Abstract
Escherichia coli strains bearing single-copy fusions between the lacZ reporter gene and the cspA, ibp, or P3rpoH stress promoters offer a simple means to detect sublethal concentrations of antibacterial agents interfering with prokaryotic translation or cell envelope integrity while simultaneously providing information on the mechanism of action of the test compound (A. A. Bianchi and F. Baneyx, Appl. Environ. Microbiol. 65:5023-5027, 1999). Here, we expand the usefulness of this system by (i) demonstrating that a fusion between the SOS-inducible sulA promoter and lacZ is a highly specific probe for the detection of antimicrobial agents that ultimately interfere with DNA replication, (ii) showing that inactivation of the tolC gene allows efficient detection of very low concentrations of model antibiotics (including aminoglycosides) whereas polymyxin B-mediated outer membrane permeabilization facilitates the identification of intermediate concentrations of hydrophobic compounds, and (iii) validating the potential of detector strains and sensitization strategies for high-throughput screening using a reproducible and internally consistent 96-well microplate assay.
The emergence of antimicrobial resistance by once-susceptible pathogens is rapidly becoming a major concern in human medicine. Although the cessation of abusive practices such as the inclusion of antibiotics in animal feed (7) and their overprescription for human diseases (14) may mitigate the problem, a need for new antimicrobial agents is likely since antibacterial resistance appears to have minor fitness costs and is slowly lost once acquired (3, 11). Furthermore, certain microorganisms such as the opportunistic pathogen Pseudomonas aeruginosa possess high intrinsic resistance to the arsenal of antibacterial agents currently in use owing to a rather impermeable outer membrane and the presence of multiple multidrug efflux pumps (18). Although the availability of nearly 30 prokaryotic genomes may allow the identification of common molecular targets and, possibly, the development of wide-spectrum antimicrobials (16), the rational, one-target route may actually limit the discovery of antibacterial compounds. Indeed, despite evidence of renewed activity by pharmaceutical companies, only one antibiotic active against a novel target class has been approved by the U.S. Food and Drug Administration in over 35 years (33).
A common approach to antimicrobial compound discovery is to screen natural products, and more recently combinatorial or biodiversity libraries, for molecules that inhibit bacterial cell growth. In traditional incarnations, these screens require high concentrations of test compounds and long incubation times, even if growth inhibition is measured by sensitive techniques (e.g., radioactivity assays). This complicates their adaptation to high-throughput platforms in which the molar concentration of candidate molecules must remain low due to multiplex format and compound precipitation at high concentration. In addition, unless specifically designed to do so (for examples, see references 8, 17, and 37), growth inhibition assays do not provide information on the molecular target of the lead compounds which, if available, greatly accelerates medicinal chemistry modifications for improved efficacy and reduced toxicity.
Recently, a simple system for the rapid detection and classification of antimicrobial agents interfering with prokaryotic translation or cell envelope integrity that does not suffer from the above shortcomings was described (2). The current system consists of three isogenic strains bearing single-copy gene fusions between the lacZ reporter gene and the promoter regions of the major cold shock protein CspA (10), the highly inducible cytoplasmic small heat shock proteins IbpA and IbpB (4), and the P3 promoter of the rpoH gene which is transcribed by σE-bound RNA polymerase upon protein misfolding in the periplasm (21). The cspA::lacZ fusion is induced by the so-called C-group translational inhibitors (e.g., chloramphenicol and tetracycline), which trigger the cold shock response and leave ribosomes with an occupied A site, but not by H-group antibiotics targeting translation (e.g., streptomycin and neomycin), which activate the cytoplasmic heat shock response and leave ribosomes with a vacant A site (2, 39). Strains containing the ibp::lacZ fusion exhibit the opposite pattern of induction when exposed to the same antibiotics. Finally, compounds that damage the outer membrane (e.g., polymyxin B) or interfere with peptidoglycan synthesis (e.g., carbenicillin) selectively activate the P3rpoH promoter. For unclear reasons, high concentrations of polymyxin B also stimulate lacZ transcription from the ibp promoter (2). An important feature of this screen is that growth inhibition—and therefore high concentrations of antimicrobial agents—is not required for promoter activation (2).
In this work, we have expanded on the above concept and the universe of molecular targets by showing that isogenic lysogens bearing a fusion between the SOS-inducible sulA promoter and lacZ are suitable for the sensitive and highly selective detection of antimicrobial compounds affecting DNA replication. We further demonstrate that inactivation of TolC, the outer membrane channel of the AcrAB and EmrAB multidrug efflux systems (22, 43) and a dominant player in multidrug resistance (36), allows high signal-to-background detection of very low concentrations of model antibiotics, while addition of the outer membrane permeabilizer polymyxin B sulfate bolsters the sensitivity of the system to intermediate concentrations of hydrophobic antimicrobial compounds. Finally, we show that stress promoter-based detection of antimicrobial agents and sensitization strategies can be reliably scaled down to microplate format.
MATERIALS AND METHODS
Strain constructions.
