-3′) and BR2 (5′-AGGAATAAGTGTTGCACGGAAATT-3′) were used in PCR to amplify the gene sequence from various Campylobacter strains derived from different animal species. The locations of these PCR primers are indicated in Fig. 1. To determine if the insertional mutation in cmeB affected the expression of Cj0364, an open reading frame (ORF) immediately downstream of the cmeABC operon encoding a hypothetical protein with unknown function, reverse transcriptase PCR (RT-PCR) was performed to assess the expression of Cj0364. For this purpose, total RNA was isolated separately from C. jejuni 81-176 and 9B6 by using the RNeasy Kit (Qiagen). RNA samples were treated with RNase-free DNase (Epicentre) at 37°C for 30 min, followed by heat inactivation at 75°C for 5 min. RT-PCR was conducted using a MasterAmp Kit (Epicentre) and a pair of Cj0364-specific primers (5′-TGGATAAAGCCAAAATTGTTCA-3′ and 5′-GCCTTGTAAATAGCAGGCAATAA-3′). Cycling conditions for the RT-PCR included an initial incubation at 60°C for 20 min, followed by 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s. An RT-PCR mixture lacking the RT was included as a negative control.
Sequence analysis and prediction of secondary structures.
PCR products were sequenced using an automated DNA sequencer (model 377; Applied Biosystems). Sequence analysis was performed with the Genetics Computer Group (GCG) Sequence Analysis software package (Oxford Molecular). Hydropathy profiles and antigenic properties of CmeABC were analyzed with the GCG package. For predicting the secondary structures of CmeABC, the Peptidestructure program of GCG was used for initial prediction. Other programs, including COILS, TMpred, SOSUI, and PHDsec (BCM Search Launcher Texas, Baylor College of Medicine [http://dot.imgen.bcm.tmc.edu:9331/seq-search/struc-predict.html]) were also used to improve prediction power.
Production of recombinant CmeABC proteins and generation of polyclonal antisera.
Histidine (His)-tagged recombinant peptides of CmeA, CmeB, and CmeC were produced in E. coli by using the pQE-30 vector in the QIAexpress expression system (Qiagen). Based on the complete sequence of cmeABC of C. jejuni 81-176 and the predicted antigenicity profiles of each gene product, three pairs of primers were designed to amplify a portion of each component of cmeABC. Primers AF (5′-TTTGGATCCTTGATGGCTAAGGCAACTTTC-3′) and AR (5′-CTCCAATTTCTTAAGCTTCGCTACCAA-3′) were used to amplify a fragment encoding a 258-amino-acid (aa) peptide (aa 108 to 365) of CmeA. Primers BF1 (5′-GCTGGATCCATAGGTCTTACAAAT-3′) and BR1 (5′-GGACAAAGCTTGTTGTATCGTAAGGAA-3′) were used to amplify a gene segment encoding a 308-aa peptide (aa 26 to 333) of CmeB. Primers CF (5′-GCTTGGATCCTTATCTTGGGAAAAA-3′) and CR (5′-TTTTTAAAGCTTTAAGGTAATTTTCTT-3′) were used to amplify a fragment encoding a 208-aa peptide (aa 41 to 248) of CmeC. The locations of these primers in relation to each ORF are indicated in Fig. 1. A restriction site (underlined in the primer sequences) was attached to the 5′ end of each primer to facilitate the cloning of the amplified PCR products into the pQE-30 vector. The amplified PCR products were digested with BamHI and HindIII and then purified by using the QIAquick PCR cleaning kit (Qiagen). The pQE-30 vector was also digested with BamHI and HindIII and then purified by gel extraction. The digested pQE-30 vector and PCR products were ligated with T4 DNA ligase. Transformation and screening for positive recombinants were performed according to the procedures supplied with the pQE vector. Each plasmid in the E. coli clone expressing a recombinant peptide was sequenced, revealing no mutations in the coding sequence (data not shown). Purification of His-tagged recombinant CmeA, CmeB, or CmeC protein under native conditions was performed as described previously (40).
Recombinant proteins resuspended in phosphate-buffered saline (PBS) were emulsified with an equal volume of incomplete Freund's adjuvant and subcutaneously injected into New Zealand White rabbits (100 μg of protein/rabbit). Each rabbit received two additional booster immunizations at 2-week intervals. Two rabbits were used for each protein. The rabbits were bled at 21 days after the last injection. Pre- and postimmune serum samples were analyzed by immunoblotting, using both recombinant proteins and membrane proteins of C. jejuni 81-176. The postimmune sera reacted specifically with the corresponding components of the CmeABC system, while the preimmune sera were negative with any of the components (data not shown).
SDS-PAGE and immunoblotting.
