Abstract
A PCR-based technique using molecular beacons was developed to detect the chloroquine resistance-associated pfcrt K76T point mutation in Plasmodium falciparum. One hundred thirty African clinical isolates were tested by the new method in comparison with the PCR-restriction fragment length polymorphism method. This rapid and inexpensive genomic assay could expand the possibilities for monitoring chloroquine resistance.
Plasmodium falciparum, the cause of the most lethal malaria in humans, is a major cause of disease and death worldwide. Every year in Africa an estimated 500 million cases of falciparum malaria occur, of which about at least one million result in death (9). This major health care concern is aggravated by the widespread diffusion of falciparum chloroquine resistance (CQR), whose emergence has been associated with a dramatic increase in malaria mortality (15). However, chloroquine remains the first-line drug for the treatment of acute uncomplicated malaria in most African countries. This is mainly due to its low cost and good tolerability and because alternative drugs are lacking or too expensive, except the combination sulfadoxine-pyrimethamine. The molecular basis of CQR is not totally elucidated. Point mutations in a gene, pfcrt, have been identified elsewhere as strongly associated with in vitro CQR in parasite lines and in natural isolates (1, 5, 19). In particular, the PfCRT K76T and A220S mutations were always detected in CQR parasites. Genetic complementation experiments have demonstrated further the role of pfcrt in CQR (7). Several in vivo studies showed absolute selection of the pfcrt K76T mutant allele in therapeutic failure (2, 12). However, the presence of parasites having the pfcrt K76T mutant allele was not always predictive of the clinical outcome in individuals due to host factors such as drug uptake, distribution, and metabolism and, probably more importantly, to preexisting immunity. In areas of high transmission, where immunity is high, subjects having pfcrt K76T mutant Plasmodium were frequently able to clear their parasitemia. In some areas where CQR rates are high, the pfcrt mutant allele was reported as ubiquitous. Nevertheless, diagnostic assays for pfcrt mutations are already used as tools for monitoring CQR, besides in vitro and in vivo studies (19). The majority of experimenters used nested PCR followed by restriction fragment length polymorphism (RFLP), a relatively more rapid method than DNA sequencing, though still labor-intensive. Other authors used nested-mutation-specific PCR assays which need strictly controlled conditions to be reproducible (3). As one of the main features of molecular assays consists in processing a great number of samples at a low cost and in a short amount of time, it appeared of interest to apply the molecular beacon technique to the detection of the pfcrt K76T mutation. Molecular beacons are hairpin-shaped probes that undergo a spontaneous fluorogenic conformational change when they hybridize to the sequence of a target DNA or RNA molecule (8, 16, 17). A fluorophore and a quencher group are covalently attached to each end of an allele-specific oligonucleotide. This brings the fluorophore and the quencher in close proximity, so that fluorescence cannot occur in the absence of the target. When the probe encounters a target molecule, a hybrid is formed that is more stable than the hairpin structure of the isolated probe, resulting in the separation of the fluorophore and quencher and the restoration of fluorescence. This study describes the development and evaluation of a new method of detection of the pfcrt K76T mutation which can be easily used for epidemiological studies.
