Abstract
Fluoroquinolones acting equally through DNA gyrase and topoisomerase IV in vivo are considered desirable in requiring two target mutations for emergence of resistant bacteria. To investigate this idea, we have studied the response of Staphylococcus aureus RN4220 to stepwise challenge with sparfloxacin, a known dual-target agent, and with NSFQ-105, a more potent sulfanilyl fluoroquinolone that behaves similarly. First-step mutants were obtained with both drugs but only at the MIC. These mutants exhibited distinctive small-colony phenotypes and two- to fourfold increases in MICs of NSFQ-105, sparfloxacin, and ciprofloxacin. No changes were detected in the quinolone resistance-determining regions of the gyrA, gyrB, grlA, or grlB gene. Quinolone-induced small-colony mutants shared the delayed coagulase response but not the requirement for menadione, hemin, or thymidine characteristic of small-colony variants, a subpopulation of S. aureus that is often defective in electron transport. Second-step mutants selected with NSFQ-105 had gyrA(S84L) alterations; those obtained with sparfloxacin carried a gyrA(D83A) mutation or a novel gyrB deletion (ΔRKSAL, residues 405 to 409) affecting a trypsin-sensitive region linking functional domains of S. aureus GyrB. Each mutation was associated with four- to eightfold increases in MICs of NSFQ-105 and sparfloxacin, but not of ciprofloxacin, which we confirm targets topoisomerase IV. The presence of wild-type grlB-grlA gene sequences in second-step mutants excluded involvement of topoisomerase IV in the small-colony phenotype. Growth revertants retaining mutant gyrA or gyrB alleles were quinolone susceptible, indicating that resistance to NSFQ-105 and sparfloxacin was contingent on the small-colony mutation. We propose that small-colony mutations unbalance target sensitivities, perhaps through altered ATP or topoisomerase levels, such that gyrase becomes the primary drug target. Breaking of target parity by genetic or physiological means eliminates the need for two target mutations and provides a novel mechanism for stepwise selection of quinolone resistance.
Staphylococcus aureus is a key gram-positive pathogen that causes life-threatening systemic infections, including pneumonia, septicemia, endocarditis, and osteomyelitis. Although several effective antistaphylococcal agents have been developed, their use has been compromised by the emergence of resistant strains. Thus, the introduction of penicillins was followed by the rapid selection of β-lactamase-expressing strains (5). Similarly, strains resistant to methicillin through expression of the mecA gene are widespread in many hospitals, and such isolates frequently express a multiple-resistance phenotype (5). In addition, recent work suggests that small-colony variants (SCVs) of S. aureus associated with persistent and refractory infections may play a role in clinical resistance (22, 37). SCVs are naturally occurring variants with a normal microscopic morphology but a small colony size and a complex phenotype, including reduced expression of coagulase and hemolysin. One important subset of SCVs has an electron transport deficiency affecting the proton motive force and ATP synthesis (37, 45) and resulting in aminoglycoside resistance (36). These various developments underscore the need for new antistaphylococcal drugs and a detailed understanding of the drug resistance mechanisms that operate in this organism.
Fluoroquinolones are a potentially useful class of agents active against S. aureus. They exert their antibacterial effects by interfering with DNA gyrase and topoisomerase IV, two essential ATP-dependent enzymes that function by a double-strand DNA breakage and rejoining to promote DNA unwinding during DNA replication and chromosome segregation at cell division (8, 15, 24, 31, 46). Gyrase, a tetramer containing two GyrA and two GyrB subunits encoded by the gyrA and gyrB genes, respectively, catalyzes negative supercoiling of DNA and is thought to counter the formation of positive supercoils that arise during DNA unwinding in replication (8, 16, 46). Topoisomerase IV is also a tetramer and is specified by the parC and parE genes, usually termed grlA and grlB in S. aureus, respectively (9). Resistance to quinolones commonly arises from mutations in the quinolone resistance-determining regions (QRDRs) of the DNA gyrase or topoisomerase IV genes (6, 9, 10, 12, 13, 15, 40, 41, 48, 49). Many quinolones, including ciprofloxacin, gatifloxacin, levofloxacin, norfloxacin, ofloxacin, pefloxacin, premafloxacin, and trovafloxacin, select mutations in grlA and then in gyrA, indicating that topoisomerase IV is the primary target and gyrase is the secondary target in S. aureus (9, 10, 17, 20, 21, 32, 47). Nadifloxacin and the des-F(6)-quinolone BMS-284756 differ from other quinolones in selecting gyrA mutants of S. aureus (7, 42).
Studies with Streptococcus pneumoniae and S. aureus have identified quinolones that appear to act with parity through both topoisomerase targets: so-called dual target agents (11, 34, 47). Two features characterize such agents. First, neither gyrA nor parC mutations alone have much effect on drug susceptibility, whereas gyrA parC mutants exhibit significant resistance. Second, it is difficult to select resistant topoisomerase mutants, which is attributed to the requirement for two target mutations. These criteria are met by clinafloxacin against S. pneumoniae (34) and by sparfloxacin against S. aureus (11, 47). Thus, Yamagishi et al. have reported an inability to obtain resistant mutants of S. aureus by challenge with sparfloxacin at four times the MIC, a level that for ciprofloxacin and other quinolones readily selects grlA mutants in a single step (47). However, with sparfloxacin at lower concentrations, Gootz et al. noted that small-colony mutants of S. aureus RN4220 were recovered, but these were not characterized further (17). Recently, Ruiz et al. employed subinhibitory levels of sparfloxacin in a continuous culture system to obtain sparfloxacin-resistant S. aureus mutants (38). A gyrA (E88K) mutation and a grlA (S80Y) change were detected in the fifth and seventh selection steps, respectively, but none of the mutants from the first four steps were characterized. These experiments confirm the difficulty of obtaining topoisomerase mutants of S. aureus with sparfloxacin in a single step but suggest the existence of alternative resistance pathways.
To understand how resistance to the dual-target fluoroquinolones may develop, we investigated the response of S. aureus RN4220 to stepwise drug challenge with sparfloxacin and with NSFQ-105 (2), a sulfanilylciprofloxacin that we show behaves similarly. Unlike ciprofloxacin, neither drug selected topoisomerase mutants in the first step. Instead, we easily recovered resistant mutants with a small-colony phenotype but only at the MIC. Surprisingly, subsequent selection and expression of gyrase-mediated resistance were conditional on the small-colony background. We have characterized the various mutants and propose a mechanism for this novel two-step resistance pathway.
MATERIALS AND METHODS
Bacterial strains and drugs.
S. aureus RN4220 (23) was kindly donated by J. Lindsay. Ciprofloxacin hydrochloride (Bayer, Newbury, United Kingdom) was dissolved in 0.1 M NaOH and stored at −20°C. Sparfloxacin was a gift from S. Nakamura, Dainippon Pharmaceutical Co., Suita, Japan. NSFQ-105 was kindly synthesized by M. R. Mazzieri and M. J. Nieto.