The Escherichia coli strains used in this study are listed in Table 1. AB734, a wild-type E. coli K-12 strain which contains a lacZ mutation but lacks antibiotic resistance markers (6), was obtained from the E. coli Genetic Stock Center. Strains GJ1922 (30) and LBB1175 (9) were the sources of λφ(sulA::lacZ) and tolC::Tn10, respectively. AB734 derivatives were constructed by P1 transduction or lambda infection using standard protocols (20, 34). The presence of the tolC::Tn10 mutation was confirmed by determining the MIC of chloramphenicol as follows. Cells from overnight cultures grown in Luria-Bertani (LB) medium were sedimented by centrifugation and resuspended in an equal volume of 10 mM MgSO4. Aliquots containing 5 × 104 cells (as determined by colony counting) were used to inoculate 1 ml of LB supplemented with serial dilutions of chloramphenicol. Cultures were incubated for 24 h at 37°C before the absorbance at 600 nm (A600) was determined. The concentration of antibiotic giving an A600 value less than 0.01 was identified as the MIC. MICs for other antibiotics were determined in a similar fashion. All experiments were carried out at least in duplicate.
TABLE 1.
E. coli strains used in this study
| Strain | Genotype | Construction detailsa or reference |
|---|---|---|
| GJ1922 | thr leu arg his ΔlexA300::(Smr Spr) sulA λφ(sulA::lacZ) | 33 |
| LBB1175 | F−lac ara mal xyl mtl gal rpsL tolC::Tn10 | 8 |
| AB734 | F−lac-6(del) | 6 |
| ES100 | AB734 tolC::Tn10 | P1(LBB1175) × AB734 → Tetr; MIC |
| ADA110 | AB734 λφ(ibp::lacZ) | 2 |
| ADA120 | AB734 λφ(ibp::lacZ) tolC::Tn10 | λ(ADA110) × ES100 → Lac+; Tetr; MIC |
| ADA310 | AB734 λφ(cspA::lacZ) | 2 |
| ADA320 | AB734 λφ(cspA::lacZ) tolC::Tn10 | λ(ADA310) × ES100 → Lac+; Tetr; MIC |
| ADA410 | AB734 λφ(P3rpoH::lacZ) | 2 |
| ADA420 | AB734 λφ(P3rpoH::lacZ) tolC::Tn10 | λ(ADA410) × ES100 → Lac+; Tetr; MIC |
| ADA510 | AB734 λφ(sulA::lacZ) | P1(GJ1922) × AB734 → Lac+ |
| ADA520 | AB734 λφ(sulA::lacZ) tolC::Tn10 | P1(GJ1922) × ES100 → Lac+; Tetr; MIC |
P1 transductions are represented as P1 (donor) × recipient → phenotypes used for selection. Bacteriophage λ infections are represented as λ(donor) × recipient → phenotypes used for selection. Tetr, tetracycline resistance; Lac+, formation of blue colonies on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates; MIC, minimum inhibitory concentration for chloramphenicol of 2 μg/ml.
Culture and induction conditions.
Shake flasks (500 ml) containing 100 ml of LB were inoculated at a 1:50 dilution by using overnight cultures, and cells were grown at 30°C [for λφ(ibp::lacZ), λφ (P3rpoH::lacZ), and λφ(sulA::lacZ) derivatives] or 37°C [for λφ(cspA::lacZ) derivative]. When A600 reached approximately 0.4, 25-ml aliquots were transferred to preheated 125-ml shake flasks and the cultures were treated with the indicated concentrations of antibiotics. Polymyxin B sulfate was added at a final concentration of 0.5 μg/ml (tolC+ strains) or 0.3 μg/ml (tolC mutants) for the experiments illustrated in Fig. 3 and 4. Control cultures received either no additive or an equal volume of 100% ethanol for experiments involving chloramphenicol or tetracycline induction. Addition of ethanol alone did not cause induction or growth inhibition at any of the concentrations used. All antibiotics were purchased from Sigma. Stock solutions of chloramphenicol and tetracycline (5 mg/ml) were prepared in 100% ethanol. Streptomycin sulfate (5 mg/ml), neomycin sulfate (5 mg/ml), carbenicillin (5 mg/ml), polymyxin B sulfate (5 mg/ml), nalidixic acid (15 mg/ml), ofloxacin (100 mg/ml), ethidium bromide (15 mg/ml), and novobiocin (10 mg/ml) were dissolved in deionized H2O. All experiments were performed at least in triplicate.
FIG. 3.
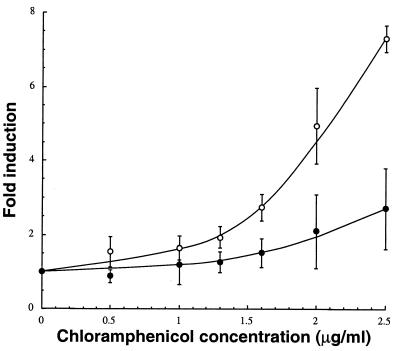
Polymyxin B-mediated outer membrane permeabilization improves the induction of the cspA::lacZ fusion at intermediate chloramphenicol concentrations. ADA310 cultures were grown to mid-exponential phase in LB medium at 37°C. Chloramphenicol was added at the indicated concentrations along with 0.5 μg of polymyxin B sulfate per ml (○) or no additive (•). Clarified extracts were assayed for β-galactosidase activity 3 h after treatment. Error bars correspond to triplicate experiments.
