Abstract
The recent emergence of pneumococcal isolates exhibiting an unusual resistance phenotype of higher amoxicillin MICs in relation to the penicillin MICs prompted an analysis of the pbp genes from three such strains isolated in France. For comparison, three amoxicillin-susceptible strains were included in the study. DNA sequence analysis of the pbp2x, pbp2b, and pbp1a genes revealed extensive sequence divergence in all six isolates compared to the sequences of the genes of penicillin-susceptible strain R6. With the exception of pbp2b, no amino acid mutations were unique to the resistant isolates. Transformation experiments with cloned pbp genes isolated from one of the resistant isolates demonstrated a stepwise development of amoxicillin resistance involving penicillin-binding proteins (PBPs) 2X, 2B, and 1A. Full resistance, equivalent to that of the donor strain, was achieved only when genomic DNA was transformed into R62x/2b/1a mutants, suggesting that full resistance development in this isolate is mediated by a non-PBP determinant. Moreover, the recently identified murMN resistance determinant does not appear to have any impact on resistance in this isolate. This determinant (from the French isolate) was, however, able to transform an R6 mutant harboring pbp2x, pbp2b, and pbp1a genes from a Hungarian clone with an extremely high level of penicillin resistance so that it had increased levels of penicillin resistance. These results indicate that the development of high-level β-lactam resistance is a complex process and that the involvement of MurMN in penicillin resistance appears to be dependent on specific mutations in PBPs 2X, 2B, and/or 1A. Furthermore, an additional (as yet unidentified) non-PBP-mediated resistance determinant is required for full resistance development in some pneumococci.
Acute otitis media (AOM) is characterized by acute inflammation of the middle ear accompanied by the presence of middle ear fluid. Episodes of otitis media can occur at any age but are most common in young children and infants, particularly those from 3 months to 3 years of age. In the United States, approximately 60% of all children contract otitis media in their first year of life and 80% contract otitis media by age 3 (13, 39). Most cases of AOM among patients in developed countries resolve spontaneously, but a selected population will develop recurrent and severe disease (3). In populations in developing countries, otitis media often commences within 3 months of birth and persists throughout childhood. This is due to high rates of carriage of multiple types of respiratory bacterial pathogens and high rates of cross-infection caused by overcrowding and poor hygiene (23, 24).
The pneumococcus is responsible for 40 to 50% of cases of AOM, whereas Haemophilus influenzae causes 20 to 30% of cases of AOM and Moraxella catarrhalis causes 10 to 15% of cases of AOM (20). In selecting an antimicrobial agent for treatment of AOM, efficacy against Streptococcus pneumoniae is the most important consideration. For this reason, amoxicillin has remained the initial drug of choice for the treatment of uncomplicated AOM, regardless of the prevalence of drug-resistant S. pneumoniae in the community. In addition, otitis media associated with H. influenzae or M. catarrhalis is more likely to resolve spontaneously (8, 9).
Amoxicillin (α-amino-p-hydroxybenzyl penicillin) was introduced in 1972 as an analogue of ampicillin (it differs structurally from ampicillin by the presence of a p-hydroxyl group). Amoxicillin has a spectrum of activity similar to that of ampicillin, but the improved absorption of amoxicillin from the gastrointestinal tract leads to higher concentrations of the drug in serum, middle ear fluid, and urine (21, 38). Besides its use in the treatment of otitis media, amoxicillin is also an excellent agent for the treatment of bacterial sinusitis (31).
NCCLS guidelines (30) state that a pneumococcal isolate that is susceptible to penicillin can be considered susceptible to other β-lactams. Consequently, it has been suggested that penicillin MICs can be used to predict the MICs of other β-lactam antimicrobials for S. pneumoniae (4). It is generally accepted that the MICs of amoxicillin and extended-spectrum cephalosporins are usually equal to or two to four times less than the MIC of benzylpenicillin (5, 33, 40). The high rate of penicillin resistance among pneumococci in France has resulted in increased rates of use of amoxicillin, cefotaxime, and ceftriaxone (17). Recently, pneumococci resistant to amoxicillin (MICs, ≥4 μg/ml) and/or extended-spectrum cephalosporins (cefotaxime MICs, ≥4 μg/ml) have been identified in France (10). In addition, the MICs of these agents for pneumococci are equal to or 1 dilution higher than the MIC of penicillin. The emergence of such strains is of concern because it restricts the selection of drugs that can be used to treat severe pneumococcal infections.
The aim of this study was to analyze the pbp genes from three French S. pneumoniae strains with intermediate to high-level resistance to amoxicillin. Three isolates with increased susceptibility to amoxicillin were included in the study for comparative purposes. It has been suggested that analysis of the DNA sequences of the pbp genes (especially pbp2b) be carried out to determine the genetic modifications responsible for the development of amoxicillin resistance (10).
MATERIALS AND METHODS
Bacterial isolates.
Six pneumococcal strains isolated in France, together with strain R6 (a penicillin-susceptible, unencapsulated laboratory strain), were used in the study. Characteristics of these strains are listed in Table 1. Penicillin and amoxicillin MICs were determined according to the specifications of NCCLS (30). The antimicrobial agents were purchased from Sigma Chemical Co., St. Louis, Mo.
TABLE 1.
Characteristics of pneumococcal strains isolated in France
| Isolate | Serotype | MIC (μg/ml)
|
Sourcea | |
|---|---|---|---|---|
| Penicillin | Amoxicillin | |||
| 18062 | 14 | 1 | 1 | Unknown |
| 9481 | 23F | 2 | 1 | CSF |
| 60702 | 6B | 2 | 1 | CSF |
| 73202 | 6B | 2 | 4 | BLD |
| 72521 | 23F | 4 | 8 | BAL |
| 73311 | 23F | 8 | 16 | NP |
BAL, bronchoalveolar lavage fluid; CSF, cerebrospinal fluid; BLD, blood; NP, nasopharyngeal swab.