Selection of quinolone-resistant S. aureus RN4220 mutants.
Approximately 109 CFU of susceptible isolate RN4220 was plated on brain heart infusion plates containing ciprofloxacin at 2 μg/ml; NSFQ-105 at 0.06, 0.125, 0.25, or 0.5 μg/ml; or sparfloxacin at 0.25, 0.5, or 1 μg/ml. Plates were incubated aerobically for 48 h at 37°C. Second-step mutants were obtained similarly with mutant strains as parents.
Characterization of mutants.
Genomic DNAs from RN4220 and its drug-resistant mutants were prepared by boiling bacteria for 5 min followed by removal of debris by centrifugation at 14,000 × g for 5 min. The supernatants were used as PCR templates to amplify topoisomerase QRDRs for selected strains. Primers used in PCR were designed using published sequences (4, 9, 27, 47) and are shown in Table 1. PCR conditions were 93°C for 30 s, 50°C for 30 s, and 74°C for 1 min. This procedure was repeated for 30 cycles. Analysis by electrophoresis in 2% low-gelling agarose confirmed the amplification of gyrA, grlA, gyrB, and grlB products as 394-, 340-, 300-, and 340-bp products, respectively.
TABLE 1.
Oligonucleotides employed to amplify or sequence QRDRs of S. aureus gyrase and topoisomerase IV genes
| Oligonucleotide (gene) | Usea | Sequence (nucleotide position, 5′ to 3′)b | Codons sequenced |
|---|---|---|---|
| SAA1 (gyrA) | FP | TCGTGCATTGCCAGATGTTCG (96-116) | |
| SAA4 (gyrA) | RP | TCGAGCAGGTAAGACTGACGG (489-469) | |
| SAA2 (gyrA) | S | AAACCAGTACATCGTCGTATA (127-147) | 50-156 |
| SAC1 (grlA) | FP | TGCCAGATGTTCGTGATGGT (92-111) | |
| SAC4 (grlA) | RP | TGGAATGAAAGAAACTGTCTC (431-411) | |
| SAC2 (grlA) | S | TACGCAATGTATTCAAGTGG (138-157) | 53-137 |
| SAB1 (gyrB) | FP | AAAAAGCGCGTGAAGTAACAC (1190-1210) | |
| SAB4 (gyrB) | RP | TCGCTAGATCAAAGTCGCCA (1489-1470) | |
| SAB2 (gyrB) | S | CGTAAATCAGCGTTAGATG (1213-1231) | 410-490 |
| SAE1 (grlB) | FP | CAAGGGAAGCTGCACGTAAAG (1154-1174) | |
| SAE4 (grlB) | RP | TACACGATTATAATTACTATC (1493-1474) | |
| SAE2 (grlB) | S | CGTGAAGATGCTCGTTCAGG (1177-1196) | 399-491 |
HinfI digestion of PCR products and gel electrophoresis in 2% agarose gels were used to test for the presence of the common QRDR mutations at codon 84 of gyrA and codon 80 of parC (41). For DNA sequence analysis, PCR products were purified using a Qiagen PCR purification kit and the DNA was sequenced directly using an ABI Prism automated sequencer.
DNA sequence analysis of the entire grlB-grlA locus in strains 2NSA5 and 2SSA3 was carried out using overlapping 1,578-, 1,270-, and 2,355-bp PCR products comprising most of the grlB gene, the 3′ end of grlB and the 5′ end of grlA, and most of the grlA gene, respectively. The primers used were based on published sequences and were as follows: SAE5 (5′-GCATTTTACGCTGATTTATAT, nucleotide positions −83 to −63 upstream of grlB) and reverse primer SAE4 (5′-TACACGATTATAATTACTATC, positions 1493 to 1474 in grlB), SAE1 (5′-CAAGGGAAGCTGCACGTAAAG, positions 1154 to 1174 in grlB) and reverse primer SAC4 (5′-TGGAATGAAAGAAACTGTCTC, positions 431 to 411 in grlA), and SAC1 (5′-TGCCAGATGTTCGTGATGGT, positions 92 to 111 in grlA) and reverse primer SAC8 (5′-CATTACTGTATTTTATCATTTA, 44 to 23 nucleotides downstream of the grlA stop codon). Chromosomal DNA was used as the template for Taq polymerase in the presence of 1.5 mM MgCl2. PCR conditions were 94°C for 1 min, 50°C for 1 min, and 74°C for 3 min for 30 cycles. PCR products were purified using Qiagen minispin columns and sequenced directly by Lark Technologies using an automated DNA sequencer and a series of nested primers. The DNA sequence was compared to that published for S. aureus grlB-grlA (4, 47).
Auxotrophy for hemin, menadione, and thymidine, either singly or in combination, was tested as described previously (22) by plating on chemically defined medium (CDM) (44) with the compounds at 1, 10 and 100 μg/ml, respectively.
S. aureus GyrB expression plasmid pPS1.
PCR was used to amplify a 1.9-kbp fragment carrying the full-length S. aureus gyrB gene with chromosomal DNA from strain 81231 as the template (27). The primers used were RK5 (5′-TAACAGAAAGCCATGGTGACTGCA [an NcoI site overlapping the ATG initiation codon is underlined]) and reverse primer PS4 (5′-AGAGTTCCTCGAGCAAAAGTTCAG [10 bp downstream of the TAA termination codon; underlining indicates an artificial XhoI site]). DNA from S. aureus 81231 and primers were incubated with Vent DNA polymerase in a standard PCR buffer containing 1.5 mM MgCl 2 under the following conditions: denaturation at 93°C for 30s, annealing at 50°C for 1 min, and extension at 74°C for 3 min. This cycle was repeated 30 times, with a final incubation at 74°C for 10 min. The PCR product was purified by phenol extraction and blunt-end ligated into plasmid pCR-Script SK(+) in the presence of SrfI as per the manufacturer's instructions prior to transformation of supercompetent XL-Blue Escherichia coli. The insert from one plasmid was recovered by digestion with NcoI and XhoI, ligated into NcoI-XhoI-cut plasmid pET19B, and used to transform BL21(λDE3)pLysE. The complete sequence of the gyrB insert in three recombinant plasmids was determined using a set of nested primers. These were identical, and one plasmid, pPS1, was used for GyrB overexpression.
Expression and purification of full-length S. aureus GyrB.