Cell envelopes of C. jejuni 81-176 and mutant 9B6 were prepared as previously described (4). To prepare whole-cell lysates, various Campylobacter strains were grown in MH broth to late logarithmic phase (≈3 × 109 cells/ml), harvested by centrifugation, and solubilized by boiling for 5 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Ten-microgram membrane fractions or approximately 108 whole cells were loaded in each lane and separated by SDS-PAGE with a 12% (wt/vol) (for CmeA and CmeC) or 9% (wt/vol) (for CmeB) polyacrylamide separating gel (16). After SDS-PAGE, the gels were equilibrated for 30 min in the transfer buffer (0.025 M Tris base and 0.192 M glycine with 20% methanol; pH 8.3). Proteins in the gels were then electrophoretically transferred to nitrocellulose membranes (Bio-Rad) at 60 V for 1 h at 4°C. The membranes were incubated with blocking buffer (5% Nestle skim milk powder in PBS) for 16 h at 4°C prior to incubation with primary antibodies (rabbit anti-CmeABC sera; 1:3,000 dilution in the blocking buffer). After incubation at 25°C for 1 h, the blots were washed three times with PBS containing 0.05% Tween 20 and subsequently incubated with secondary antibodies (1:1,000 dilution of goat anti-rabbit immunoglobulin G-horseradish peroxidase; Kirkegaard & Perry) at 25°C for 1 h. After washing, the blots were developed with the 4 CN Membrane Peroxidase Substrate System (Kirkegaard & Perry). Prestained molecular mass markers (Bio-Rad) were coelectrophoresed and blotted to allow estimation of the sizes of the proteins.
Susceptibility tests.
The MICs were determined using the standard microtiter broth dilution method (30) in MH broth with an inoculum of 106 bacteria/ml. Microtiter plates were incubated for 2 days under microaerophilic conditions at 42°C. The antibiotics and other compounds used in this study were purchased from Sigma Chemical Co. (nalidixic acid, norfloxacin, erythromycin, cefotaxime, rifampin, trimethoprim, cycloheximide, ampicillin, tetracycline, chloramphenicol, gentamicin, polymyxin B, protamine, cholic acid, chenodeoxycholic acid, taurocholic acid, deoxycholic acid, CoCl2, CuCl2, and ZnSO4); ICN Biomedicals Inc. (ciprofloxacin); EM Science (SDS); and AMRESCO (ethidium bromide [EB]).
Accumulation assays.
Accumulation of EB and ciprofloxacin in C. jejuni was determined as described previously (5, 11) with some modifications. To measure the accumulation of EB, the bacteria were grown in MH broth to the late logarithmic phase, harvested, washed once in 15 mM PBS (pH 7.2), and resuspended in PBS (pH 7.2) to an A600 of 0.2. The cells were incubated for 10 min at 37°C. Then, EB was added to the cell suspension at a final concentration of 2 μg/ml. Fluorescence of the cell suspension was used as an indicator of the amount of EB taken up by the cells (the fluorescence of EB increased greatly upon binding to intracellular components) and was directly recorded at different time points with a Perkin-Elmer (Norwalk, Conn.) spectrofluorometer at excitation and emission wavelengths of 530 and 600 nm, respectively. The efflux pump inhibitor carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added to the cell suspension at a final concentration of 200 μM at 7 min after EB addition. The natural fluorescence of the Campylobacter cells was eliminated by adjusting the zero point prior to the addition of EB, and the measured fluorescence intensity was expressed in arbitrary units. To study the accumulation of ciprofloxacin in C. jejuni, bacteria were grown in MH broth to the late logarithmic phase, harvested and washed once in 15 mM PBS (pH 7.2), and adjusted to 1011 CFU/ml. The cell suspension was incubated for 10 min at 37°C. Then, ciprofloxacin was added to a final concentration of 10 μg/ml. After addition of ciprofloxacin, 0.5-ml samples were removed at different time points. At 7 min after ciprofloxacin addition, CCCP was added to one-half of the reaction mixture to a final concentration of 200 μM and the other half was used as control (no CCCP). Accumulation measurements of both fractions were continued until 20 min. Each of the collected samples at different time points was immediately diluted in 2.5 ml of ice-cold PBS and then centrifuged for 5 min at 6,000 × g at 4°C. The pellets were washed once with 2 ml of ice-cold PBS, resuspended in 2 ml of 0.1 M glycine hydrochloride (pH 3.0), and shaken at 25°C for 16 h. The samples were then centrifuged at 6,000 × g for 15 min. The fluorescence of the supernatant was measured with a Perkin-Elmer spectrofluorometer at excitation and emission wavelengths of 279 and 447 nm, respectively. The concentrations of ciprofloxacin in the supernatant were calculated by comparison with a standard curve of ciprofloxacin in 0.1 M glycine hydrochloride (pH 3.0). The results were expressed as nanograms of ciprofloxacin per milligram (wet weight) of bacteria. Three independent experiments were performed to measure the accumulation of EB and ciprofloxacin.