PCR and DNA sequencing.
Genomic DNAs were extracted from the six isolates and strain R6 by a standard phenol-chloroform method, as described by Ausubel et al. (1). Their pbp2x, pbp2b, and pbp1a genes were amplified by PCR, as described previously (34). The pbp1b, pbp2a, pbp3, and murMN genes from isolate 72521 (penicillin and amoxicillin MICs, 4 and 8 μg/ml, respectively) were also amplified by PCR. The sequences of the PCR primers are listed in Table 2. PCR products were purified by adding 0.6 volume of a 20% polyethylene glycol-2.5 M NaCl solution (32). The DNA strands were sequenced with an ABI Prism Big Dye Terminator Cycle Sequencing kit (Perkin-Elmer Applied Biosystems, Foster City, Calif.), and electrophoresis was performed on a 310 Genetic Analyzer (Perkin-Elmer Applied Biosystems).
TABLE 2.
Primers used for amplification of pbp and murMN genes
| Genea | Primer sequence (5′→3′)b | Product length (kb) |
|---|---|---|
| pbp2x-F | CGTGGGACTATTTATGACCGAAATGG | 2.0 |
| pbp2x-R | AATTCCAGCACTGATGGAAATAAACATATTA | |
| pbp2b-F | GATCCTCTAAATGATTCTCAGGTGG | 1.5 |
| pbp2b-R | CAATTAGCTTAGCAATAGGTGTTGG | |
| pbp1a-F | CGGCATTCGATTTGATTCGCTTCT | 2.4 |
| pbp1a-R | TCGTACTATTATTTGTGCTTGGAGTc | |
| pbp1b-F | TGGCAAGAAAGGTTCAAGTACCAAA | 2.2 |
| pbp1b-R | AATACTAGACCAAGCATTCTGATAA | |
| pbp2a-F | AACAAGTGAACTAGAGGACTCTGA | 2.0 |
| pbp2a-R | CTTAATCTTCGCATCTGAGATAGC | |
| pbp3-F | TTATTTCTACGAAACATGAGTTACC | 1.4 |
| pbp3-R | TTGATTTTGAAGATGAACTTGTATC | |
| murMN-F | TTCAAACGAAAGTAGTAGd | 2.6 |
| murMN-R | GCATGTCTCTCCACCTTTCTAGCd |
F, forward (upstream) primer; R, reverse (downstream) primer.
Primers for amplification of the pbp2x, pbp2b, and pbp1a (with the exception of pbp1a-R) genes were the same as those used by Munoz et al. (29), Dowson et al. (11), and Coffey et al. (6), respectively.
Primer pbp1a-R binds to an area slightly upstream of primer Pn1Adown used by Coffey et al. (6).
Cloning and transformation studies.
The pbp and murMN genes from isolate 72521 were amplified by PCR (with the primers mentioned above) and purified with Geneclean (Bio 101, Inc., La Jolla, Calif.). The entire gene was amplified for pbp1b, pbp2a, and pbp3, whereas 90 to 95% of the gene was amplified for pbp2x, pbp2b, and pbp1a. The amplified products were cloned into the SmaI site of plasmid pGem-3Zf (+) (Promega Corp., Madison, Wis.) and transformed into Escherichia coli JM109 (Promega) according to the instructions of the manufacturer. Penicillin-susceptible pneumococcal strain R6 was used as the recipient in transformation studies. Transformation experiments were carried out by the method described by Smith and Klugman (35). Penicillin and amoxicillin were used for the selection of transformants. Sequencing and subsequent analysis of the pbp genes of the transformants were performed predominantly for sequences within the penicillin-binding domain since this area is known to interact with β-lactam drugs. This region extends 60 amino acid residues upstream of the first conserved amino acid motif to 60 amino acid residues downstream of the last conserved motif, comprising approximately 40% of the entire gene.
DNA fingerprint analysis.
The pbp2x, pbp2b, pbp1a, pbp1b, pbp2a, and pbp3 genes from the six isolates and strain R6 were amplified by PCR and digested with HinfI (pbp2x, pbp2b, pbp2a, pbp1a, pbp1b, pbp3), StyI (pbp1b, pbp2a), MseI plus DdeI (pbp2x), StyI plus DdeI (pbp1a, pbp3), and HaeIII (pbp2b), as described previously (2). All restriction enzymes with the exception of MseI were purchased from Roche Molecular Biochemicals, Mannheim, Germany; MseI was supplied by New England Biolabs, Beverly, Mass. The clonal relatedness of the six isolates was determined by BOX-PCR and pulsed-field gel electrophoresis (PFGE) by previously described methods (26).
Nucleotide sequence accession numbers.
Partial gene sequences for the pbp2x, pbp2b, and pbp 1a genes of isolates 73311, 72521, 73202, 9481, 18062, and 60702 appear in GenBank under the following accession numbers: pbp2x, AF468662 to AF468667, respectively; pbp2b, AF467811 to AF467816, respectively; and pbp1a, AF467817 to AF467822, respectively.
RESULTS AND DISCUSSION
BOX-PCR revealed that isolates 72521, 73311, and 9481 are genotypically indistinguishable. Further analysis by PFGE showed only slight variations between the isolates and showed that isolates 73311 and 9481 are almost identical. Isolates 73202 and 60702 appear to be closely related by BOX-PCR, yet PFGE yielded distinctly different profiles for the two isolates. Isolate 18062 exhibited profiles different from those of the other five isolates. Previous work by Doit et al. (10) also revealed close homology between isolates 72521, 73311, and 9481 (although they used DNA ribotyping and restriction fragment length polymorphism [RFLP] analysis), and it was shown that these three isolates are closely related to the serotype 23F multiresistant Spanish clone (>70% homology). Isolates 73202 and 60702 were shown to be almost identical to each other (>95% homology) and indistinguishable from the serotype 6B Spanish clone.