The purification of S. aureus GyrB will be described in detail elsewhere. Briefly, Luria-Bertani medium containing 50 μg of carbenicillin per ml and 34 μg of chloramphenicol per ml was inoculated with a culture of E. coli BL21(λDE3)pLysE carrying expression plasmid pPS1 and grown at 37°C to an optical density at 600 nm of 0.6. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to induce expression of the S. aureus gyrB gene, and the cells were incubated at 37°C for a further 3 h prior to harvesting, freezing in liquid nitrogen, and storage at −70°C. Cells were lysed by thawing and centrifuged, and S. aureus GyrB was purified to 95% homogeneity from the supernatant by a protocol involving streptomycin sulfate precipitation and ammonium sulfate fractionation, followed by column chromatography sequentially on heparin-agarose, phenyl-Sepharose, and MonoQ. Fractions were frozen immediately in liquid nitrogen and stored at −70°C.
Trypsin cleavage.
S. aureus GyrB protein (180 μg) in 20 mM Tris-HCl (pH 7.5)-50 mM NaCl-10% glycerol-0.2 mM Na3EDTA-5 mM dithiothreitol (100-μl final volume) was incubated with trypsin at 0.4 μg/ml at 25°C. Samples (15 μl) were removed at intervals, added to an equal volume of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis loading buffer, boiled for 2 min, and stored on ice. Proteins were analyzed by electrophoresis on SDS-10% polyacrylamide gels.
Coagulase response.
Coagulase testing was done by the standard tube assay.
RESULTS
Response of grlA and grlA gyrA mutants of S. aureus RN4220 to NSFQ-105 and sparfloxacin.
To generate defined mutants that would be informative about quinolone action, we first selected ciprofloxacin-resistant strains of S. aureus RN4220. Approximately 109 CFU of RN4220 was plated onto each of two brain heart infusion agar plates containing ciprofloxacin at 2 μg/ml, i.e., two to four times the MIC. After aerobic incubation at 37°C for 48 h, mutants 1CSA1 and 1CSA2 were obtained. Second-step mutants 2CSA1 to 2CSA4 and 2CSA5 to 2CSA8 were derived from 1CSA1 and 1CSA2, respectively, by selection with ciprofloxacin at 16 μg/ml (Fig. 1) at a frequency of 5 × 10−9. All of the mutants exhibited a normal colony size and were characterized in terms of quinolone susceptibility and the status of their gyrA, gyrB, grlA, and grlB QRDRs (Table 1) by HinfI-restriction fragment length polymorphism (RFLP) analysis and DNA sequencing of appropriate PCR products.
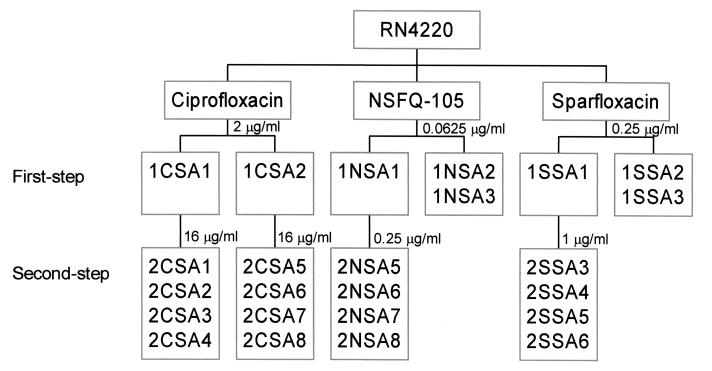
FIG. 1.
Relationships of S. aureus mutants selected stepwise with quinolones. Single numbers above the boxes indicate the concentrations of ciprofloxacin, sparfloxacin, and NSFQ-105 used in each step of selection.
First-step mutants 1CSA1 and 1CSA2 exhibited ciprofloxacin MICs of 8 μg/ml, i.e., 8- to 16-fold higher than that for the parental strain, associated with the acquisition of grlA mutations encoding respective S80F and S80Y alterations at the protein level (Table 2). Second-step mutants 2CSA1 to 2CSA8 exhibited ciprofloxacin MICs of 64 μg/ml, and all had acquired a mutation at gyrA codon 84 as revealed by HinfI-RFLP analysis of PCR products. For 2CSA1 and 2CSA5, the gyrA mutation resulted in an S84L GyrA change (Table 2). These particular grlA and gyrA changes are commonly associated with quinolone resistance, and their order of appearance confirms that ciprofloxacin acts through topoisomerase IV in S. aureus (10).
TABLE 2.
Fluoroquinolone resistance profiles of isogenic S. aureus mutants
| Strain | MIC (μg/ml) ofa:
|
Mutation in the QRDR ofb:
|
|||||
|---|---|---|---|---|---|---|---|
| CIP | NSFQ-105 | SPX | GyrA | GyrB | GrlA | GrlB | |
| RN4220 | 0.5-1 | 0.06 | 0.125-0.25 | None | None | None | None |
| 1CSA1c | 8 | 0.125 | 0.5 | None | NDd | S80F | ND |
| 1CSA2c | 8 | 0.125 | 0.5 | None | ND | S80Y | ND |
| 2CSA1 | 64 | 2-4 | 32 | S84L | ND | S80F | ND |
| 2CSA5 | 64 | 2-4 | 32 | S84L | ND | S80Y | ND |
| 1NSA1c | 1-2 | 0.125 | 0.5 | None | None | None | None |
| 1NSA2 | 1 | 0.06 | 0.25 | − | − | ||
| 2NSA5 | 2 | 0.5 | 2 | S84L | None | None | None |
| 2NSA6 | 2 | 1 | 2 | S84L | None | None | None |
| 2NSA7 | 2 | 1 | 2 | + | − | ||
| 2NSA8 | 2 | 1 | 2 | + | − | ||
| 1SSA1c | 2 | 0.125 | 0.5 | None | None | None | None |
| 1SSA2 | 2 | 0.125 | 0.5 | − | − | ||
| 1SSA3 | 1 | 0.06-0.125 | 0.25 | − | − | ||
| 2SSA3 | 2 | 0.5 | 2 | D83A | None | None | None |
| 2SSA4 | 2 | 0.5 | 2 | None | Δ405-409 | None | None |
| 2SSA5 | 2 | 0.5 | 2 | D83A | None | None | None |
| 2SSA6 | 2 | 0.5 | 2 | None | Δ405-409 | None | None |
CIP, ciprofloxacin; NSFQ-105, 4-(4-aminophenylsulfonyl)-1-piperazinylciprofloxacin; SPX, sparfloxacin.
GrlA mutations resulted from the following nucleotide changes: S80F, TCC to TTC; S80Y, TCC to TAC. GyrA changes were as follows: D83A, GAC to GCC; S84L, TCA to TTA. + and −, presence and absence of a mutation at gyrA codon 84 or parC codon 80 detected by HinfI digestion of PCR products (41).
Strain used in selection of second-step mutants.
ND, not determined.