Nucleotide sequence accession number.
The cmeABC gene sequences of C. jejuni 81-176 determined in this study were deposited in GenBank under accession number AF466820.
RESULTS
Identification and features of the cmeABC operon in C. jejuni 81-176.
The sequence of a continuous 5,939-bp DNA fragment amplified from C. jejuni 81-176 by using primers F and R (Fig. 1) revealed three ORFs (designated cmeA, cmeB, and cmeC, respectively). The three ORFs were located in the same coding strand and tandemly positioned on the chromosome of 81-176 (Fig. 1). cmeA (nucleotides 207 to 1307), cmeB (nucleotides 1310 to 4429), and cmeC (nucleotides 4425 to 5900) encoded 367 aa, 1,040 aa, and 492 aa, respectively. The amino acid sequences of CmeA, CmeB, and CmeC were 98.4, 99.2, and 99.6% identical to the encoded products of Cj0367c, Cj0366c, and Cj0365c of C. jejuni NCTC 11168, respectively. The transposon insertion in mutant 9B6 occurred within the codon encoding aa 360 of CmeB. The stop codon (TAA) of cmeA overlaps with the start codon (ATG) of cmeB, while cmeB and cmeC overlap by eight nucleotides. The overlaps between the three genes and the lack of predicted stem-loop structures between the ORFs suggested that the three genes are organized into an operon. Potential ribosome-binding sites occurred immediately upstream of each of the three ORFs. Several inverted repeats, characteristic of binding sites for transcription factors, occurred upstream of cmeA, suggesting that the cmeABC operon is potentially regulated by other proteins.
CmeA (37.9 kDa) showed significant similarities to membrane fusion proteins, including MtrC (U14993; 29.9% aa identity) of Neisseria gonorrhoeae; MexA (L11616; 29.2% aa identity) and MexC (U57969; 29.6% aa identity) of Pseudomonas aeruginosa; and AcrA (U00734; 31.4% aa identity) of E. coli. CmeB, with a calculated molecular mass of 114 kDa, exhibited a significant sequence homology to inner membrane transporters, including AcrB (U00734; 41% aa identity), AcrD (U10436; 42% aa identity), and AcrF (M96848; 41% aa identity) of E. coli as well as MexB (L11616; 41% aa identity) and MexF (X99514; 40% aa identity) of P. aeruginosa. CmeC was found to be similar to the outer membrane proteins OprM (A49937; 25% aa identity) and OprN (X99514; 24% aa identity) of P. aeruginosa and TolC (X54049; 24% aa identity) of E. coli. The predicted secondary structures of CmeA, CmeB, and CmeC were also consistent with the known features of the three members in tripartite multidrug efflux pumps of gram-negative bacteria (14, 29, 36, 39).
An ORF homologous to Cj0368c of NCTC 11168 occurred immediately upstream of cmeA (Fig. 1). A BLAST search indicated that the deduced amino acid sequence of Cj0368c shares similarities to the members of the TetR/AcrR family of transcriptional repressors of efflux systems. The MOTIF program (http://www.motif.genome.ad.jp) also identified a helix-turn-helix DNA binding motif, a signature sequence of the TetR family, at the N-terminal region of the product encoded by Cj0368c. Together, these observations suggest that Cj0368c likely encodes a transcriptional regulator of the cmeABC operon. Another ORF that is homologous to Cj0364 of NCTC 11168 occurred immediately downstream of cmeC (Fig. 1). Cj0364, encoding a hypothetical protein with unknown function, is transcribed in the opposite direction from the cmeABC operon (Fig. 1). RT-PCR demonstrated that Cj0364 was transcribed at comparable levels in both mutant 9B6 and wild-type 81-176 (data not shown), indicating that the insertional mutation in the cmeB gene did not cause a polar effect on the gene downstream of the cmeABC operon.
Transposon insertion in cmeB impaired the expression of cmeB and cmeC.