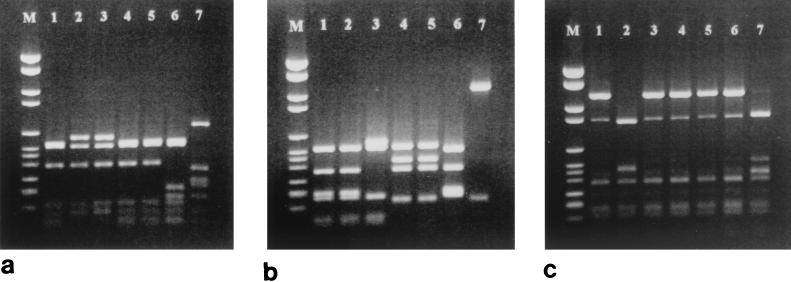
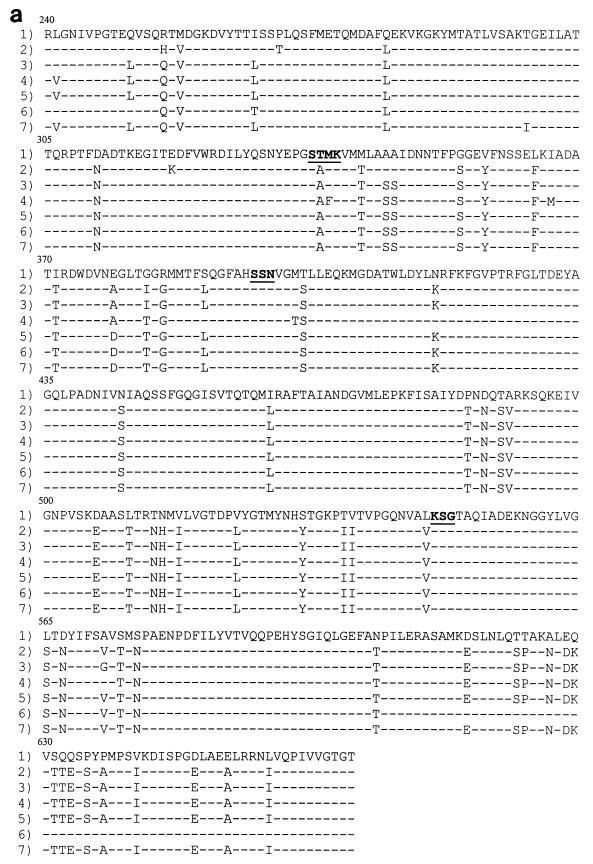
DNA fingerprinting analysis revealed three different gene profiles for pbp2x, with isolates 73202 and 73311 and isolates 18062, 9481, and 72521 sharing the same patterns (Fig. 1a). DNA sequence analysis of the pbp2x penicillin-binding domain from the six isolates revealed extensive divergence in their sequences compared with that of strain R6 (up to 19% diversity), resulting in up to 59 amino acid substitutions. None of the isolates carried an identical set of mutations, and no mutations were common to the amoxicillin-resistant isolates only (Fig. 2a). The substitutions of Thr338 for Ala (present in all six isolates) and Gln633 for Glu (present in all isolates except isolate 60702) have been documented previously by Mouz et al. (27). They suggested that these two mutations are putative determinants for resistance to β-lactams. Thr338 is located within the Ser337-Thr-Met-Lys active site, and the effect of a change at this position in modulating the affinity of this conserved motif toward β-lactams has been confirmed (28). PBP 2X most likely contributes to low-level amoxicillin resistance, but it is unlikely that it plays a role in the development of high-level resistance in these isolates. Three amino acid mutations, namely, Met339 to Phe (located within the Ser337-Thr-Met-Lys motif), Ile366 to Met, and Met400 to Thr (adjacent to the Ser395-Ser-Asn motif), were unique to isolate 73311. Since this isolate displays increased levels of resistance to cefotaxime (MIC, 8 μg/ml) and since PBP 2X has been shown to be a target for this antibiotic, these alterations may contribute to the development of cefotaxime resistance rather than amoxicillin resistance. Coffey et al. (7) have observed two of these mutations (Met339 to Phe and Met400 to Thr) in two U.S. strains with high-level resistance to cefotaxime (MICs, 8 and 32 μg/ml, respectively).
FIG. 1.