NSFQ-105 and sparfloxacin were more active than ciprofloxacin against RN4220 and its mutants (Table 2), displaying MICs against RN4220 of 0.06 and 0.125 to 0.25 μg/ml, respectively. The grlA mutation in 1CSA1 and 1CSA2 increased the MICs of both drugs only about 2-fold, compared with 8- to 16-fold for ciprofloxacin (Table 2). However, the grlA gyrA strains 2CSA1 and 2CSA5 were highly resistant, displaying respective NSFQ-105 and sparfloxacin MICs of 2 to 4 and 32 μg/ml, values that were, respectively, 32- to 64-fold and 128- to 256-fold higher than those for the parental strain. These data are in line with earlier work showing that grlA or gyrA mutations alone have little effect on sparfloxacin MIC (11, 47), consistent with dual targeting in S. aureus. NSFQ-105 exhibited similar behavior (Table 2).
First-step mutants selected with NSFQ-105 or sparfloxacin exhibit a small-colony phenotype, and second-step mutants carry D83A or S84L gyrA changes or a novel gyrB(Δ405-409) deletion.
Two series of mutants were selected stepwise by challenge with NSFQ-105 and with sparfloxacin on brain heart infusion agar (Fig. 1). RN4220 (109 CFU) was exposed to NSFQ-105 at 0.0625, 0.125, 0.25, and 0.5 μg/ml. Mutants were recovered only at 0.0625 μg/ml at a frequency of 2 × 10−7, and all colonies were one-quarter the size seen for RN4220. No mutants were obtained on plates containing NSFQ-105 at 0.125 μg/ml and above, even when 1011 CFU was plated. First-step mutant 1NSA1 (MIC, 0.125 μg/ml) was exposed to drug at 0.25 μg/ml, yielding second-step mutants (at a frequency of 4 × 10−8), of which 2NSA5 to 2NSA8 were examined in detail.
First-step mutants 1NSA1 and 1NSA2 exhibited a twofold decrease in susceptibility to ciprofloxacin, NSFQ-105, and sparfloxacin and had wild-type topoisomerase QRDRs (Table 2). Second-step mutants 2NSA5 to 2NSA8 exhibited NSFQ-105 MICs of 0.5 to 1 μg/ml, i.e., 8- to 16-fold higher than that of the parental strain (Table 2). The sparfloxacin MICs for these mutants were increased by a similar factor, but the ciprofloxacin MIC was only twofold higher. HinfI-RFLP analysis detected a codon 84 gyrA change in each of the four strains, which in 2NSA5 and 2NSA6 resulted in an S84L GyrA alteration.
Exposure of RN4220 to sparfloxacin at 0.25, 0.5, and 1 μg/ml allowed recovery of mutants only at the MIC (0.25 μg/ml) at a frequency of 5 × 10−7; no mutants were obtained at higher drug concentrations even when 1010 CFU was screened. All mutants exhibited a pinpoint colony morphology, and 1SSA1 to 1SSA3 were chosen for further study (Fig. 1). 1SSA1 was exposed to sparfloxacin at 1 μg/ml (two times the MIC), producing second-step mutants at a frequency of 6.5 × 10−8, from which strains 2SSA3 to -6 were characterized. No mutants were obtained with drug at 2 μg/ml.
Similar to 1NSA1 and 1NSA2, 1SSA1 to 1SSA3 were twofold more resistant than RN4220 to ciprofloxacin, NSFQ-105, and sparfloxacin, and none had mutations in topoisomerase QRDRs (Table 2). Second-step mutants 2SSA3 to -6 displayed an 8- to 16-fold increase in sparfloxacin and NSFQ-105 MICs over those of the parental strain, with little effect on ciprofloxacin MIC. 2SSA3 and 2SSA5 each carried a gyrA mutation (detectable by HinfI-RFLP) resulting in an unusual D83A mutation at the protein level. Surprisingly, strains 2SSA4 and 2SSA6 had each acquired a 15-bp in-frame deletion in gyrB, resulting in the loss of residues RKSAL at positions 405 to 409 in GyrB. Both the D83A GyrA and Δ405-409 GyrB mutations are novel, and they indicate that sparfloxacin, like NSFQ-105, selects gyrase mutants of S. aureus.
Second-step mutants carry wild-type grlB and grlA genes, and therefore topoisomerase IV is not implicated in the small-colony phenotype.
Some quinolones select mutations in topoisomerase IV that lie outside the conventional QRDRs (20, 21). To examine this possibility, we sequenced the entire 4.5-kb grlB-grlA region from strains 2NSA5 and 2SSA3, which in each case was amplified as three overlapping PCR products constituting sequence from bp −83 upstream of the grlB start codon to bp 43 downstream of the grlA stop codon. For each strain, the inferred amino acid sequences for the GrlB and GrlA proteins were identical to those of wild-type topoisomerase IV (9) except at three positions in GrlA: Gly267 (GGT), Arg567 (CGA), and Val688 (GTT) in the sequence reported by Ferrero et al. (9) were replaced by Ser (AGT), Ala (GCA), and Ala (GCT), respectively. However, DNA sequence analysis of the RN4220 grlB and grlA genes revealed that these were polymorphisms that were also present in the parental RN4220 strain. Given that the grlB and grlA genes are wild type in second-step mutants 2NSA5 and 2SSA3, it follows they must also be unchanged in the precursor strains 1NSA1 and 1SSA1, which were used in the selection of all second-step mutants in this study (Fig. 1). Thus, changes in topoisomerase IV genes and promoters are not implicated in the small-colony phenotype.
Small-colony mutants exhibit a delayed coagulase response but not hemin, menadione, or thymidine auxotrophy characteristic of classical SCVs.
It was possible that small-colony mutants were SCVs, a subpopulation of S. aureus often characterized by an electron transport deficit (37, 45). These SCVs have a complex phenotype characterized by auxotrophy for hemin, menadione, or thiamine (factors involved in cytochrome and menaquinone biosynthesis) or thymidine (for SCVs recovered from patients undergoing trimethoprim treatment) (22), lack of pigmentation, and an altered coagulase response (reduced protein synthesis due to lowered ATP levels) (37). To determine whether we had recovered SCVs, we tested all of the mutants described in Table 2 for the relevant auxotrophies by plating on CDM (44) by a standard disk method (22). In fact, even in the absence of supplements, NSFQ-105-selected mutants grew as well on CDM as the parental strain, indicating that the small-colony phenotype seen on brain heart infusion medium is not a classical SCV effect. For the sparfloxacin-selected mutants, inclusion of hemin, menadione, or thymidine, either singly or in combination, did not rescue the small-colony phenotype, again suggesting that these mutants are not conventional SCVs. In terms of the other SCV phenotypes, RN4220 is naturally lacking in pigmentation and therefore both parent and mutant strains had the same (uninformative) color. Although quinolone-derived mutants exhibited a delayed coagulase response (see below), the data overall suggest that we had not isolated electron transport-deficient SCVs.