FIG. 4.
Individual and combined effects of polymyxin B supplementation and tolC inactivation. AB734 derivatives harboring the indicated fusions and containing (+) or lacking (−) an intact tolC gene were grown to mid-exponential phase in LB medium at either 30°C [λφ (ibp::lacZ) and λφ(sulA::lacZ) lysogens] or 37°C [λφ(cspA::lacZ) lysogens]. Cultures were treated with 1 μg of chloramphenicol per ml, 4 μg of streptomycin per ml, or 5 μg of nalidixic acid per ml together with no additive (−) or 0.5 μg of polymyxin B per ml for tolC+ cells or 0.3 μg of polymyxin B per ml for tolC strains (+). Clarified extracts were assayed for β-galactosidase activity 1 h [λφ(ibp::lacZ) lysogens] or 3 h [λφ(sulA::lacZ) and λφ(cspA::lacZ) lysogens] after treatment. Error bars correspond to at least triplicate experiments.
β-Galactosidase assays.
Culture samples (2 ml) were harvested immediately before culture division and at the indicated time points. The A600 was recorded, and cells were sedimented by centrifugation at 7,000 × g for 10 min. The pellet was resuspended in 2 ml of 50 mM potassium phosphate monobasic (pH 6.5), and cells were lysed with a French press at 10,000 lb/in2. Lysates were clarified by centrifugation at 10,000 × g for 10 min, and supernatants were assayed in duplicate for β-galactosidase activity by using o-nitrophenyl-β-d-galactopyranoside (ONPG) according to the method of Miller (19). β-Galactosidase specific activities are reported in Miller units (1,000 × ΔA420/A600 of culture per milliliter of culture per minute of reaction).
Microplate assays.
Lysogens were grown to mid-exponential phase (A600 ≈ 0.4) in LB medium at either 30°C (ADA110 and ADA520) or 37°C (ADA310). Culture aliquots (90 μl) were inoculated into the wells of sterile 96-well microtiter plates, and 10 μl of appropriately diluted antibiotic stock was added to give the indicated final concentration. ADA310 cells were further supplemented with 0.5 μg of polymyxin B per ml. Plates were incubated at 30 or 37°C with shaking for 1 h (ADA110) or 2 h (ADA310 and ADA520). The A600 was measured in a thermostated Molecular Dynamics VersaMax microplate reader, and 25 μl of B-PER II bacterial protein extraction reagent (Pierce, Rockford, Ill.) was rapidly added to the wells by using a multichannel pipetter. The plate was agitated for 15 s before addition of 50 μl of ZOB buffer (1) prepared by mixing Z buffer (74 mM NaH2PO4, 126 mM Na2HPO4, 2 mM MgSO4, 0.4 mM MnSO4·H2O, 399 mg of hexadecyltrimethylammonium bromide per liter, 199.5 mg of sodium deoxycholate per liter, 174 mM β-mercaptoethanol) with 8 mg of ONPG per ml in T-base [15.1 mM (NH4)2SO4, 80 mM K2HPO4, 44 mM KH2PO4, and 1 g of Na3C6H5O7·2H2O per liter] at a 4:1 ratio. The A420 was read immediately after ZOB buffer addition and after 5 min (ADA520), 10 min (ADA310), or 30 min (ADA110) of incubation at room temperature. β-Galactosidase specific activities are reported in units (U) calculated using the following formula: (final A420 − initial A420)/A600.
RESULTS AND DISCUSSION
sulA::lacZ is a highly specific probe for the detection of antimicrobial compounds interfering with DNA replication.
Although AB734 derivatives carrying single-copy lacZ fusions to the cspA, ibp, and P3rpoH promoters allow selective detection of sub-MIC concentrations of antibiotics interfering with prokaryotic translation and cell envelope integrity (2), they are not induced by molecules interfering with DNA replication. This precludes the detection of an important class of antibacterial agents. A hallmark of the exposure of E. coli to conditions or agents that lead to DNA damage is the induction of the SOS response (42). Because SOS induction occurs primarily at the transcriptional level, fusions between the recA, uvrA, sulA, dinD, or cda promoters and the Vibrio fischeri luxCDABE operon or the E. coli lacZ gene have been used to screen restriction endonuclease mutants (12) and for the detection of bioantimutagens (35), UV irradiation, and genotoxins (5, 25, 27, 28, 41). Here, we selected the SOS-inducible promoter of the cell division inhibitor SulA since a λ-borne sulA::lacZ fusion is available (30) and because sulA is the most tightly repressed and highly inducible SOS gene characterized to date (32). By contrast, RecA is present at as many as 7,000 copies per cell (31) and the uvrA promoter experiences only a four- to fivefold induction following DNA damage (32).