One hundred thirty clinical isolates of P. falciparum, having parasite densities ranging from 80 to 7,100 trophozoites/μl, were obtained in 2000 and 2001 from malaria-infected travelers returning to France from mostly African countries. Parasite DNA was extracted from venous blood as previously described (6). The amplification was performed on 2 μl of DNA solution by a PCR assay which was developed in order to obtain an amplicon of 199 bp centered on codon 76. The primers used were synthesized based on the published full-length pfcrt sequence of P. falciparum (clone DIV30) (GenBank accession no. AF233064) and with the use of Primer 3 software (Steve Rozen and Helen J. Skaletsky, 1996, 1997; Primer 3 code available at http://www.genome.wi.mit.edu/cgi-bin/primer/primer3.cgi). To genotype the alleles, two molecular beacons were designed, one specific for the K76 wild-type allele and 5′ labeled with a green fluorophore (fluorescein) and the other specific for the K76T mutant allele and 5′ labeled with a red fluorophore (tetramethylrhodamine). Both oligonucleotides were 3′ labeled with the quencher 4-(4′-dimethylaminophenylazo)benzoic acid (DABCYL). The PCR conditions were as follows. A mixture of 15 pmol each of sense primer 5′-TTTAGGTGGAGGTTCTTGTC-3′, hybridizing to positions 170 to 190 of the pfcrt gene, and antisense primer 5′-AATAAAGTTGTGAGTTTCGGA-3′, hybridizing to positions 368 to 348; 200 μM concentrations of each deoxynucleoside triphosphate; buffer (15 mM Tris-HCl [pH 8.0], 50 mM KCl, 6 mM MgCl2); 2.5 U of ampliTaq Gold DNA polymerase; and 10 pmol each of fluorescein-5′-GCGACGTGTAATGAATAAAATTTTTGGTCG-3′-DABCYL (wild-type beacon) which hybridizes to positions 283 to 303 of the gene pfcrt, and tetramethylrhodamine-5′-GCGACGTGTAATTGAAACAATTTTGTCGC-3′-DABCYL (mutant beacon) (Eurogentec, S.A., Seraing, Belgium), which hybridizes to positions 283 to 301 of the pfcrt gene of isolates originating from Southeast Asia and Africa, was incubated in a 50-μl volume for 7 min at 95°C followed by 40 reaction cycles (95°C, 40 s; 52°C, 40 s; 72°C, 40 s). A further step was added to enable the final hybridization of the probes (95°C, 2 min; 53°C, 6 min; 20°C, 2 min). After completion of the PCR, sealed tubes were placed on a 96-well Costar microplate in a Fluostar 403 (BMG Labotechnologies GmbH) spectrofluorometer. Fluorescence measurements were carried out at an excitation wavelength of 480 nm, an emission wavelength of 520 nm for fluorescein, and emission wavelengths of 544 and 590 nm for tetramethylrhodamine. For each well, a ratio was calculated as previously described (4).
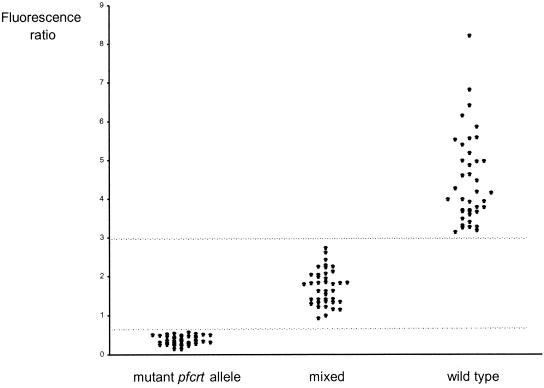
The determination of the thresholds was previously done on known samples of a limited series (data not shown). K76T mutant isolates had a ratio of <0.70, and K76 wild-type isolates had a ratio of >2.90. Between these two values were those for mixed isolates (K76-K76T). For comparison, the same isolates were processed blindly in a PCR-RFLP method according to the following conditions: 17 μl of the 199-bp amplified DNA fragments was incubated with 0.5 U of the restriction enzyme ApoI (New England Biolabs) at 50°C for 8 h according to the manufacturer's protocol. PCR products with the wild-type codon were cleaved into two fragments (126 and 73 bp) by ApoI, as PCR products with the mutant allele were not cut. Following enzymatic digestion, the PCR products were separated by 3% agarose gel electrophoresis. Eighty isolates displayed the K76T mutation, either alone (n = 40) or in mixed isolates (n = 40); 50 isolates had the K76 codon alone (wild type). For all isolates, results obtained with the fluorescent method (Fig. 1) were consistent with the data of the PCR-RFLP assay.
FIG. 1.
Distribution of fluorescence ratios of K76T, mixed K76-K76T, and K76 clinical P. falciparum isolates. Fluorescence ratios express fluorescence measurements at 520 and 590 nm. Data represent the means of three independent experiments; ranges of fluorescence ratios were 0 to 0.09 for wild-type isolates, 0.02 to 0.36 for mixed isolates, and 0.04 to 1.15 (for highest mean value) for mutant isolates. Isolates having fluorescence ratios above 9 are not shown in the figure.
To validate the thresholds further, a study of artificial mixtures of the clones 3D7 (wild-type allele) and W2 (mutant allele) was performed. P. falciparum clones 3D7 and W2 were originally obtained from D. Walliker (University of Edinburgh, Edinburgh, Scotland, United Kingdom). Stock cultures of 3D7 and W2 clones were grown as previously described (14). Cultures having a parasitemia of 1% were mixed in order to have the following W2/3D7 ratios: 1:1, 1:10, 1:50, 1:100, 1:1,000, 10:1, 50:1, 100:1, and 1,000:1. For all ratios, results were obtained from simultaneous assays based on triplicate determinations. The ability of the fluorescent method to detect minority clones appeared identical to that of the PCR-RFLP method (Table 1).