DNA fingerprinting analysis of the pbp2x, pbp2b, and pbp1a genes from S. pneumoniae strains isolated in France. Lane M, molecular size marker VI (Roche); lanes 1 to 7, isolates 72521, 73311, 73202, 18062, 9481, 60702, and R6, respectively. (a) pbp2x digested with HinfI; (b) pbp2b digested with HaeIII; (c) pbp1a digested with StyI plus DdeI
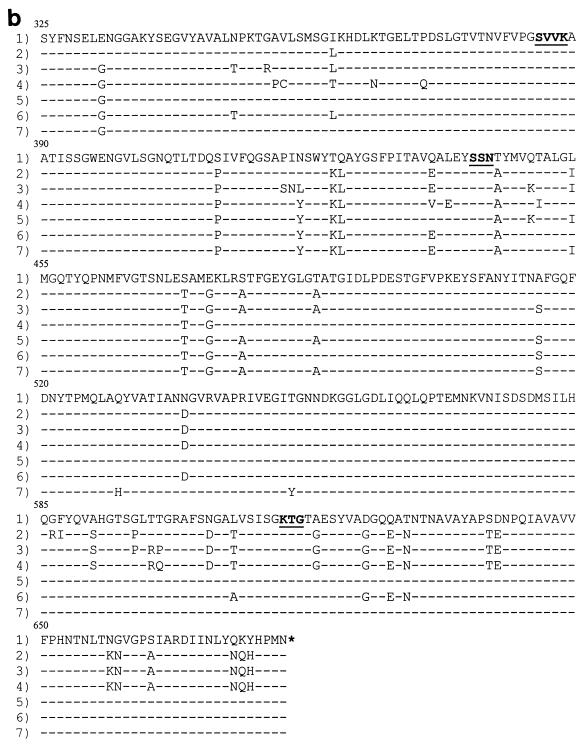
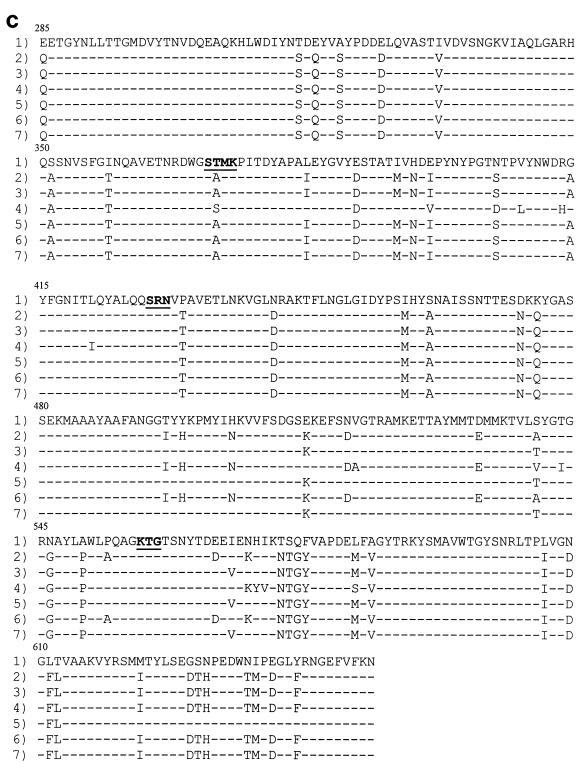
FIG. 2.
Amino acid sequences of the penicillin-binding domain of the PBP 2X (a), PBP 2B (b), and PBP 1A (c) proteins. Row 1, the sequence from penicillin-susceptible strain R6. The conserved amino acid motifs are underlined and set in boldface type. Rows 2 through 7, isolates 73202, 72521, 73311, 18062, 60702, and 9481, respectively. Only those amino acids that differ from the R6 sequence are shown. Amino acids are numbered according to their positions in the gene.
DNA fingerprint analysis of pbp2b revealed four different gene profiles, with isolates 72521 and 73311 and isolates 9481 and 18062 sharing the same gene profiles. The remaining two isolates (isolates 60702 and 73202) exhibited RFLP patterns distinct from those of the other isolates and each other (Fig. 1b). DNA sequence analysis of the pbp2b penicillin-binding domain revealed several nucleotide substitutions (up to 13% nucleotide sequence divergence from strain R6). The least number of amino acid substitutions (n = 13) occurred in isolate 18062 (penicillin and amoxicillin MICs, 1 μg/ml), while isolate 72521 (penicillin and amoxicillin MICs, 4 and 8 μg/ml, respectively) displayed the highest number of substitutions (n = 40) (Fig. 2b). The previously documented substitution of Thr445 for Ala (adjacent to the Ser442-Ser-Asn conserved motif) was noted in all six isolates. Dowson et al. (11) have previously noted the importance of this conserved motif in interacting with penicillin. Piperacillin-resistant laboratory mutants were also found to harbor this substitution (14).
Amoxicillin-resistant isolates 73311, 72521, and 73202 (amoxicillin MICs, ≥4 μg/ml) had 12 amino acid substitutions within the locality of the conserved motif Lys614-Thr-Gly (Fig. 2b). These mutations were absent in isolates 9481, 18062, and 60702 (MICs, 1 μg/ml). Two additional substitutions were found to occur in isolates 72521 and 73311 only (MICs, 8 and 16 μg/ml, respectively), namely, Thr598 to Arg and Thr599 to Gln in isolate 73311 and Pro in isolate 72521. This area has previously been found to be important for the interaction of β-lactams with penicillin-binding proteins (PBPs). Piperacillin-resistant laboratory mutants were found to have a single substitution of either Gly616 to Ala (which is part of the Lys614-Thr-Gly triad) or Gly659 to Asp in their PBP 2B proteins (18). Interestingly, mutations within the same region of PBP 2X were observed in independently selected cefotaxime-resistant laboratory mutants (22). Although the mutations are not identical, the fact that they map within the same region suggests that although PBPs 2X and 2B differ in important aspects of their design, this area of the PBP is crucial for the interaction with β-lactams. Once more, isolate 73311 displayed six mutations not observed in any of the other isolates (Ala353 to Pro, Val354 to Cys, Lys365 to Asn, Pro371 to Gln, Leu439 to Glu, and Thr450 to Ile). The first four mutations occur just prior to the Ser385-Val-Val-Lys motif, while the remaining two occur within the vicinity of the Ser442-Ser-Asn motif. Since PBP 2B is not a target for the extended-spectrum cephalosporins, these mutations may contribute to the high level of amoxicillin resistance found in this isolate.