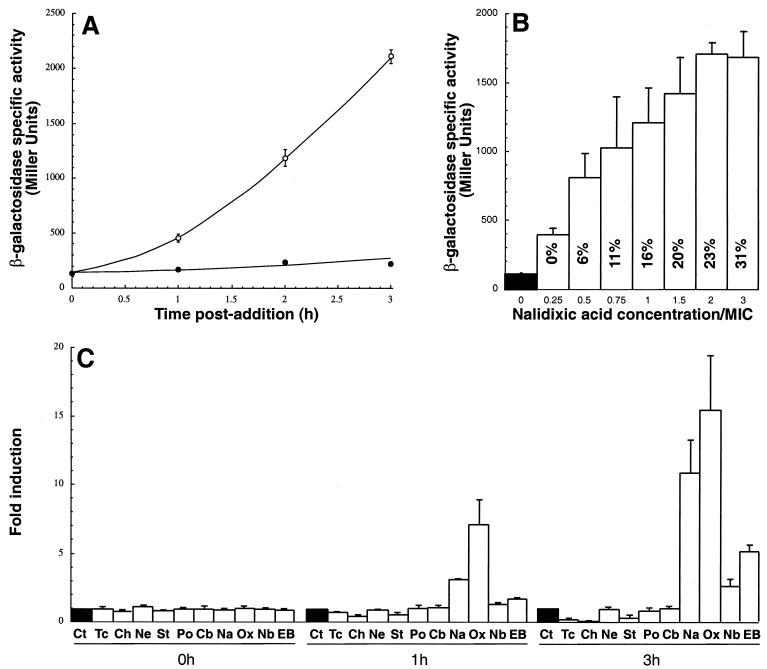
The sulA::lacZ fusion was moved from GJ1922 to AB734 by P1 transduction, yielding ADA510 (Table 1). The basal levels of β-galactosidase specific activity in cultures grown in LB medium at 30°C and sampled for up to 3 h after mid-exponential phase (A600 ≈ 0.4) remained at a constant value of about 150 U (Fig. 1A), indicating that lacZ transcription from the sulA promoter is well repressed under our experimental conditions. As expected, addition of 15-μg/ml concentration of the SOS-inducing agent nalidixic acid to mid-exponential-phase cells led to a progressive increase in β-galactosidase specific activity, and about 10-fold more enzyme was present in treated cultures than in the control after 3 h of incubation at 30°C (Fig. 1A). This time point was selected for all subsequent shake flask experiments.
FIG. 1.
Induction characteristics and specificity of λφ(sulA::lacZ) lysogens. (A) Time course of induction. Mid-exponential-phase cultures of ADA510 growing in LB medium at 30°C were supplemented with 15 μg of nalidixic acid per ml (○) or no additive (•). Samples were withdrawn at the indicated time points, and clarified lysates were assayed for β-galactosidase activity. (B) Effects of nalidixic acid concentration on growth and induction ratios. ADA510 cultures grown as above were supplemented with the indicated concentrations of nalidixic acid, and β-galactosidase activities were determined 3 h postaddition. Values inside bars correspond to the percentages of growth inhibition relative to control cultures at the time of sample collection. (C) Specificity of induction. ADA510 cultures grown as above were treated with no additive (Ct), 5 μg of tetracycline (Tc) per ml, 5 μg of chloramphenicol (Ch) per ml, 16 μg of neomycin (Ne) per ml, 8 μg of streptomycin (St) per ml, 1 μg of polymyxin B sulfate (Po) per ml, 8 μg of carbenicillin (Cb) per ml, 15 μg of nalidixic acid (Na) per ml, 0.2 μg of ofloxacin (Ox) per ml, 100 μg of novobiocin per ml, or 50 μg of ethidium bromide (EB) per ml. Clarified extracts were assayed for β-galactosidase activities immediately before and 1 and 3 h after addition. Typical percentages of growth inhibition after 3 h of incubation with the above concentrations of antibacterial agents were 60% for tetracycline, 30% for chloramphenicol, 15% for neomycin, 25% for streptomycin, 20% for polymyxin B, 15% for carbenicillin, 15% for nalidixic acid, 45% for ofloxacin, 25% for novobiocin, and 20% for ethidium bromide. Error bars correspond to triplicate experiments.
The sensitivity of the system was assessed by exposing λφ(sulA::lacZ) lysogens to various concentrations of nalidixic acid. Figure 1B shows that addition of 1.25 μg of nalidixic acid per ml (0.25 times the MIC; Table 2) was sufficient to cause an about fourfold increase in β-galactosidase specific activity relative to untreated cultures, and that maximum signal-to-background ratio was obtained with a 10-μg/ml concentration (twice the MIC) of the quinolone. At 15 μg/ml, nalidixic acid did not further increase induction ratios although it consistently caused higher growth inhibition (Fig. 1B).
TABLE 2.
Susceptibility of wild-type and tolC cells to various antibacterial agents
| Compound | MIC (μg/ml)
|
|
|---|---|---|
| AB734 (tolC+) | ES100 (tolC) | |
| Nalidixic acid | 5.0 | 1.0 |
| Ofloxacin | 0.3 | 0.2 |
| Novobiocin | 85 | 1.0 |
| Ethidium bromide | 500 | 220 |
| Chloramphenicol | 6.0 | 1.75 |
| Tetracycline | 1.0 | NDa |
| Streptomycin | 6.0 | 6.0 |
| Neomycin | 10 | 10 |
| Carbenicillin | 12 | 6.0 |
| Polymyxin B | 0.2 | 0.2 |
ND, not determined, owing to the fact that the tolC mutation is linked to Tn10, which encodes tetracycline resistance.