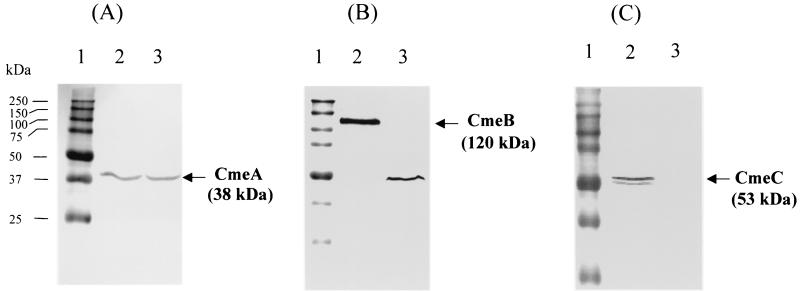
To determine if cmeABC is expressed in wild-type 81-176 and what the effect of the transposon insertion in cmeB is on the expression of cmeABC, cell envelope proteins of mutant 9B6 and wild-type 81-176 were analyzed by immunoblotting, using specific anti-CmeA, anti-CmeB, and anti-CmeC antibodies. As shown in Fig. 2A, the anti-CmeA antibody detected a band of approximately 38 kDa in both C. jejuni 81-176 and 9B6, which was consistent with the calculated molecular mass of mature CmeA. The anti-CmeB antibody reacted with a protein band of 120 kDa in wild-type 81-176 that was comparable to the deduced molecular mass of CmeB (Fig. 2B). However, the anti-CmeB antibody only detected a band of 49 kDa in 9B6 (Fig. 2B), which was much smaller than the intact CmeB protein and was likely a truncated form of CmeB resulting from the transposon insertion. The anti-CmeC antibody did not react with any proteins in isogenic mutant 9B6, but it detected two bands in wild-type 81-176 that were 53 and 50 kDa (Fig. 2C). The major 53-kDa band was consistent with the deduced molecular mass of mature CmeC, while the minor 51-kDa band could represent a partially degraded or unmodified CmeC. These results indicated that (i) cmeABC was expressed in wild-type 81-176 at a level that could be readily detected with specific antibodies; and (ii) transposon insertion in cmeB impaired the production of both CmeB and CmeC, but it did not have any effect on the expression of cmeA. Since the three members of a tripartite efflux system function together (39), disruption of CmeB and CmeC was expected to cause malfunction of the CmeABC system.
FIG. 2.
Immunoblotting analysis of CmeABC expression in mutant 9B6 and wild-type 81-176. Cell envelopes prepared from C. jejuni 81-176 (lane 2) and mutant 9B6 (lane 3) were blotted with specific antibodies against CmeA (A), CmeB (B), and CmeC (C). Prestained molecular mass markers (lane1; Bio-Rad) were coelectrophoresed and blotted to allow estimation of the sizes of the proteins.
CmeABC contributes to resistance to structurally unrelated antimicrobial compounds.
To determine if CmeABC contributes to multidrug resistance, we tested the susceptibilities of wild-type 81-176 and the isogenic mutant 9B6 to structurally unrelated antibiotics and other antimicrobial compounds. Compared to wild-type 81-176, 9B6 showed significantly increased susceptibilities to many antimicrobial compounds (Table 1). In mutant 9B6, the MICs of fluoroquinolones were decreased 8-fold (ciprofloxacin) and 2-fold (norfloxacin and nalidixic acid); the MICs of two β-lactams were decreased 256-fold (cefotaxime) and 32-fold (ampicillin); and the MICs of erythromycin, rifampin, tetracycline, and EB were decreased 4-fold, 128-fold, 8-fold, and 8-fold, respectively. Interestingly, rifampin is often used as a selective agent in culture medium for isolating C. jejuni because this organism is intrinsically resistant to this antibiotic. The intrinsic resistance to rifampin can now be attributed, at least in part, to the function of CmeABC. MICs of chloramphenicol, gentamicin, the antimicrobial peptide protamine, and the heavy metals CoCl2 and CuCl2 were slightly (twofold) but reproducibly decreased in 9B6. The mutant 9B6 demonstrated a wild-type level of resistance to trimethoprim, cycloheximide, polymyxin B, and ZnSO4, suggesting that these compounds are not the substrates of the CmeABC efflux system. Notably, the insertional mutation in CmeB conferred hypersusceptibility to bile salts, a group of detergent-like compounds produced in liver and secreted into the bile for dispersion and digestion of fats in small intestines, on mutant 9B6. The MICs of selected bile salts were decreased 4,000-fold (chenodeoxycholic acid), 1,000-fold (deoxycholic acid), and 64-fold (cholic acid and taurocholic acid). Wild-type 81-176 was resistant to tetracycline because it carries the pTet plasmid, which contains the tet(O) gene (1). Transposon insertion in cmeB resulted in an eightfold decrease in the MIC of tetracycline in mutant 9B6, even though the pTet plasmid was still present in this mutant, as determined by PCR (data not shown), suggesting that cmeABC functions synergistically with tet(O) to contribute to the acquired resistance to tetracycline.
TABLE 1.