DNA fingerprint analysis of the pbp1a genes from the strains isolated in France showed that all isolates except isolate 73311 had identical gene profiles (Fig. 1c). DNA sequence analysis of the penicillin-binding domain again revealed an extensive divergence in sequence compared with that of susceptible strain R6, with the number of amino acid substitutions ranging from 37 for isolate 18062 to 52 for isolate 73311 (Fig. 2c). The previously documented mutation of Thr371 to Ser or Ala and the four consecutive substitutions at residues 574 to 577 were present in all six isolates. Smith and Klugman (34) have confirmed the significance of these residues in contributing to the development of high-level penicillin resistance. The substitution of Ala for Thr at residue 371 is homologous to the Thr338-to-Ala substitution in PBP 2X (within the Ser337-Thr-Met-Lys conserved motif).
Seven amino acid substitutions were present only in isolate 73311 (Val408 to Leu, Arg413 to His, Leu421 to Ile, Val518 to Ala, Thr543 to Ile, His571 to Tyr, and Ile572 to Val). These mutations are not present in the pbp1a genes from 11 penicillin-resistant South African isolates (34). However, all substitutions with the exception of Ile572 to Val were found to occur in another South African penicillin-resistant isolate (MIC, 2 μg/ml) harboring an antigenically distinct pbp1a gene with an unusually low affinity for β-lactams (25). The different mutations observed in the pbp1a gene from isolate 73311 (compared to the mutations observed in the other isolates) may contribute to high-level amoxicillin resistance; however, since resistant isolates have been shown to harbor allelic variants of pbp genes (11, 25), it may be that pbp1a from isolate 73311 is just another variant. This divergence of gene sequences among resistant isolates appears to be due to recombination events involving multiple transfer events with DNA from different pneumococci and other streptococcal species.
Fingerprint analysis of the pbp1b, pbp2a, and pbp3 genes revealed that the pbp1b and pbp3 genes of all isolates had identical profiles and the pbp2a genes of all isolates except isolate 73311 had the same profile. The pbp1b, pbp2a, and pbp3 genes from isolate 72521 were sequenced, and few amino acid alterations were observed (one, four, and two substitutions in pbp1b, pbp2a, and pbp3, respectively). These genes did not display the typical mosaic gene patterns frequently observed in the pbp2x, pbp2b, and pbp1a genes of β-lactam-resistant pneumococci. Interestingly, DNA sequence analysis of the pbp2a gene from isolate 73311 revealed 40 nucleotide substitutions (compared to 11 nucleotide substitutions in isolate 72521) in the penicillin-binding domain, resulting in seven amino acid alterations. Six of these were different from the substitutions present in isolate 72521.
Penicillin-susceptible strain R6 was used as the recipient in transformation studies. The results of these experiments are summarized in Table 3. Cloned pbp2x, pbp2b, pbp2a, pbp1b, pbp1a, pbp3, and murMN genes from isolate 72521 were used as the transforming DNA. Strain R6 was transformed with individual as well as different combinations of pbp genes. DNA fingerprinting and sequencing were used to confirm that these genes had been introduced into strain R6. Mutants were selected with both penicillin and amoxicillin, with the drug concentrations ranging from 0.03 to 16 μg/ml. Transformation of strain R6 with the pbp2x gene resulted in transformants that carried an altered pbp2x gene (penicillin and amoxicillin MICs, 0.03 μg/ml). Subsequent transformation of R62x mutants with pbp2b resulted in the selection of transformants with altered pbp2x and pbp2b genes (penicillin and amoxicillin MICs, 0.12 and 0.25 μg/ml, respectively). A third transformation step, in which R62x/2b mutants were transformed with pbp1a, selected for transformants for which penicillin and amoxicillin MICs were 0.5 and 1 μg/ml, respectively. Transformation of the pbp2x and pbp1a genes into strain R6 yielded transformants with integrated pbp2x genes only (penicillin and amoxicillin MICs, 0.03 μg/ml). Transformation of R62x/2b/1a mutants with the pbp1b, pbp2a, and pbp3 genes yielded no transformants. Full resistance, equivalent to that of the donor strain, was achieved when R62x/2b/1a mutants were transformed with genomic DNA, suggesting that a non-PBP-mediated mechanism is responsible for the development of full penicillin and amoxicillin resistance in these isolates.
TABLE 3.
Transformation of pneumococcal strain R6 with DNA from isolate 72521a
| Recipient | Transforming DNA | MIC (μg/ml) for transformants
|
Altered pbp gene(s) in transformants | |
|---|---|---|---|---|
| Penicillin | Amoxicillin | |||
| R6 | pbp2x | 0.03 | 0.03 | pbp2x |
| R6 | pbp2x and pbp1a | 0.03 | 0.03 | pbp2x |
| R672521/2x | pbp2b | 0.12 | 0.25 | pbp2x, pbp2b |
| R672521/2x/2b | pbp1a | 0.5 | 1 | pbp2x, pbp2b, pbp1a |
| R672521/2x/2b/1a | pbp1b, pbp2a, and pbp3 | —b | — | |
| R672521/2x/2b/1a | genomic DNA | 4 | 8 | pbp2x, pbp2b, pbp1a |
| R672521/2x/2b/1a | murMN | — | — | |
| R63191/2x/2b/1a | murMN | 12 | 8 | pbp2x, pbp2b, pbp1a, MurMN |
Penicillin and amoxicillin were used as selective antibiotics.
—, no transformants were selected.