To evaluate the specificity of induction, ADA510 cultures were supplemented with model antibacterial agents targeting the ribosome (chloramphenicol, tetracycline, neomycin, and streptomycin) or affecting outer membrane integrity (polymyxin), peptidoglycan synthesis (carbenicillin), or DNA gyrase and/or DNA replication (nalidixic acid, ofloxacin, novobiocin, and ethidium bromide). Concentrations causing comparable degrees of growth inhibition were used for these experiments. Figure 1C shows that, among the panel of compounds tested, only SOS-inducing agents activated the sulA::lacZ fusion. Ofloxacin, a fluoroquinolone that is more hydrophilic than nalidixic acid and diffuses more efficiently across the membranes (26), led to the highest level of induction. At the concentrations used, the DNA-intercalating agent ethidium bromide and the coumarin glycoside antibiotic novobiocin caused significant induction (5- and 2.5-fold, respectively) only at the 3 h time point. Two C-group antibiotics (tetracycline and chloramphenicol) and one H-group antibiotic (streptomycin) downregulated β-galactosidase synthesis from the sulA promoter (Fig. 1C). A similar effect was previously observed when λφ(P3rpoH::lacZ) cells were exposed to chloramphenicol or streptomycin (2). While the mechanisms responsible for downregulation remain unclear, it is interesting that another H-group antibiotic (neomycin) had no influence on the levels of enzymatic activity, despite the fact that it led to a degree of growth inhibition comparable to that of streptomycin (20 to 25%) and that the two aminoglycosides have similar MICs for wild-type cells (Table 2). Finally, the levels of β-galactosidase specific activity were unchanged in cultures treated with the cell envelope-active compounds carbenicillin and polymyxin. Overall, these results indicate that λφ(sulA::lacZ) lysogens respond with high signal-to-background ratios and specificity to antibacterial agents that directly or indirectly interfere with DNA replication.
Inactivation of TolC allows efficient detection of low concentrations of antibacterial agents.
Although the concentrations of antibacterial agents needed to activate the stress promoters-lacZ fusions are far lower than those required in growth inhibition assays, a threshold amount must still be added to the cultures to achieve full induction (Fig. 1B) (2). This could prevent the detection of library compounds present at very low concentrations or having poor antibacterial activity. The recently crystallized TolC protein (13) acts as an outer membrane channel for the export of a variety of antimicrobial agents by the AcrAB and EmrAB efflux systems (15). Due to their inability to detoxify the cell, tolC mutants are hypersensitive to dyes, detergents, and lipophilic antibiotics (43). This phenotype has been exploited to enhance the sensitivity of whole-cell biosensors to pollutants (40) and genotoxic compounds (5).
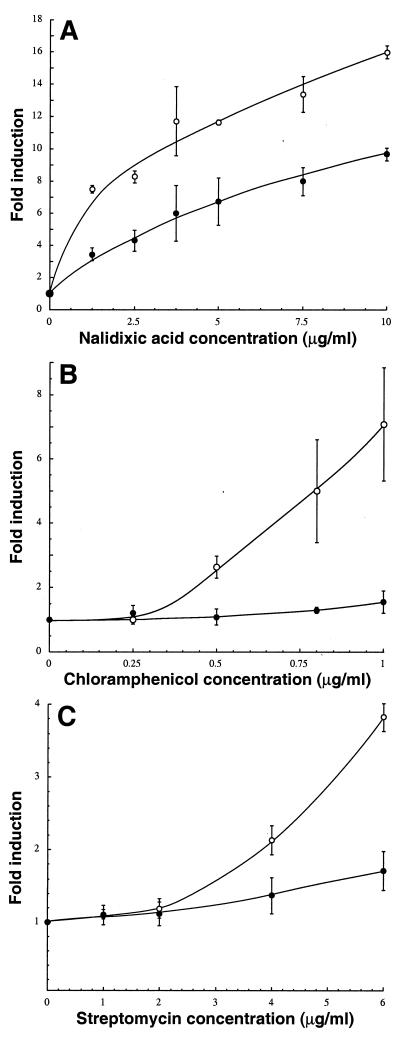
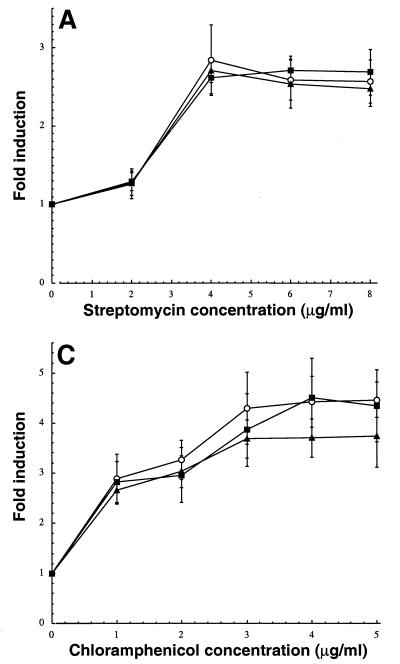
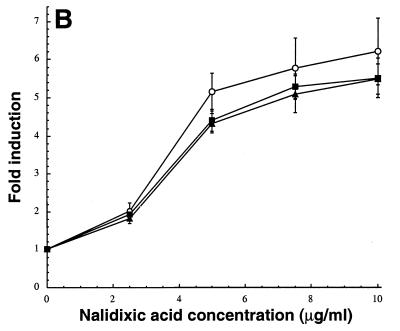