Susceptibilities of C. jejuni 81-176, 21190, and their cmeB mutants to different antimicrobials
| Antimicrobial | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| 81-176 | 9B6 | Fold differenced | 21190 | cmeB isogenic mutant | Fold differencee | |
| Ciprofloxacin | 0.313 | 0.039 | 8 | 0.156 | 0.020 | 8 |
| Norfloxacin | 0.078 | 0.039 | 2 | 0.078 | 0.039 | 2 |
| Nalidixic acid | 1.25 | 0.625 | 2 | 1.25 | 0.625 | 2 |
| Erythromycin | 0.078 | 0.020 | 4 | 0.625 | 0.04 | 16 |
| Ampicillin | 0.312 | 0.010 | 32 | 62.5b | 62.5b | — |
| Cefotaxime | 1.60 | 0.006 | 256 | 1.56 | 0.097 | 16 |
| Rifampin | 100 | 0.78 | 128 | 50 | 0.78 | 64 |
| Trimethoprim | >200 | >200 | —c | >200 | >200 | — |
| Cycloheximide | >500 | >500 | — | >500 | >500 | — |
| Tetracycline | 50a | 6.25a | 8 | 0.313 | 0.039 | 8 |
| Chloramphenicol | 0.850 | 0.425 | 2 | 4.25 | 2.1 | 2 |
| Gentamicin | 0.265 | 0.133 | 2 | 0.39 | 0.2 | 2 |
| Polymyxin B | 3.0 | 3.0 | — | 3.0 | 3.0 | — |
| Protamine | 25.0 | 12.5 | 2 | 25 | 12.5 | 2 |
| Ethidium bromide | 0.625 | 0.078 | 8 | 1.25 | 0.078 | 16 |
| CoCl2 | 312 | 156 | 2 | 312 | 312 | — |
| CuCl2 | 391 | 196 | 2 | 391 | 391 | — |
| ZnSO4 | 78 | 78 | — | 156 | 156 | — |
| Sodium dodecyl sulfate | 250 | 62.5 | 4 | 125 | 31.3 | 4 |
| Cholic acid | 6,250 | 98 | 64 | 3,125 | 98 | 32 |
| Chenodeoxycholic acid | 50,000 | 12.5 | 4,000 | 1,560 | 6.1 | 256 |
| Taurocholic acid | >50,000 | 780 | >64 | >50,000 | 780 | >64 |
| Deoxycholic acid | 10,000 | 10 | 1,000 | 1,250 | 19.5 | 64 |
Both wild-type 81-176 and 9B6 contain the pTet plasmid that carries tet(O) (1).
Both wild-type 21190 and its cmeB mutant produce β-lactamase as determined by Cefinase disk assay.
—, no MIC difference was observed.
Difference in MICs for 81-176 and 9B6.
Difference in MICs for 21190 and cmeB isogenic mutant.
To further confirm that the observed susceptibility changes in mutant 9B6 were associated with the cmeB mutation, the insertional mutant created in strain 21190 was also tested with various antimicrobial agents. Compared with wild-type 21190, the isogenic cmeB mutant of 21190 also showed substantial increases in the susceptibility to different antimicrobials (Table 1). For the majority of compounds examined in this study, the MIC differences caused by the cmeB mutation in the 21190 background were identical or similar to those in the 81-176 background. However, some unique observations were made with 21190 and its cmeB mutant. Firstly, wild-type strain 21190 was resistant to ampicillin and positive with β-lactamase as determined by Cefinase disk assay (Becton Dickinson) (data not shown). The cmeB knockout in strain 21190 did not change the MIC of ampicillin and had less of an impact on the change in MIC of cefotaxime compared to that in 81-176. Secondly, wild-type 21190 was relatively more resistant to erythromycin, and the cmeB mutation resulted in a greater difference in the MIC of erythromycin than that in strain 81-176. In addition, wild-type 21190 was less resistant to chenodeoxycholic acid and deoxycholic acid, and consequently the cmeB mutation-mediated decreases in the MICs of the two compounds in 21190 were not as great as those in 81-176. Despite these strain-related variations, these results further confirmed the role of CmeABC in the intrinsic resistance to various antimicrobial agents in Campylobacter.
CmeABC is an energy-dependent efflux pump.
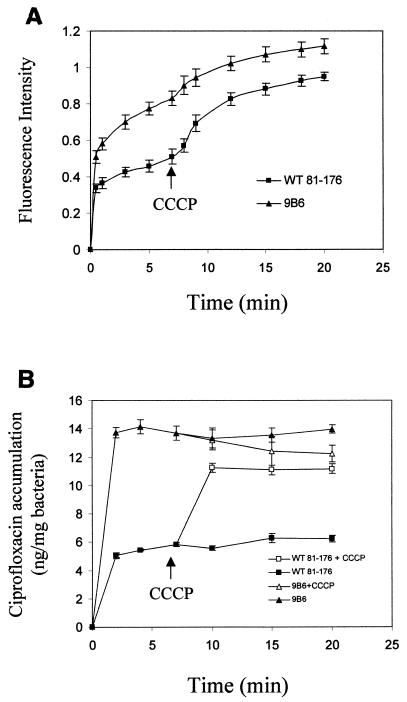
To further establish CmeABC as an active efflux system, accumulations of EB and ciprofloxacin were measured in wild-type 81-176 and mutant 9B6. We first investigated the accumulation of the fluorescent dye EB, a common substrate for bacterial efflux pumps (27). As shown in Fig. 3A, the wild-type 81-176 cells accumulated EB at a much lower level than mutant 9B6. Addition of CCCP, a proton conductor, increased EB uptake in 81-176. This finding indicated that CmeABC contributes significantly to the efflux of EB. To further compare the efflux capability of 9B6 to that of wild-type 81-176, ciprofloxacin was used as a substrate for the second accumulation assay. As shown in Fig. 3B, at the steady state reached within 2 min following addition of ciprofloxacin, 9B6 accumulated about threefold more ciprofloxacin than wild-type 81-176 did. Addition of CCCP resulted in a very rapid and dramatic increase in cell-associated ciprofloxacin in wild-type 81-176 but not in mutant 9B6. At the steady state following addition of CCCP, the accumulation levels of ciprofloxacin were similar between 81-176 and 9B6. These results suggested that CmeABC is the main pump (if not the only one) for the efflux of ciprofloxacin in Campylobacter. Together, these findings firmly established that CmeABC functions as an active efflux system in C. jejuni.