The GyrB(Δ405-409) mutation occurs at a trypsin cleavage site demarcating functional domains of GyrB protein.
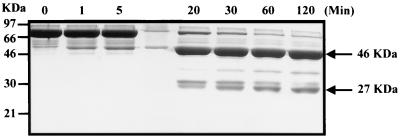
Quinolone resistance usually accrues from missense mutations in topoisomerase genes. It was therefore interesting to observe a gyrB(Δ405-409) mutation in second-step sparfloxacin-resistant mutants. The region in E. coli GyrB equivalent to S. aureus GyrB residues 405 to 409 forms a protease-sensitive linker that connects ATPase- and GyrA-interacting domains (1, 14). To determine if the Δ405-409 deletion might confer resistance by affecting communication between gyrase domains, we examined whether the RKSAL sequence also lies in a linker region of the S. aureus GyrB protein. The full-length 72-kDa S. aureus GyrB protein was overexpressed in E. coli and purified to 95% homogeneity (see Materials and Methods for details). Figure 2 shows a time course for trypsin digestion of the protein. It can be seen that proteolysis generated two major protease-resistant fragments that were approximately 46 and 27 kDa in size, consistent with cleavage at an internal trypsin-sensitive site. Both fragments and full-length GyrB protein were electroblotted onto polyvinylidine difluoride membrane, and their N-terminal sequences were determined by Edman degradation (28). The N-terminal sequence of the 46-kDa fragment was identical to that of the full-length protein, (M)VTALDSDVNNTDNYGAGQIQ, and except for the N-terminal methionine, which appears to be removed in E. coli, was that predicted from the nucleotide sequence of the gyrB gene. The 27-kDa fragment had the N-terminal sequence R--ALD, in which the second and third Edman cycles gave indistinct signals. This sequence and the size of the tryptic fragment are consistent with enzyme cleavage after the first arginine (R404) of the sequence RRKSALD. Thus, the RKSAL sequence deleted in second-step mutants 2SSA4 and 2SSA6 lies at a tryptic cleavage site defining 46-kDa N-terminal ATPase and 27-kDa C-terminal GyrA interaction domains of the S. aureus GyrB protein. These results suggest that quinolone resistance associated with the GyrB deletion may occur by a novel mechanism.
FIG. 2.
Trypsin digestion of S. aureus GyrB protein generates 46-kDa N-terminal and 27-kDa C-terminal fragments. GyrB protein was incubated with trypsin, and samples were removed at the time points indicated. Reactions were quenched in SDS-gel loading buffer and applied to an SDS-10% polyacrylamide gel. After electrophoresis, protein bands were visualized by staining with Coomassie blue dye.
Second-step growth revertants retain gyrase mutations but exhibit quinolone susceptibility and a normal coagulase response.
When sparfloxacin-resistant S. aureus mutants were grown on drug-free brain heart infusion agar, we noted the occasional presence of normal-size colonies. These spontaneous revertants occurred at a frequency of 5 × 10−8, consistent with a single mutational step. To determine if the pinpoint colony phenotype was essential for expression of a delayed coagulase response and quinolone resistance, we characterized revertants 1SSA1-R, 2SSA3-R, and 2SSA4-R, which were derived from sparfloxacin-selected first-step mutant 1SSA1 and second-step gyrA and gyrB mutants 2SSA3 and 2SSA4, respectively (Table 3). Growth reversion was associated with reversion to the wild-type coagulase response. Thus, whereas the original mutants exhibited a delayed coagulase response showing positivity at 24 h, with strain 2SSA4 positive at 4 h, the revertant strains all behaved like wild-type RN4220 in showing coagulase positivity at 2 h. Second, revertant 1SSA1-R exhibited wild-type susceptibility to sparfloxacin, NSFQ-105, and ciprofloxacin compared with first-step mutant 1SSA1. These results indicate that low-level quinolone resistance, delayed coagulase effects, and small colony size are likely conferred by the same mutation. Surprisingly, revertant strains 2SSA3-R and 2SSA4-R also reverted to near-wild-type susceptibility to all three quinolones (Table 3). DNA sequence analysis confirmed the retention of mutant gyrA(D83A) and gyrB(Δ405-409) genes in 2SSA3-R and 2SSA4-R, respectively. Thus, it appears that expression of gyrase-mediated resistance to NSFQ-105 and sparfloxacin is contingent on the first-step mutation responsible for the small-colony phenotype and altered coagulase response.
TABLE 3.
Phenotypes of small-colony S. aureus mutants and their revertants
| Strain | MIC (μg/ml) ofa:
|
SCb | Time (h) of coagulase positivity | Mutation in the QRDR ofc:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| CIP | NSFQ-105 | SPX | GyrA | GyrB | GrlA | GrlB | |||
| RN4220 | 0.5-1 | 0.06 | 0.125-0.25 | No | 2 | None | None | None | None |
| 1NSA1 | 1-2 | 0.125 | 0.5 | Yes | 24 | None | None | None | None |
| 1SSA1 | 2 | 0.125 | 0.5 | Yes | 24 | None | None | None | None |
| 1SSA1-Rd | 0.5-1 | 0.06 | 0.125-0.25 | No | 2 | ND | ND | ND | ND |
| 2SSA3 | 2 | 0.5 | 2 | Yes | 24 | D83A | None | None | None |
| 2SSA3-R | 0.5-1 | 0.125 | 0.25 | No | 2 | D83A | ND | ND | ND |
| 2SSA4 | 2 | 0.5 | 2 | Yes | 4 | None | Δ405-409 | None | None |
| 2SSA4-R | 0.5-1 | 0.125 | 0.125-0.25 | No | 2 | ND | Δ405-409 | ND | ND |
| 2NSA5 | 2 | 0.5 | 2 | Yes | 24 | S84L | None | None | None |
The presence of gyrase mutations in revertants allows selection of grlA mutants with sparfloxacin.
Gyrase mutants 2SSA3-R and 2SSA4-R, like grlA mutants, were susceptible to NSFQ-105 and sparfloxacin, consistent with the dual-target action of these drugs against wild-type S. aureus (Tables 2 and 3). We were interested in examining whether the presence of a gyrase mutation then allowed the selection of topoisomerase IV mutants. Strains 2SSA3-R and 2SSA4-R were plated on brain heart infusion agar containing sparfloxacin at 0.5 and at 1 μg/ml. Mutants with a normal colony size were readily isolated at frequencies of 1.2 × 10−7 and 6.7 × 10−8, respectively. By HinfI-RFLP analysis of grlA PCR products, six of seven mutant strains derived from both 2SSA3-R and 2SSA4-R had acquired a mutation at the codon 80 hot spot. It appears that once a gyrA mutation has been acquired, sparfloxacin readily selects topoisomerase IV mutants of S. aureus.