In an effort to achieve reliable detection of low concentrations of antimicrobial agents, we constructed an isogenic set of λφ(ibp::lacZ), λφ(cspA::lacZ), λφ(P3rpoH::lacZ), and λφ(sulA::lacZ) lysogens lacking TolC. All mutants were more susceptible to chloramphenicol and other antibacterial compounds exported by the AcrAB-TolC system (Table 2). In the case of λφ(sulA::lacZ) lysogens, the tolC mutation conferred an about twofold increase in the magnitude of induction ratios over a broad range of nalidixic acid concentrations (Fig. 2A) and it was possible to detect the quinolone with a sevenfold signal-to-background ratio at concentrations as low as 1.25 μg/ml. The increase in sensitivity brought about by tolC inactivation was even more pronounced in the case of λφ(cspA::lacZ) lysogens (Fig. 2B). Whereas chloramphenicol did not activate the cspA promoter at a concentration below 1 μg/ml in tolC+ bacteria, appreciable induction was observed in the 0.5- to 1-μg/ml range with the mutant strain. λφ(ibp::lacZ) lysogens exhibited a similar pattern: streptomycin could be readily detected at 4- to 6-μg/ml in tolC mutants, while the same concentrations had little inducing effect in isogenic tolC+ cells (Fig. 2C). Induction ratios were also improved 30 to 40% in the cases of neomycin (at 6 μg/ml) and kanamycin (at 9 μg/ml). The latter set of results was surprising, since aminoglycosides do not appear to be natural substrates of the AcrAB-TolC or EmrAB-TolC efflux pumps (22). Rather, they are preferentially exported by AcrD, an AcrB homolog belonging to the resistance nodulation division family, which has no known membrane fusion protein or outer membrane channel partners (29). Based on the observation that tolC null mutants do not exhibit increased sensitivity to aminoglycosides in MIC tests (Table 2) (29, 36), it has been concluded that AcrD does not use the TolC outer membrane factor. However, our observation that induction of the ibp::lacZ fusion by streptomycin is enhanced in tolC mutants (a test much more sensitive than MIC assays) suggests that TolC is directly or indirectly implicated in aminoglycosides efflux.
FIG. 2.
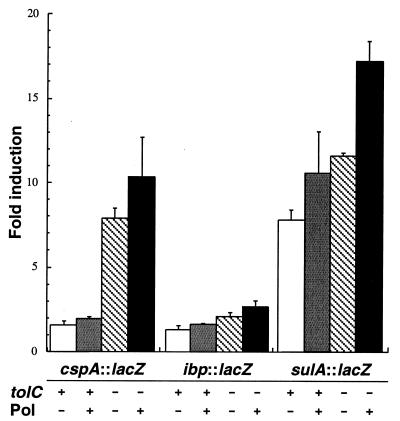
Influence of the tolC::Tn10 allele on the induction of λφ(sulA::lacZ) (A), λφ(cspA::lacZ) (B), and λφ(ibp::lacZ) (C) lysogens by model antibacterial agents. Wild-type (tolC+) strains (•) and their isogenic tolC derivatives (○) were grown to mid-exponential phase in LB medium at either 30°C (panels A and C) or 37°C (panel B). Cultures were supplemented with the indicated concentrations of antimicrobial compounds, and clarified extracts were assayed for β-galactosidase activity 1 h (panel C) or 3 h (panels A and B) after treatment. Error bars correspond to triplicate experiments.
Finally, although AcrAB-TolC is known to be involved in the detoxification of β-lactams (22), we observed only a slight (≈30%) improvement in enzymatic activity when a tolC derivative of the λφ(P3rpoH::lacZ) lysogen was exposed to 0.8 μg of carbenicillin per ml and the mutant strain was very sensitive to carbenicillin above 1 μg/ml. This result underscores the fact that, although tolC strains are very useful for the detection of low concentrations of antibacterial agents, they will be more rapidly killed by active or concentrated compounds that may be present in certain libraries (22).
Outer membrane permeabilization increases sensitivity to intermediate concentrations of hydrophobic antibacterial agents.