TABLE 1.
pfcrt point mutation at position 76 determined by molecular beacons and PCR-RFLP in artificial mixtures of P. falciparum clones
| Clone or mixture | Fluorescence ratioa | Codon 76 by assay:
|
|
|---|---|---|---|
| Fluorescence | PCR-RFLP | ||
| W2 alone | 0.23 (0.21-0.28) | Thr | Thr |
| 3D7 alone | 3.88 (3.61-4.17) | Lys | Lys |
| W2/3D7 ratio | |||
| 10:1 | 0.98 (0.93-1.06) | Lys/Thr | Lys/Thr |
| 50:1 | 0.40 (0.37-0.42) | Thr | Thr |
| 100:1 | 0.31 (0.30-0.32) | Thr | Thr |
| 1,000:1 | 0.22 (0.20-0.23) | Thr | Thr |
| 1:1 | 1.46 (1.21-1.55) | Lys/Thr | Lys/Thr |
| 1:10 | 1.64 (1.51-1.82) | Lys/Thr | Lys/Thr |
| 1:50 | 3.23 (3.13-3.52) | Lys | Lys |
| 1:100 | 3.20 (3.11-3.47) | Lys | Lys |
| 1:1,000 | 3.86 (3.71-4.26) | Lys | Lys |
Ratios express fluorescence measurements at 520 and 590 nm. Values are medians, with ranges shown in parentheses.
Molecular beacons have been already used for mutation detection with humans (8) and with various microorganisms such as Mycobacterium tuberculosis or retroviruses (11, 18). The main advantages of the technique are well known. Unlike the previously used methods that create the risk of contaminating untested samples by the handling of the amplified DNA, the amplicon detection by molecular beacons is carry out in a hermetically sealed tube. This avoids the possibility of having false-positive signals. Results are obtained very rapidly because there is no electrophoresis step, and the cost is relatively low. The present study confirmed the absolute specificity of the interaction of molecular beacons with their target strands. Results were perfectly matched with those obtained on the same isolate by PCR-RFLP. A high sensitivity was also observed. In particular minority alleles whose presence was equal to or greater than about 10% were detected by the study of artificial mixtures.
The molecular beacon technique has already been applied successfully to the detection of the dihydrofolate reductase S108N mutation in P. falciparum (4). With different fluorophores and a multiplex PCR assay (18), the simultaneous determination of the dihydrofolate reductase 108 and pfcrt 76 alleles could be performed in a single sealed reaction tube. Automated mutation detection with molecular beacons is currently under development and may result in methods suitable for high-throughput analysis (13).
Among the eight point mutations described for pfcrt resistance alleles, we chose codon 76 as the target of the probes because pfcrt K76T was always reported in resistant isolates originating from all four resistance foci which are known today, i.e., the focus in Southeast Asia that has spread into Africa, the two foci in South America, and the focus in Papua New Guinea (19). However, due to the polymorphism in codons 74 and 75 reported for resistance alleles from South America and Papua New Guinea, the present test is not suitable for isolates originating from these areas.
Drug-resistant malaria represents today a major health care concern in countries where malaria is endemic. Making therapy and prophylaxis policies requires current and comprehensive data on resistance. The limitations of available in vivo and in vitro methods for monitoring resistance highlight the need for developing molecular assays. Today, molecular markers are used in many studies focusing on malaria drug resistance (10, 19). The molecular surveillance of CQR may rely on the determination of the proportion of the pfcrt alleles of a population in a given geographical area. Results may provide a rationale for considering the withdrawal of chloroquine as a first-line treatment of uncomplicated malaria. Conversely, in areas where the shift to sulfadoxine-pyrimethamine has been done, a decrease of the proportion of CQR isolates could lead to a reintroduction of chloroquine, maybe in combination with another drug. The rapid and inexpensive genomic assay that is now available to detect the pfcrt K76T mutation could expand the possibilities for monitoring resistance.