DISCUSSION
We have investigated the stepwise selection of resistance in S. aureus to the dual-target agents sparfloxacin and NSFQ-105, a potent sulfanilyl derivative of ciprofloxacin. Resistance to both fluoroquinolones developed by a novel mechanism in which first-step mutants with distinctive small-colony phenotypes preceded second-step gyrase mutants. Surprisingly, analysis of growth revertants revealed that gyrase-mediated resistance to both drugs was conditional on the small-colony mutation. These results are in striking contrast to those obtained with ciprofloxacin and many other quinolones, which at two to four times the MIC select grlA and then gyrA mutants of S. aureus. We show that the small-colony mutants may be related to SCVs, a subpopulation of S. aureus that includes electron transport mutants defective in ATP synthesis. These properties allow a plausible model in which small-colony mutants facilitate the emergence of resistance to dual-target fluoroquinolones by relaxing the need for two target alterations. These observations could have important clinical consequences.
Sparfloxacin-selected mutants exhibited a pinpoint colony size that was considerably smaller than that of mutants selected with NSFQ-105. This observation and differences in reversion frequencies suggest that the two drugs select genetically different mutants in the first step. At present, we do not know the identities of the genes involved. Although grlA mutations would predispose to selection of gyrase mutants by sparfloxacin (47), we found no alteration of topoisomerase IV QRDRs in first-step mutants recovered with either NSFQ-105 or sparfloxacin (Table 2). Moreover, two independent second-step mutants had wild-type topoisomerase IV grlB-grlA genes and promoters and therefore so must first-step mutants 1NSA1 and 1SSA1, the precursor strains used to select all second-step mutants (Fig. 1; Table 2). It follows that mutations in topoisomerase IV are not responsible for the observed small-colony phenotypes. Second, based on the definition of a colony size 1/10 of that of the wild-type strain (37), mutants selected with sparfloxacin (but not NSFQ-105) would qualify as SCVs, an important subset of which are thought to have an ATP deficit due to defective electron transport (37). These particular variants exhibit lack of pigmentation, delayed coagulase response, and auxotrophy for hemin or menadione, two factors required for the synthesis of cytochrome and menaquinone components of the respiratory chain. An additional thymidine auxotrophy has been reported for S. aureus SCVs isolated from cystic fibrosis patients treated with trimethoprim-sulfamethoxazole (22). Our small-colony mutants were nonpigmented (as was the parental RN4220 strain) and exhibited the delayed coagulase response seen for SCVs (Table 3), but they were not hemin, menadione, or thymidine auxotrophs. Conceivably, the quinolone-derived mutants represent a different subset of SCVs with mutations in other pathways affecting ATP, e.g., the citric acid cycle enzymes, defects in which confer nalidixic acid resistance in E. coli (19, 25).
In contrast to wild-type S. aureus, gyrase mutants were readily selected from the first-step small-colony strains. Mutants isolated with NSFQ-105 all carried gyrA mutations affecting the S84 hot spot, which in two cases were identified as the well-known S84L alteration (Table 2). These mutants were cross-resistant to sparfloxacin but not ciprofloxacin, which operates through topoisomerase IV. On the other hand, sparfloxacin selected gyrA(D83A) or gyrB(Δ405-409) mutants which were cross-resistant to NSFQ-105 but not ciprofloxacin (Table 2). To our knowledge, the D83A GyrA mutation has not been previously reported for S. aureus. However, the equivalent D82A (or D82G) change in E. coli GyrA has been shown to confer quinolone resistance (12, 43). The GyrB(Δ405-409) change is unusual in being a deletion and in lying just outside the conventional GyrB QRDR (bounded by EGDSA and PLRGK motifs) in a region that we have shown by proteolysis forms a linker between the ATPase- and GyrA-interacting domains of GyrB (Fig. 2). Although we did not examine the properties of the GyrB(Δ405-409) protein, several lines of evidence suggest that the deletion does confer resistance. First, it was acquired in a single-step challenge with sparfloxacin. Second, similar to observations for the revertant 2SSA3-R (gyrA D83A), the presence of the deletion in strain 2SSA4-R allowed ready selection of grlA changes in the secondary target topoisomerase IV (see Results). Third, it is known that alterations in the equivalent region of E. coli GyrB affect quinolone susceptibility. For example, the compensatory mutation in the ΔtopA strain DM800 involves a two-amino-acid insertion in the GyrB linker region (29) that confers hypersensitivity to fluoroquinolones in vivo (18). It is known that the GyrB C-terminal domain binds GyrA (14) and likely forms part of the quinolone binding pocket (6). Moreover, cleavable complex formation is affected by ATP binding at the ATPase domain (26). Therefore, the GyrB linker deletion could confer quinolone resistance through effects on GyrB folding or on mutual interactions with GyrA, quinolones, and DNA. These data implicate gyrase as the primary target of NSFQ-105 and sparfloxacin, at least in the small-colony background.
Analysis of revertants yielded the surprising result that small-colony mutations not only precede changes in gyrase but indeed are essential for expression of resistance (Table 3). As a working hypothesis, we propose that they act by breaking the parity of topoisomerase sensitivities such that gyrase becomes the primary target of NSFQ-105 and sparfloxacin. Evidence in support of this model comes from comparison of small-colony mutants carrying mutations in gyrase (strains 2NSA5 to 2NSA8 and 2SSA3 to 2SSA6 in Table 2) or in grlA. The required grlA small-colony mutants (ciprofloxacin MICs of 8 to 16 μg/ml) bearing GrlA S80Y or D79N changes were generated from 1NSA1 and 1SSA1 by single-step ciprofloxacin exposure (X.-S. Pan and L. M. Fisher, unpublished results). In the small-colony background, the MICs of both NSFQ-105 and sparfloxacin were increased fourfold by gyrase mutations (Table 2), whereas grlA mutations produced no change in susceptibility to NSFQ-105 and a twofold increase for sparfloxacin (Pan and Fisher, unpublished results). These differential effects suggest that small-colony mutations unbalance the target sensitivities for NSFQ-105 and sparfloxacin in favor of gyrase.
Currently, we do not know the mechanism underlying the breaking of target parity. Although SCVs have altered accumulation of drugs, including quinolones (30, 37), it is difficult to envisage how changes in efflux could differentially affect drug targeting. Instead, we envisage that small-colony mutations either alter quinolone-target interactions at the level of cleavable-complex formation with gyrase and topoisomerase IV or, alternatively, influence the processing of such complexes to give the lethal lesion. Altered ATP levels could be responsible. Thus, it is known that ATP stimulates cleavable-complex formation by quinolones with E. coli gyrase and, conversely, that depletion of intracellular ATP protects against quinolone killing (26). Differential binding of subsaturating ATP to gyrase and topoisomerase IV could serve to modulate cleavable-complex formation in favor of gyrase. However, other parity-breaking mechanisms can be envisaged, e.g., changes in topoisomerase gene expression or in other metabolites known to affect enzyme activity, e.g., glutamate. Aside from gene mutations, these changes might also occur physiologically through alterations in growth or environmental conditions. Once a gyrase change has been acquired, subsequent selection of topoisomerase IV mutations would occur at normal frequency and result in the emergence of highly resistant mutants. This process may be facilitated by the reversibility of the small-colony phenotype (Table 3), a feature observed for some, but not all, SCVs (36, 37).