To date, no β-lactamase-mediated resistance has been described in pneumococci. Two non-PBP-mediated resistance mechanisms have been described so far, although these have been described only in laboratory mutants (15, 16). Resistance in these mutants was low level and was due to mutations in CiaH (a protein kinase) and CpoA (a putative glycosyltransferase). Smith and Klugman (35, 36) have recently identified a non-PBP-mediated resistance determinant associated with high-level penicillin and cefotaxime resistance (MICs, 16 and 4 μg/ml, respectively) in a pneumococcal strain originating from Hungary. This determinant has been identified as altered MurM, an enzyme involved in the biosynthesis of branched stem cell wall peptides. Earlier findings by Filipe and Tomasz (12) verify that the murMN operon is critical for the expression of penicillin resistance. DNA sequence analysis of the murMN operon from isolate 72521 revealed an altered murM gene whose sequence diverged from the sequence of the gene from strain R6 by 6.5%, resulting in 20 amino acid substitutions. Alterations were also observed in murN, but to a lesser degree (4% divergence in the nucleotide sequence compared with that of strain R6). Smith and Klugman (36) noted that a common area of alteration (from amino acid residues 42 to 131) could be identified among all R63191/2x/2b/1a/MurMN transformants, suggesting that mutations within this region are important in the development of resistance.
Sequencing of the murMN genes from R672521/2x/2b/1a mutants transformed with genomic DNA showed that the genes were unaltered. Also, transformation of an R672521/2x/2b/1a mutant with murMN yielded no resistant transformants. Interestingly, murMN from isolate 72521 was able to transform mutant R63191/2x/2b/1a to increased levels of penicillin and amoxicillin resistance (MICs, 12 and 8 μg/ml, respectively). These results suggest that the impact of MurMN on the expression of high-level resistance is complex and appears to be dependent on the nature of the pbp gene alterations in the recipient strain. The PBP 2X, 2B, and 1A proteins from the Hungarian isolate allow R6 to express high-level resistance to penicillin and amoxicillin in the presence of MurMN from either the Hungarian isolate (isolate 3191) or the French isolate (isolate 72521). The PBPs of the French isolate do not appear to allow the expression of altered MICs due to MurMN. Full resistance to both penicillin and amoxicillin is, however, caused by some other, as yet unidentified gene that is present in isolate 72521, as transformation with genomic DNA was able to transform R6 to this phenotype. Further studies will be conducted in an attempt to identify this resistance determinant.
The results of transformation studies with various combinations of pbp genes from isolate 72521 proved to be similar when selection was with either penicillin or amoxicillin. The stepwise development of amoxicillin resistance appears to involve altered PBPs 2X, 2B, and 1A, in much the same way that penicillin resistance develops. Transformation of strain R6 with pbp2x only yielded transformants for which the penicillin and amoxicillin MICs were identical (0.03 μg/ml), whereas subsequent transformations with pbp2b yielded transformants for which the amoxicillin MICs were greater by 1 dilution. Interestingly, analysis of the DNA sequence of pbp2b showed that isolates 73311, 72521, and 73202 (amoxicillin MICs, ≥4 μg/ml) had additional amino acid substitutions clustered around the region of the Lys614-Thr-Gly motif. These mutations were not present in the pbp2b gene of Hungarian strain 3191 with high-level penicillin resistance (penicillin and amoxicillin MICs, 16 and 8 μg/ml, respectively). These results indicate that PBP 2B may be instrumental in triggering the expression of the unusual phenotype of higher amoxicillin MICs in relation to the penicillin MICs.
Previous studies have suggested that PBP 2B may play a significant role in contributing to the development of amoxicillin resistance. Doit et al. (10) examined the gene profiles of the pbp2x, pbp2b, and pbp1a genes from 29 clinical isolates for which the amoxicillin MICs were ≥4 μg/ml. For comparison, 11 isolates with various levels of susceptibility to amoxicillin (MICs, <4 μg/ml) were also studied. Great heterogeneity was observed in the pbp2x and pbp1a RFLP patterns; however, for pbp2b, those isolates for which the amoxicillin MICs were ≥4 μg/ml had RFLP patterns distinct from those for isolates for which the MICs were ≤2 μg/ml. Ikeda et al. (19) compared the levels of binding of five structurally diverse β-lactams (amoxicillin, cefaclor, cefotiam, cefpodoxime, and cefdinir) to the PBPs of two penicillin-resistant clinical isolates of S. pneumoniae (penicillin and amoxicillin MICs, 0.5 and 1 μg/ml and 0.25 and 0.5 μg/ml, respectively) and a susceptible strain. A common feature of the PBP patterns of these two resistant strains was the absence of PBP 1A, which was detected in the susceptible strain. Furthermore, PBP 2B (from the resistant strains) was the least sensitive to β-lactam antibiotics, showing a strong affinity only for amoxicillin.
Previous work has shown that resistance to β-lactam antibiotics in S. pneumoniae is mediated by successive alterations in essential PBPs, with specific mutations in PBPs only being expressed in combination with mutations in other genes. The final resistance phenotype would therefore be dependent on the collective actions of multiple PBPs. According to NCCLS guidelines, a pneumococcal isolate that is susceptible to penicillin can be considered susceptible to other β-lactams; however, penicillin-susceptible clinical isolates with increased levels of resistance to other β-lactams have been reported. Coffey et al. (7) reported the occurrence of a high-level cefotaxime-resistant clinical isolate (MIC, 32 μg/ml, respectively) that exhibited only intermediate resistance to penicillin (MIC, 0.25 μg/ml). Smith et al. (37) recently reported on the isolation of two clinical isolates that had high-level resistance to cefotaxime and ceftriaxone (MICs, 4 μg/ml) yet that were susceptible to penicillin (MICs, 0.04 μg/ml). Analysis of the DNA sequences of the pbp genes revealed alterations in pbp2x and pbp1a, whereas the pbp2b gene remained unaltered. This would explain the penicillin susceptibilities of these strains since penicillin resistance is dependent on alterations in PBP 2B.