REFERENCES
- 1.Basco, L. K., and P. Ringwald. 2001. Analysis of the key pfcrt point mutation and in vitro and in vivo response to chloroquine in Yaoudé, Cameroon. J. Infect. Dis. 183:1828-1831. [DOI] [PubMed] [Google Scholar]
- 2.Djimde, A., O. K. Doumbo, J. F. Cortese, K. Kayentao, S. Doumbo, Y. Diourté, A. Dicko, X. Z. Su, T. Nomura, D. A. Fidock, T. E. Wellems, and C. V. Plowe. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N. Engl. J. Med. 344:257-263. [DOI] [PubMed] [Google Scholar]
- 3.Dorsey, G., M. R. Kamya, A. Singh, and P. J. Rosenthal. 2001. Polymorphisms in the Plasmodium falciparum pfcrt and pfmdr-1 genes and clinical response to chloroquine in Kampala, Uganda. J. Infect. Dis. 183:1417-1420. [DOI] [PubMed] [Google Scholar]
- 4.Durand, R., J. Eslahpazire, S. Jafari, J. F. Delabre, A. Marmorat-Khuong, J. P. Di Piazza, and J. Le Bras. 2000. Use of molecular beacons to detect an antifolate resistance-associated mutation in Plasmodium falciparum. Antimicrob. Agents Chemother. 44:3461-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durand, R., S. Jafari, J. Vauzelle, J. F. Delabre, Z. Jesic, and J. Le Bras. 2001. Analysis of pfcrt point mutations and chloroquine susceptibility in isolates of Plasmodium falciparum. Mol. Biochem. Parasitol. 114:95-102. [DOI] [PubMed] [Google Scholar]
- 6.Durand, R., O. Ramiliarisoa, Y. Sécardin, P. Eldin de Pécoulas, L. K. Basco, and J. Le Bras. 1997. DHFR gene point mutation as a predictor of Plasmodium falciparum resistance to cycloguanil in malaria cases from Africa imported to France. Trans. R. Soc. Trop. Med. Hyg. 85:33-34. [DOI] [PubMed] [Google Scholar]
- 7.Fidock, D. A., T. Nomura, A. K. Talley, R. A. Cooper, S. M. Dzekunov, M. T. Ferdig, L. M. Ursos, A. B. Sidhu, B. Naudé, K. W. Deitsch, X. Z. Su, J. C. Wootton, P. D. Roepe, and T. E. Wellems. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell 6:861-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giesendorf, B. A., J. A. Vet, S. Tyagi, E. J. Mensink, F. J. Trijbels, and H. J. Blom. 1998. Molecular beacons: a new approach for semiautomated mutation analysis. Clin. Chem. 44:482-486. [PubMed] [Google Scholar]
- 9.Greenwood, B., and T. Mutabingwa. 2002. Malaria in 2002. Nature 415:670-672. [DOI] [PubMed] [Google Scholar]
- 10.Kublin, J. G., F. K. Dzinjalamala, D. D. Kamwendo, E. M. Malkin, J. F. Cortese, L. M. Martino, R. A. G. Mukadam, S. J. Rogerson, A. G. Lescano, M. E. Molyneux, P. A. Winstanley, P. Chimpeni, T. E. Taylor, and C. V. Plowe. 2002. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J. Infect. Dis. 185:380-388. [DOI] [PubMed] [Google Scholar]
- 11.Piatek, A. S., S. Tyagi, A. C. Pol, A. Telenti, L. P. Miller, F. R. Kramer, and D. Alland. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359-363. [DOI] [PubMed] [Google Scholar]
- 12.Pillai, D. R., A. C. Labbé, V. Vanisaveth, B. Hongvangthong, S. Pomphida, S. Inkathone, K. Zhong, and K. C. Kain. 2001. Plasmodium falciparum malaria in Laos: chloroquine treatment outcome and predictive value of molecular markers. J. Infect. Dis. 183:789-795. [DOI] [PubMed] [Google Scholar]
- 13.Smit, M. L., B. A. Giesendorf, J. A. Vet, F. J. Trijbels, and H. J. Blom. 2001. Semiautomated DNA mutation analysis using a robotic workstation and molecular beacons. Clin. Chem. 47:739-744. [PubMed] [Google Scholar]
- 14.Trager, W., and J. B. Jensen. 1976. Human malaria parasites in continuous culture. Science 193:673-675. [DOI] [PubMed] [Google Scholar]
- 15.Trape, J. F., G. Pison, M. P. Preziosi, C. Enel, A. Desgrees du Lou, V. Delaunay, B. Samb, E. Lagarde, J. F. Molez, and F. Simondon. 1998. Impact of chloroquine resistance on malaria mortality. C. R. Acad. Sci. III 321:689-697. [DOI] [PubMed] [Google Scholar]
- 16.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]
- 17.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 18.Vet, J. A., A. R. Majithia, S. A. Marras, S. Tyagi, S. Dube, B. J. Poiesz, and F. R. Kramer. 1999. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl. Acad. Sci. USA 96:6394-6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wellems, T., and C. V. Plowe. 2001. Chloroquine-resistant malaria. J. Infect. Dis. 184:770-776. [DOI] [PubMed] [Google Scholar]