FIG. 3.
Accumulation of EB (A) and ciprofloxacin (B) by C. jejuni wild-type 81-176 and isogenic mutant 9B6. (A) Fluorescence intensity in the y axis represents the levels of accumulated EB in C. jejuni. Fluorescence recording started once EB was added to the bacterial suspension, and CCCP was added to the cell suspension after 7 min of incubation. Each data point represents the mean ± standard deviation of three independent experiments. (B) Ciprofloxacin was added to the bacterial suspension to a final concentration of 10 μg/ml. At 7 min, CCCP was added to one-half of the reaction mixture, and the accumulation measurements continued. Each data point represents the mean ± standard deviation of three independent experiments.
cmeABC is broadly distributed and expressed among C. jejuni isolates.
To determine if cmeABC is present in different C. jejuni strains, cmeB-specific primers (BF2 and BR2) were used in PCR to amplify an 820-bp sequence of cmeB from various strains (including 81-176, S2b, M36292, H49024, 33291, H30769, T48768, W52546, E46972, F26747, 11084571, 15046764, 19094451, X77136, and 21190) of C. jejuni. The cmeB-specific sequence was amplified from every isolate examined in this study (data not shown), indicating its common presence in different strains. To determine if cmeB was expressed in these strains, whole-cell lysates were analyzed by immunoblotting using anti-CmeB antibody, which showed the presence of the CmeB protein in the various isolates (data not shown). Together, these results suggest that cmeABC is widely distributed in different Campylobacter strains and is constitutively expressed in wild-type strains.
DISCUSSION
This work demonstrates that CmeABC is an energy-dependent efflux system contributing to the intrinsic resistance of Campylobacter to diverse antimicrobial agents. This conclusion is based on several lines of evidence. Firstly, CmeABC shares significant sequence and structural homology with many known tripartite efflux systems in gram-negative bacterial pathogens. The genomic organization of cmeABC is also similar to other well-characterized bacterial efflux pumps, such as the MexAB-OprM system of P. aeruginosa (22, 28). Secondly, inactivation of the CmeABC pump by insertional mutagenesis substantially increased the susceptibility of C. jejuni to structurally diverse antimicrobial agents (Table 1). Thirdly, disruption of the CmeABC system resulted in significantly more accumulation of EB and ciprofloxacin within Campylobacter cells (Fig. 3). Together, these findings formally define the active role of CmeABC in the intrinsic resistance of C. jejuni to antimicrobials.
As shown in Fig. 2, transposon insertion in cmeB impaired the production of CmeB and abolished the production of CmeC. It is possible that the 49-kDa band detected by the anti-CmeB antibody in mutant 9B6 is a truncated version of CmeB, caused by the transposon insertion in this gene. Since the recombinant peptide used for producing anti-CmeB antiserum was generated from the N-terminal portion of CmeB (Fig. 1), the anti-CmeB antibody should be able to react with the truncated product of CmeB if it is expressed. It is unlikely that the 49-kDa band detected by anti-CmeB represents a nonspecific reaction, because this band was not detected in wild-type 81-176 when the same antibody was used (Fig. 2). Apparently, the insertional mutation in cmeB yielded a polar effect on expression of cmeC, eliminating the production of the CmeC protein. This finding is not surprising, as sequence analysis suggests that cmeC is likely cotranscribed with cmeA and cmeB, possibly from a promoter upstream of the cmeA gene. RT-PCR indicated that the cmeB mutation did not affect the transcription of Cj0364, which is located downstream of the cmeABC operon and transcribed in the opposite direction from the cmeABC operon. Based on these observations and the finding that the isogenic cmeB mutant of strain 21190 also showed a similar phenotype as mutant 9B6, it can be said with confidence that the phenotypic changes of mutant 9B6 were indeed due to the inactivation of the CmeABC pump.