The results presented here suggest that amoxicillin resistance is mediated by alterations in multiple PBPs in conjunction with an as yet unidentified non-PBP-mediated determinant. Furthermore, the amoxicillin-resistant isolates display an unusual phenotype in that the MICs of amoxicillin are greater than those of penicillin. In particular, PBP 2B appears to play a significant role in mediating the expression of this phenotype. Two distinct groups of amino acid substitutions were observed in the pbp2b genes from the strains isolated in France. The first group occurred within the region of the Ser442-Ser-Asn triad, and these mutations were observed in all six isolates. Mutations occurring within this area, in particular, the substitution of Ala for Thr445, have been shown to mediate the interaction of PBPs with β-lactams and may contribute to the development of low-level amoxicillin resistance. The second group of mutations was clustered around the Lys614-Thr-Gly triad. Twelve of these were unique to those isolates for which the amoxicillin MICs were ≥4 μg/ml (isolates 73311, 72521, and 73202). Two additional mutations were unique to isolates 72521 and 73311 only (MICs, 8 and 16 μg/ml, respectively), suggesting that they may contribute to the development of high-level amoxicillin resistance. Further analysis of these mutations is required to confirm their participation in the development of resistance and their contribution to this unusual phenotype. This may only be possible once the identification of the non-PBP resistance determinant has been established.
Acknowledgments
This work was supported by grants from the Medical Research Council, National Health Laboratory Service, and the University of the Witwatersrand.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Short protocols in molecular biology: a compendium of methods, p. 55-57. In Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Beall, B., R. R. Facklam, D. M. Jackson, and H. H. Starling. 1998. Rapid screening for penicillin susceptibility of systemic pneumococcal isolates by restriction enzyme profiling of the pbp2B gene. J. Clin. Microbiol. 36:2359-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Block, S. L., C. J. Harrison, J. A. Hedrick, R. D. Tyler, R. A. Smith, E. Keegan, and S. A. Chartrand. 1995. Penicillin-resistant Streptococcus pneumoniae in acute otitis media: risk factors, susceptibility patterns and antimicrobial management. Pediatr. Infect. Dis. J. 14:751-759. [DOI] [PubMed] [Google Scholar]
- 4.Brueggemann, A. B., M. A. Pfaller, and V. Doern. 2001. Use of penicillin MICs to predict in vitro activity of other β-lactam antimicrobial agents against Streptococcus pneumoniae. J. Clin. Microbiol. 39:367-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler, D. L., R. C. Gagnon, L. A. Miller, J. A. Poupard, D. Felmingham, and R. N. Gruneberg. 1999. Differences between the activity of penicillin, amoxycillin and co-amoxclav against 5,252 Streptococcus pneumoniae isolates tested in the Alexander Project, 1992-1996. J. Antimicrob. Chemother. 43:777-782. [DOI] [PubMed] [Google Scholar]
- 6.Coffey, T. J., C. G. Dowson, M. Daniels, J. Zhou, C. Martin, B. G. Spratt, and J. M. Musser. 1991. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol. Microbiol. 5:2255-2260. [DOI] [PubMed] [Google Scholar]
- 7.Coffey, T. J., M. Daniels, L. M. McDougal, C. G. Dowson, F. C. Tenover, and B. G. Spratt. 1995. Genetic analysis of clinical isolates of Streptococcus pneumoniae with high-level resistance to expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 39:1306-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig, W. A., and D. Andes. 1996. Pharmakokinetics and pharmacodynamics of antibiotics in otitis media. Pediatr. Infect. Dis. J. 15:255-259. [DOI] [PubMed] [Google Scholar]
- 9.del Castillo, F., F. Bacquero-Artigao, and A. Garcia-Perea. 1998. Influence of recent antibiotic therapy on antimicrobial resistance of Streptococcus pneumoniae in children with acute otitis media in Spain. Pediatr. Infect. Dis. J. 17:94-97. [DOI] [PubMed] [Google Scholar]
- 10.Doit, C., C. Loukil, F. Fitoussi, P. Geslin, and E. Bingen. 1999. Emergence in France of multiple clones of clinical Streptococcus pneumoniae isolates with high-level resistance to amoxicillin. Antimicrob. Agents Chemother. 43:1480-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowson, C. G., A. Hutchinson, and B. G. Spratt. 1989. Extensive re-modeling of the transpeptidase domain of penicillin-binding protein 2B of a penicillin-resistant South African isolate of Streptococcus pneumoniae. Mol. Microbiol. 3:95-102. [DOI] [PubMed] [Google Scholar]
- 12.Filipe, S. R., and A. Tomasz. 2000. Inhibition of the expression of penicillin resistance in Streptococcus pneumoniae by inactivation of cell wall muropeptide branching genes. Proc. Natl. Acad. Sci. USA 97:4891-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giebink, G. S. 1984. Epidemiology and natural history of otitis media, p. 5-9. In D. Lim, C. Bluestone, J. Klein, and J. Nelson (ed.), Recent advances in otitis media with effusion. B. C. Decker Inc., Philadelphia, Pa.