An alternate approach to improve detection sensitivity is to destabilize the outer membrane to facilitate the access of candidate compounds to their cellular targets. In this study, we made use of the cationic lipopeptide polymyxin B as a permeabilizer since it has been reported to increase the susceptibility of E. coli to a range of antimicrobial compounds (38) and does not induce the ibp, sulA, or cspA stress promoters at low concentrations (Fig. 1C) (2). Addition of 0.5 μg of polymyxin B per ml to λφ(cspA::lacZ) lysogens significantly enhanced chloramphenicol induction ratios at intermediate concentrations (1.5 to 2.5 μg/ml) but not below 1.5 μg/ml (Fig. 3 and 4). This suggests that the AcrAB-TolC system and the MdfA (Cmr) H+ antiporter, which are both involved in chloramphenicol efflux (23, 24), become unable to efficiently detoxify the cell once a threshold intracellular concentration (probably in the 1.5-μg/ml range) has been exceeded. Polymyxin supplementation was also quite effective in the case of nalidixic acid, a hydrophobic molecule that does not readily gain access to the cytoplasm compared to fluoroquinolones (26). In fact, under our experimental conditions, the contribution of enhanced passive diffusion to detection sensitivity was comparable to inactivation of TolC-dependent active efflux (Fig. 4). On the other hand, polymyxin B treatment had little beneficial effect on the induction of the ibp::lacZ fusion by 4 or 6 μg of streptomycin per ml (Fig. 4; data not shown). This result was not unexpected, since small hydrophilic antibiotics should efficiently penetrate the cell via porin channels. Finally, we did not observe any improvement in the induction of the P3rpoH::lacZ fusion following combined addition of 0.5 μg of polymyxin B per ml and 0.8 μg of carbenicillin per ml (data not shown).
Possible additive or synergistic effects were assessed by treating tolC lysogens with polymyxin B at the time of antibiotic addition. In these experiments, the polymyxin concentration was reduced to 0.3 μg/ml, which remains sufficient to cause efficient outer membrane permeabilization (38) but does not significantly increase growth inhibition for the compounds and concentrations tested. Figure 4 shows that combining genetic and chemical approaches had the most beneficial effect when λφ(sulA::lacZ) lysogens were challenged with 5 μg of nalidixic acid per ml, with an additive improvement in induction ratios. Addition of polymyxin also improved the response of tolC λφ(cspA::lacZ) lysogens to 1 μg of chloramphenicol per ml and slightly increased the enzymatic activity in ADA120 cells treated with 4 μg of streptomycin per ml. From a practical standpoint, the above data indicate that polymyxin B-mediated outer membrane permeabilization expands the usefulness of the system by allowing the detection of hydrophobic compounds whose concentrations are too low to efficiently activate stress responses in wild-type cells but high enough to be toxic to tolC mutants.
Adapting the screen to a microplate format.
To be of practical value in the identification of novel antimicrobials from large libraries, the performance of the detector strains should remain satisfactory in microplate format and sensitization strategies should be amenable to scale-down. Alksne and coworkers recently described a 96-well microplate assay for the identification of compounds interfering with E. coli secretion using a detector strain bearing a chromosomal secA::lacZ fusion (1). We conducted initial experiments using their protocol. However, we failed to achieve efficient and reliable signal detection, presumably because cell lysis was incomplete (data not shown). A single step modification, i.e., addition of the B-PER II bacterial protein extraction reagent prior to colorimetric detection, corrected this problem. The reproducibility of the microplate assay is illustrated in Fig. 5A, which shows induction ratios for λφ(ibp::lacZ) lysogens exposed to increasing concentrations of streptomycin in three independent experiments. Internal consistency was assessed by averaging enzymatic activities across one microplate column (eight wells) and calculating standard deviations. The main contributor to the error was edge effects. We also confirmed the performance of tolC strains in microplate format by using ADA520 cells and nalidixic acid challenge (Fig. 5B) and the usefulness of polymyxin B supplementation by using λφ(cspA::lacZ) lysogens and chloramphenicol induction (Fig. 5C). The fact that both assay and sensitization strategies are amenable to microplate format suggests that this approach may hold promise for high-throughput identification of novel antibacterial agents from large libraries.
FIG. 5.
Induction in microplates. (A) Mid-exponential cultures of ADA110 grown in LB medium at 30°C were aliquoted in a 96-well microplate, treated with the indicated concentrations of streptomycin, and assayed for β-galactosidase activity after 1 h of incubation at 30°C as described in Materials and Methods. (B) ADA520 cultures grown as above were supplemented with the indicated concentrations of nalidixic acid and assayed for β-galactosidase activity after 2 h of incubation at 30°C. (C) Mid-exponential cultures of ADA310 grown in LB medium at 37°C were aliquoted in a 96-well microplate, treated with the indicated concentrations of chloramphenicol together with 0.5 μg of polymyxin B per ml, and assayed for β-galactosidase activity after 2 h of incubation at 37°C. Each symbol represents a separate microplate experiment. Error bars correspond to the standard deviations of induction ratios across one microplate column (eight wells). Note that activities were measured 2 h after induction rather than the usual 3 h in the cases of λφ(sulA::lacZ) and λφ(cspA::lacZ) lysogens. While the additional hour of incubation does not significantly affect the levels of enzymatic activity for strains bearing the cspA::lacZ fusion (2), it leads to lower induction ratios in the case of ADA510 cells (Fig. 1A).
Acknowledgments
We thank Joe Fralick and J. Gowrishankar for generously providing bacterial strains.