Active extrusion of FQs via efflux pumps is an important mechanism for the resistance of gram-negative bacteria to this class of antibiotics (18, 23, 26, 37). RND-type efflux systems not only contribute to the intrinsic resistance to FQs but also confer acquired resistance to FQs by overexpression of the efflux proteins (26) or by synergetic interplay with nonefflux FQ resistance mechanisms (such as gyrA mutations) (18, 23, 37). In this study, it was found that CmeABC played a major role in the efflux of ciprofloxacin in C. jejuni (Fig. 3). It was also observed that inactivation of CmeABC increased the susceptibility of strain 81-176 to FQs, especially ciprofloxacin (Table 1). These findings indicate that CmeABC contributes significantly to the intrinsic resistance of FQs in Campylobacter. However, the contribution of CmeABC to the acquired FQ resistance and its interplay with other resistance mechanisms (e.g., gyrA mutations) in Campylobacter are unknown and need to be determined in future studies.
A key feature of these MDR efflux systems is their ability to extrude a broad spectrum of substrates, including various antimicrobial agents. Although a given microorganism can have multiple efflux transporters of different families with overlapping substrate spectra (27, 29), individual efflux systems also show certain levels of selectivity for substrates. For example, multiple tripartite efflux systems have been identified in P. aeruginosa, and each of them extrudes a broad range of antimicrobial agents (20). However, the MexAB-OprM system is the only one responsible for efflux of β-lactams, while efflux of aminoglycosides is mainly mediated by the MexXY-OprM pump (20, 33). Besides CmeABC, there may be additional efflux systems functioning in C. jejuni, because the insertional mutation in cmeB had little or no effect on MICs of some of the antimicrobial agents examined in this study (Table 1). The possible existence of other efflux pumps in Campylobacter is also supported by analysis of the genomic sequence of C. jejuni NCTC 11168 (24), which identified another putative RND family efflux system comprised of Cj1031, Cj1032, and Cj1033 and several putative non-RND-type drug efflux transporters. However, the contributions of these systems to antibiotic resistance in Campylobacter are unknown at present. Functional evaluation of these uncharacterized systems will certainly improve our understandings of the antibiotic resistance mechanisms of C. jejuni.
One striking finding in this study is that the CmeABC efflux pump greatly contributes to C. jejuni resistance to bile, a group of bactericidal detergents present in the intestinal tracts of animals. Inactivation of CmeABC resulted in up to 4,000-fold decreases in the MICs of bile salts (Table 1). Detergent-like bile salts kill bacterial cells by destroying the lipid bilayer of membrane (12). Thus, resistance to bile salts is important for enteric pathogens to survive in the intestinal tract. To diminish the action of bile salts, many enteric pathogens utilize the MDR efflux pumps, such as the AcrAB and EmrAB efflux pumps of E. coli (10, 34) and VceAB of Vibrio cholerae (7), to extrude them out of cells. Inactivation of the acrB gene in Salmonella enterica serovar Typhimurium reduced the colonization of this organism in the intestinal tracts of mice (15). A recent study by Bina and Mekalanos (2) suggested that TolC-mediated bile resistance contributed to the intestinal colonization of V. cholerae in an infant mouse colonization model. These observations suggest that bacterial MDR efflux systems not only play an important role in antibiotic resistance but also contribute to bacterial pathogenesis. Since CmeABC contributes significantly to bile resistance, it is tempting to speculate that CmeABC may also be required for successful colonization of C. jejuni in animal intestines. This possibility remains to be examined in future studies.
Acknowledgments
This work was supported in part by the Research Enhancement Competitive Grants Program of OARDC at the Ohio State University and USDA CSREE competitive grant 00-51110-9741.
DNA sequences were determined at the Molecular, Cellular, and Imaging Center of OARDC.