- 14.Grebe, T., and R. Hakenbeck. 1996. Penicillin-binding proteins 2b and 2x from Streptococcus pneumoniae are primary resistance determinants for different classes of β-lactam antibiotics. Antimicrob. Agents Chemother. 40:829-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grebe, T., J. Paik, and R. Hakenbeck. 1997. A novel resistance mechanism against β-lactams in Streptococcus pneumoniae involves CpoA, a putative glycosyltransferase. J. Bacteriol. 179:3342-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guenzi, E., A. M. Gasc, M. A. Sicard, and R. Hakenbeck. 1994. A two component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol. 12:505-515. [DOI] [PubMed] [Google Scholar]
- 17.Guillemot, D., P. Maison, C. Carbon, B. Balkau, Vauzelle-Kervroëdan, C. Sermet, G. Bouvenot, and E. Eschwège. 1998. Trends in antimicrobial drug use in the community—France, 1981-1992. J. Infect. Dis. 177:492-497. [DOI] [PubMed] [Google Scholar]
- 18.Hakenbeck, R., C. Martin, C. Dowson, and T. Grebe. 1994. Penicillin-binding protein 2b of Streptococcus pneumoniae in piperacillin-resistant laboratory mutants. J. Bacteriol. 176:5574-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda, F., Y. Yokota, A. Ikemoto, N. Teratani, K. Shunomura, and H. Kanno. 1995. Interaction of beta-lactam antibiotics with the penicillin-binding proteins of penicillin-resistant Streptococcus pneumoniae. Chemotherapy 41:159-164. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs, M. R. 1996. Increasing importance of antibiotic-resistant Streptococcus pneumoniae in acute otitis media. Pediatr. Infect. Dis. J. 15:940-943. [DOI] [PubMed] [Google Scholar]
- 21.Klimek, J. J., C. Nightingale, W. B. Lehmann, and R. Quintiliani. 1977. Comparison of concentrations of amoxicillin and ampicillin in serum and middle ear fluid of children with chronic otitis media. J. Infect. Dis. 135:999-1002. [DOI] [PubMed] [Google Scholar]
- 22.Laible, G., and R. Hakenbeck. 1991. Five independent combinations of mutations can result in low-affinity penicillin-binding protein 2x of Streptococcus pneumoniae. J. Bacteriol. 173:6986-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leach, A. J. 1999. Otitis media in Australian aboriginal children: an overview. Int. J. Pediatr. Otorhinolaryngol. 49(Suppl. 1):S173-S178. [DOI] [PubMed] [Google Scholar]
- 24.Leach, A. J., J. B. Boswell, V. Asche, T. G. Nienhuys, and J. D. Mathews. 1994. Bacterial colonization of the nasopharynx predicts very early onset and persistence of otitis media in Australian aboriginal infants. Pediatr. Infect. Dis. J. 13:983-989. [DOI] [PubMed] [Google Scholar]
- 25.Martin, C., C. Sibold, and R. Hakenbeck. 1992. Relatedness of penicillin-binding protein 1a genes from different clones of penicillin-resistant Streptococcus pneumoniae isolated in South Africa and Spain. EMBO J. 11:3831-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefévre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the Pneumococcal Molecular Epidemiology Network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mouz, N., A.-M. Di Guilmi, E. Gordon, R. Hakenbeck, O. Dideberg, and T. Vernet. 1999. Mutations in the active site of penicillin-binding protein PBP2x from Streptococcus pneumoniae. J. Biol. Chem. 274:19175-19180. [DOI] [PubMed] [Google Scholar]
- 28.Mouz, N., E. Gordon, A.-M. Di Guilmi, I. Petit, Y. Petillot, Y. Dupont, R. Hakenbeck, T. Vernet, and O. Dideberg. 1998. Identification of a structural determinant for resistance to β-lactam antibiotics in gram-positive bacteria. Proc. Natl. Acad. Sci. USA 95:13403-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munoz, R., T. J. Coffey, M. Daniels, C. G. Dowson, G. Laible, J. Casal, R. Hakenbeck, M. Jacobs, J. M. Musser, B. G. Spratt, and A. Tomasz. 1991. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J. Infect. Dis. 164:302-306. [DOI] [PubMed] [Google Scholar]
- 30.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5, 5th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 31.Neu, H. C. 1979. Diagnosis and treatment: drugs five years later. Amoxicillin. Ann. Intern. Med. 90:356-360. [DOI] [PubMed] [Google Scholar]
- 32.Paithankar, K. R., and K. S. N. Prasad. 1991. Precipitation of DNA by polyethylene glycol and ethanol. Nucleic Acids Res. 19:1346.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pankuch, G. A., M. R. Jacobs, and P. C. Appelbaum. 1995. Comparative activity of ampicillin, amoxycillin, amoxycillin/clavulanate and cefotaxime against 189 penicillin-susceptible and -resistant pneumococci. J. Antimicrob. Chemother. 35:883-888. [DOI] [PubMed] [Google Scholar]
- 34.Smith, A. M., and K. P. Klugman. 1998. Alterations in PBP 1A essential for high-level penicillin resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:1329-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, A. M., and K. P. Klugman. 2000. Non-penicillin-binding protein mediated high-level penicillin and cephalosporin resistance in a Hungarian clone of Streptococcus pneumoniae. Microb. Drug Resist. 6:105-110. [DOI] [PubMed] [Google Scholar]
- 36.Smith, A. M., and K. P. Klugman. 2001. Alterations in MurM, a cell wall muropeptide branching enzyme, increase high-level penicillin and cephalosporin resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:2393-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, A. M., R. F. Botha, H. J. Koornhof, and K. P. Klugman. 2001. Emergence of a pneumococcal clone with cephalosporin resistance and penicillin susceptibility. Antimicrob. Agents Chemother. 45:2648-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sutherland, R., E. A. P. Croydon, and G. N. Rolinson. 1972. Amoxycillin: a new semisynthetic penicillin. Br. Med. J. 3:13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teele, D. W., J. O. Klein, and B. Rosner. 1989. Epidemiology of otitis media during the first seven years of life in children in greater Boston: a prospective, cohort study. J. Infect. Dis. 160:83-94. [DOI] [PubMed] [Google Scholar]
- 40.Thornsberry, C., P. Ogilvy, J. Kahn, J. Mauriz, and the Laboratory Investigator Group. 1997. Surveillance of antimicrobial resistance in Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in the United States in 1996-1997 respiratory season. Diagn. Microbiol. Infect. Dis. 29:249-257. [DOI] [PubMed] [Google Scholar]