Conditional resistance through small-colony mutants is a new resistance mechanism in S. aureus, in addition to altered efflux through the NorA pump, selection of first-step gyrase mutants by nadifloxacin, and recovery of first-step grlA mutants by ciprofloxacin and many other quinolones. For any particular quinolone, we presume that the resistance mechanism will depend on the molecular structure of the drug, which in turn determines intracellular target preference. This principle is already well established for S. pneumoniae, in which NSFQ-105 and sparfloxacin target gyrase whereas ciprofloxacin acts through topoisomerase IV (3, 33, 35). As NSFQ-105 is identical to ciprofloxacin except for the addition of a benzylsulfonamido group to the C-7 piperazinyl ring, it appears that the C-7 moiety can influence target preference in S. pneumoniae. NSFQ-105 is a dual-target agent in S. aureus, whereas ciprofloxacin acts through topoisomerase IV, suggesting that the C-7 substituent also affects drug targeting in this organism. It is interesting to ask whether small-colony mutants can be selected by quinolones other than NSFQ-105 and sparfloxacin and acquire further mutational resistance. It has been reported that small-colony mutants are induced in liquid culture at the MIC by pazufloxacin, a putative dual-target agent (42), and by nalidixic acid, which is thought to target gyrase, but not by ciprofloxacin, norfloxacin, fleroxacin, enoxacin, pipemidic acid, or tosufloxacin, which act through topoisomerase IV (30). More work will be needed to establish whether failure to recover small-colony mutants in vitro is due to a chemical feature of the quinolone or, more likely, a narrow selection window given their low-level (twofold) resistance. For clinically isolated SCVs of S. aureus, ciprofloxacin will select more resistant mutants following multiple rounds of exposure in liquid culture (39).
In summary, we have shown that resistance of S. aureus to NSFQ-105 and sparfloxacin develops through small-colony mutants as obligate intermediates. These mutants share certain similarities with but have some differences from SCVs that have been identified clinically with persistent systemic disease. SCVs have diminished pathogenicity due to low expression of hemolysin and coagulase and can survive in an intracellular environment. Clearly, further studies will be needed to define the exact functional and clinical relationship between SCVs and quinolone-derived small-colony mutants.
Acknowledgments
X.-S.P. was supported by grants from Pfizer (Warner-Lambert) and the BBSRC. P.J.H. was funded by an MRC Ph.D. studentship. R.T.-V. was supported by a visiting fellowship from the Conselleria de Cultura i Educacio, Generalitat Valenciana, Spain. F.L.A. was funded by the FOMEC Project (Facultad de Ciencias Quimicas, Universidad Nacional de Cordoba, Cordoba, Argentina).
We thank Aodhan Breathnach and Julie Johnson for carrying out coagulase testing.
REFERENCES
- 1.Adachi, T., M. Mizuuchi, E. A. Robinson, E. Appella, M. H. O'Dea, M. Gellert, and K. Mizuuchi. 1987. DNA sequencing of the Escherichia coli gyrB gene: application of a new sequencing strategy. Nucleic Acids Res. 15:771-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alovero, F. L., M. Nieto, M. R. Mazzieri, R. Then, and R. H. Manzo. 1998. Mode of action of sulfanilyl fluoroquinolones. Antimicrob. Agents Chemother. 42:1495-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alovero, F. L., X.-S. Pan, J. E. Morris, R. H. Manzo, and L. M. Fisher. 2000. Engineering the specificity of antibacterial fluoroquinolones: benzenesulfonamide modifications at C-7 of ciprofloxacin change its primary target in Streptococcus pneumoniae from topoisomerase IV to gyrase. Antimicrob. Agents Chemother. 44:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brockbank, S. M., and P. T. Barth. 1993. Cloning, sequencing, and expression of the DNA gyrase genes from Staphylococcus aureus. J. Bacteriol. 175:3269-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers, H. F. 1997. Methicillin resistance in staphylococci: biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullen, M. E., A. W. Wyke, R. Kuroda, and L. M. Fisher. 1989. Cloning and characterization of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob. Agents Chemother. 33:886-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Driscotta, L. F., L. E. Lawrence, K. L. Denbleyker, and J. F. Barrett. 2001. Staphylococcus aureus mutants selected by BMS-284756. Antimicrob. Agents Chemother. 45:3273-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrero, L., B. Cameron, B. Manse, D. Lagneux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. [DOI] [PubMed] [Google Scholar]
- 10.Ferrero, L., B. Cameron, and J. Crouzet. 1995. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 39:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fournier, B., and D. C. Hooper. 1998. Mutations in topoisomerase IV and DNA gyrase of Staphylococcus aureus: novel pleiotropic effects on quinolone and coumarin activity. Antimicrob. Agents Chemother. 42:121-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman, S. M., T. Lu, and K. Drlica. 2001. Mutation of the DNA gyrase A gene of Escherichia coli that expands the quinolone resistance-determining region. Antimicrob. Agents Chemother. 45:2378-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda, H., S. Hori, and K. Hiramatsu. 1998. Antibacterial activity of gatifloxacin (AM-1155, CG5501, BMS-206586), a newly developed fluoroquinolone, against sequentially acquired quinolone-resistant mutants and the norA transformant of Staphylococcus aureus. Antimicrob. Agents Chemother. 42:1917-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gellert, M., L. M. Fisher, and M. H. O'Dea. 1979. DNA gyrase: purification and catalytic properties of a fragment of gyrase B protein. Proc. Natl. Acad. Sci. USA 76:6289-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gellert, M., K. Mizuuchi, M. H. O'Dea, T. Itoh, and J.-I. Tomizawa. 1977. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. USA 74:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gellert, M., K. Mizuuchi, M. H. O'Dea, and H. A. Nash. 1976. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. USA 73:3872-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gootz, T. D., R. P. Zaniewski, S. L. Haskell, F. S. Kaczmarek, and A. E. Maurice. 1999. Activities of trovafloxacin compared with those of other quinolones against purified topoisomerases and gyrA and grlA mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 43:1845-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heddle, J. G., T. Liu, Z. Zhao, K. Drlica, and A. Maxwell. 2001. gyrB225, a mutation of DNA gyrase that compensates for topoisomerase I deficiency: investigation of its low activity and quinolone hypersensitivity. J. Mol. Biol. 309:1219-1231. [DOI] [PubMed] [Google Scholar]