REFERENCES
- 1.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bina, J. E., and J. J. Mekalanos. 2001. Vibrio cholerae tolC is required for bile resistance and colonization. Infect. Immun. 69:4681-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 4.Blaser, M. J., J. A. Hopkins, R. M. Berka, M. L. Vasil, and W. L. Wang. 1983. Identification and characterization of Campylobacter jejuni outer membrane proteins. Infect. Immun. 42:276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charvalos, E., Y. Tselentis, M. M. Hamzehpour, T. Kohler, and J. C. Pechere. 1995. Evidence for an efflux pump in multidrug-resistant Campylobacter jejuni. Antimicrob. Agents Chemother. 39:2019-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman, K., M. Athalye, A. Clancey, M. Davison, D. J. Payne, C. R. Perry, and I. Chopra. 1994. Bacterial resistance mechanisms as therapeutic targets. J. Antimicrob. Chemother. 33:1091-1116. [DOI] [PubMed] [Google Scholar]
- 7.Colmer, J. A., J. A. Fralick, and A. N. Hamood. 1998. Isolation and characterization of a putative multidrug resistance pump from Vibrio cholerae. Mol. Microbiol. 27:63-72. [DOI] [PubMed] [Google Scholar]
- 8.Dever, L. A., and T. S. Dermody. 1991. Mechanisms of bacterial resistance to antibiotics. Arch. Intern. Med. 151:886-895. [PubMed] [Google Scholar]
- 9.Engberg, J., F. M. Aarestrup, D. E. Taylor, P. Gerner-Smidt, and I. Nachamkin. 2001. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7:24-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giraud, E., A. Cloeckaert, D. Kerboeuf, and E. Chaslus-Dancla. 2000. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 44:1223-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunn, J. S. 2000. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2:907-913. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, J. M., and F. C. Tenover. 1997. Approaches to limiting emergence of antimicrobial resistance in bacteria in human populations. Clin. Infect. Dis. 24(Suppl. 1):S131-S135. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, J. M., and G. M. Church. 1999. Alignment and structure prediction of divergent protein families: periplasmic and outer membrane proteins of bacterial efflux pumps. J. Mol. Biol. 287:695-715. [DOI] [PubMed] [Google Scholar]
- 15.Lacroix, F. J., A. Cloeckaert, O. Grepinet, C. Pinault, M. Y. Popoff, H. Waxin, and P. Pardon. 1996. Salmonella typhimurium acrB-like gene: identification and role in resistance to biliary salts and detergents and in murine infection. FEMS Microbiol. Lett. 135:161-167. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Levy, S. B. 2000. Antibiotic and antiseptic resistance: impact on public health. Pediatr. Infect. Dis. J. 19:S120-S122. [DOI] [PubMed] [Google Scholar]
- 18.Lomovskaya, O., A. Lee, K. Hoshino, H. Ishida, A. Mistry, M. S. Warren, E. Boyer, S. Chamberland, and V. J. Lee. 1999. Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:1340-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1993. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 175:6299-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nachamkin, I., B. M. Allos, and T. Ho. 1998. Campylobacter species and Guillain-Barre syndrome. Clin. Microbiol. Rev. 11:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakajima, A., Y. Sugimoto, H. Yoneyama, and T. Nakae. 2000. Localization of the outer membrane subunit OprM of resistance-nodulation-cell division family multicomponent efflux pump in Pseudomonas aeruginosa. J. Biol. Chem. 275:30064-30068. [DOI] [PubMed] [Google Scholar]
- 23.Oethinger, M., W. V. Kern, A. S. Jellen-Ritter, L. M. McMurry, and S. B. Levy. 2000. Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob. Agents Chemother. 44:10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 25.Paulsen, I. T., J. H. Park, P. S. Choi, and M. H. Saier, Jr. 1997. A family of gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from gram-negative bacteria. FEMS Microbiol. Lett. 156:1-8. [DOI] [PubMed] [Google Scholar]
- 26.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob. Agents Chemother. 44:2233-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole, K. 2001. Multidrug resistance in gram-negative bacteria. Curr. Opin. Microbiol. 4:500-508. [DOI] [PubMed] [Google Scholar]
- 28.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sahm, D. F., and J. A. Washington II. 1991. Antimicrobial susceptibility test: dilution methods, p. 1105-1116. In A. Balows, W. J. Hausler, Jr., K. L. Herramann, H. D. Isenberg, and H. J. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 31.Slutsker, L., S. F. Altekruse, and D. L. Swerdlow. 1998. Foodborne diseases. Emerging pathogens and trends. Infect. Dis. Clin. North Am. 12:199-216. [DOI] [PubMed] [Google Scholar]
- 32.Smith, K. E., J. M. Besser, C. W. Hedberg, F. T. Leano, J. B. Bender, J. H. Wicklund, B. P. Johnson, K. A. Moore, and M. T. Osterholm. 1999. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992-1998. N. Engl. J. Med. 340:1525-1532. [DOI] [PubMed] [Google Scholar]
- 33.Srikumar, R., X. Z. Li, and K. Poole. 1997. Inner membrane efflux components are responsible for beta-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J. Bacteriol. 179:7875-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thanassi, D. G., L. W. Cheng, and H. Nikaido. 1997. Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trieber, C. A., and D. E. Taylor. 2000. Mechanisms of antibiotic resistance in Campylobacter, p. 441-454. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.
- 36.Van Bambeke, F., E. Balzi, and P. M. Tulkens. 2000. Antibiotic efflux pumps. Biochem. Pharmacol. 60:457-470. [DOI] [PubMed] [Google Scholar]
- 37.Wang, H., J. L. Dzink-Fox, M. Chen, and S. B. Levy. 2001. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob. Agents Chemother. 45:1515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, Q., J. C. Meitzler, S. Huang, and T. Morishita. 2000. Sequence polymorphism, predicted secondary structures, and surface-exposed conformational epitopes of Campylobacter major outer membrane protein. Infect. Immun. 68:5679-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]