- 19.Helling, R. B., and J. S. Kukora. 1971. Nalidixic acid-resistant mutants of Escherichia coli deficient in isocitrate dehydrogenase. J. Bacteriol. 105:1224-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ince, D., and D. C. Hooper. 2000. Mechanisms and frequency of resistance to premafloxacin in Staphylococcus aureus: novel mutations suggest novel drug target interactions. Antimicrob. Agents Chemother. 44:3344-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ince, D., and D. C. Hooper. 2001. Mechanisms and frequency of resistance to gatifloxacin in comparison to AM-1121 and ciprofloxacin in Staphylococcus aureus. Antimicrob. Agents Chemother. 45:2755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kahl, B., M. Herrmann, A. S. Everding, H. G. Koch, K. Becker, E. Harms, R. A. Proctor, and G. Peters. 1998. Persistent infection with small colony variant strains of Staphylococcus aureus in patients with cystic fibrosis. J. Infect. Dis. 177:1023-1029. [DOI] [PubMed] [Google Scholar]
- 23.Kreisworth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature (London) 305:709-712. [DOI] [PubMed] [Google Scholar]
- 24.Kreuzer, K. N., and N. R. Cozzarelli. 1979. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase A subunit: effects on deoxyribonucleic acid replication, transcription and bacteriophage growth. J. Bacteriol. 140:424-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakshmi, T. M., and R. B. Helling. 1976. Selection of citrate synthase deficiency in icd mutants of Escherichia coli. J. Bacteriol. 127:76-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, T.-K., and L. F. Liu. 1998. Modulation of gyrase-mediated DNA cleavage and cell killing by ATP. Antimicrob. Agents Chemother. 42:1022-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margerrison, E. E., R. Hopewell, and L. M. Fisher. 1992. Nucleotide sequence of the Staphylococcus aureus gyrB-gyrA locus encoding the DNA gyrase A and B proteins. J. Bacteriol. 174:1596-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsudaira, P. 1987. Sequence from picomole quantities of proteins electroblotted on to polyvinylidine difluoride membranes. J. Biol. Chem. 262:10035-10038. [PubMed] [Google Scholar]
- 29.McEachern, F., and L. M. Fisher. 1989. Regulation of DNA supercoiling in Escherichia coli: genetic basis of a compensatory mutation in DNA gyrase. FEBS Lett. 253:67-70. [DOI] [PubMed] [Google Scholar]
- 30.Mitsuyama, J. H., H. Yamada, J. Maehana, Y. Fukuda, S. Kurose, S. Minami, Y. Todo, and H. Narita. 1997. Characteristics of quinolone-induced small-colony variants of Staphylococcus aureus. J. Antimicrob. Chemother. 39:697-707. [DOI] [PubMed] [Google Scholar]
- 31.Mizuuchi, K., L. M. Fisher, M. H. O'Dea, and M. Gellert. 1980. DNA gyrase action involves the introduction of transient double strand breaks into DNA. Proc. Natl. Acad. Sci. USA 77:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng, E. Y., M. Trucksis, and D. C. Hooper. 1996. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan, X.-S., and L. M. Fisher. 1997. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob. Agents Chemother. 41:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan, X.-S., and L. M. Fisher. 1998. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan, X.-S., G. Yague, and L. M. Fisher. 2001. Quinolone resistance mutations in Streptococcus pneumoniae GyrA and ParC proteins: mechanistic insights into quinolone action from enzymatic analysis, intracellular levels, and phenotypes of wild-type and mutant proteins. Antimicrob. Agents Chemother. 45:3140-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelletier, L. L. 1979. Virulent gentamicin-induced small colony variants of Staphylococcus aureus. J. Lab. Clin. Med. 94:324-334. [PubMed] [Google Scholar]
- 37.Proctor, R. A., B. Kahl, C. von Eiff, P. E. Vaudaux, D. P. Lew, and G. Peters. 1998. Staphylococcal small colony variants have novel mechanisms for antibiotic resistance. Clin. Infect. Dis. 27(Suppl. 1):S68-S74. [DOI] [PubMed] [Google Scholar]
- 38.Ruiz, J., J. P. Sierra, M. T. Jimenez de Anta, and J. Vila. 2001. Characterization of sparfloxacin-resistant mutants of Staphylococcus aureus obtained in vitro. Int. J. Antimicrob. Agents 118:107-112. [DOI] [PubMed] [Google Scholar]
- 39.Schmitz, F. J., A. C. Fluit, A. Beeck, M. Perdikouli, and C. von Eiff. 2001. Development of chromosomally-encoded resistance mutations in small colony variants of Staphylococcus aureus. J. Antimicrob. Chemother. 47:113-115. [DOI] [PubMed] [Google Scholar]
- 40.Sreedharan, S., M. Oram, B. Jensen, L. R. Peterson, and L. M. Fisher. 1990. DNA gyrase gyrA mutations in ciprofloxacin-resistant strains of Staphylococcus aureus: close similarity with quinolone resistance mutations in Escherichia coli. J. Bacteriol. 172:7260-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sreedharan, S., L. R. Peterson, and L. M. Fisher. 1991. Ciprofloxacin resistance in coagulase-positive and -negative staphylococci: role of mutations at serine-84 in the DNA gyrase A protein of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 35:2151-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takei, M., H. Fukuda, R. Kishii, and M. Hosaka. 2001. Target preference of 15 quinolones against Staphylococcus aureus, based on antibacterial activities and target inhibition. Antimicrob. Agents Chemother. 45:3544-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Truong, Q. C., J.-C. N. Van, D. Shlaes, L. Gutmann, and N. J. Moreau. 1997. A novel, double mutation in DNA gyrase A of Escherichia coli conferring resistance to quinolone antibiotics. Antimicrob. Agents Chemother. 41:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van de Rijn, I., and R. E. Kessler. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 27:444-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Von Eiff, C., C. Heilman, R. A. Proctor, C. Woltz, G. Peters, and F. Gotz. 1997. A site-directed Staphylococcus aureus hemB mutant is a small-colony variant which persists intracellularly. J. Bacteriol. 179:4706-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, J. C. 1996. DNA topoisomerases. Annu. Rev. Biochem. 65:635-692. [DOI] [PubMed] [Google Scholar]
- 47.Yamagishi, J.-I., T. Kojima, Y. Oyamada, K. Fujimoto, H. Hattori, S. Nakamura, and M. Inouye. 1996. Alterations in the DNA topoisomerase IV grlA gene responsible for quinolone resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1157-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida, H., M. Bogaki, M. Nakamura, L. M. Yamanaka, and S. Nakamura. 1991. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother. 35:1647-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]