E.S. gratefully acknowledges NSF for a graduate fellowship. This work was supported by NSF award BES-9707729 and Research Project Grant MBC-99-335-01 from the American Cancer Society.
REFERENCES
- 1.Alksne, L. E., P. Burgio, W. Hu, B. Feld, M. P. Singh, M. Tuckman, P. J. Petersen, P. Labthavikul, M. McGlynn, L. Barbieri, L. McDonald, P. Bradford, R. G. Dushin, D. Rothstein, and S. J. Projan. 2000. Identification and analysis of bacterial protein secretion inhibitors utilizing a SecA-LacZ reporter fusion system. Antimicrob. Agents Chemother. 44:1418-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bianchi, A. A., and F. Baneyx. 1999. Stress responses as a tool to detect and characterize the mode of action of antibacterial agents. Appl. Environ. Microbiol. 65:5023-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Björkman, J., I. Nagaev, O. G. Berg, D. Hughes, and D. I. Andersson. 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287:1479-1482. [DOI] [PubMed] [Google Scholar]
- 4.Chuang, S. E., V. Burland, G. Plunkett III, D. L. Daniels, and F. R. Blattner. 1993. Sequence analysis of four new heat-shock genes constituting the hslUV/ibpAB and hslVU operons in Escherichia coli. Gene 134:1-6. [DOI] [PubMed] [Google Scholar]
- 5.Davidov, Y., R. Rozen, D. R. Smulski, T. K. Van Dyk, A. C. Vollmer, D. A. Elsemore, R. A. LaRossa, and S. Belkin. 2000. Improved bacterial SOS promoter::lux fusions for genotoxicity detection. Mutat. Res. 466:97-107. [DOI] [PubMed] [Google Scholar]
- 6.DeWitt, S. K., and E. A. Adelberg. 1962. Transduction of the attached sex factor of Escherichia coli. J. Bacteriol. 83:673-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferber, D. 1998. New hunt for roots of resistance. Science 280:27.. [DOI] [PubMed] [Google Scholar]
- 8.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, G. C. Kedar, P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Z. Y. Zhu, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387-1400. [DOI] [PubMed] [Google Scholar]
- 9.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein, J., N. S. Pollitt, and M. Inouye. 1990. Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87:283-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heineman, J. A. 1999. How antibiotics cause antibiotic resistance. Drug Discov. Today 4:72-79. [DOI] [PubMed] [Google Scholar]
- 12.Heitman, J., and P. Model. 1991. SOS induction as an in vivo assay of enzyme-DNA interactions. Gene 103:1-9. [DOI] [PubMed] [Google Scholar]
- 13.Koronakis, V., A. Scharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 14.Kunin, C. M. 1985. The responsibility of the infectious disease community for the optimal use of antimicrobial agents. J. Infect. Dis. 151:388-398. [DOI] [PubMed] [Google Scholar]
- 15.Lewis, K. 2000. Translocases: a bacterial tunnel for drugs and proteins. Curr. Biol. 10:R678-R681. [DOI] [PubMed] [Google Scholar]
- 16.Loferer, H., A. Jacobi, A. Posch, C. Gauss, S. Meier-Ewert, and B. Seizinger. 2000. Integrated bacterial genomics for the discovery of novel antimicrobials. Drug Discov. Today 5:107-114. [DOI] [PubMed] [Google Scholar]
- 17.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandell, G. L., J. E. Bennett, and R. Dolin. 2000. Principles and practice of infectious diseases, 5th ed. Churchill Livingstone, Philadelphia, Pa.
- 19.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Miller, J. H. 1993. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Missiakas, D., and S. Raina. 1998. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 28:1059-1066. [DOI] [PubMed] [Google Scholar]
- 22.Nikaido, H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nilsen, I. W., I. Bakke, A. Vader, Ø. Olsvik, and M. R. El-Gewely. 1996. Isolation of cmr, a novel Escherichia coli chloramphenicol resistance gene encoding a putative efflux pump. J. Bacteriol. 178:3188-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orser, C. S., F. C. F. Foong, S. R. Capaldi, J. Nalezny, W. MacKay, and S. B. Farr. 1995. Use of prokaryotic stress promoters as indicators of the mechanisms of chemical toxicity. In Vitro Toxicol. 8:71-85. [Google Scholar]
- 26.Piddock, L. J. V., Y.-F. Jin, V. Ricci, and A. E. Asuquo. 1999. Quinolone accumulation by Pseudomonas aeruginosa, Staphylococcus aureus and Escherichia coli. J. Antimicrob. Chemother. 43:61-70. [DOI] [PubMed] [Google Scholar]
- 27.Ptitsyn, L. R., G. Horneck, O. Komova, S. Kozubek, E. A. Kravasin, M. Bonev, and P. Rettberg. 1997. A biosensor for environmental genotoxin screening based on an SOS lux assay in recombinant Escherichia coli cells. Appl. Environ. Microbiol. 63:4377-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosen, R., Y. Davidof, R. A. LaRossa, and S. Belkin. 2000. Microbial sensors of ultraviolet radiation based on recA′::lux fusions. Appl. Biochem. Biotechnol. 89:151-160. [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg, E. Y., D. Ma, and H. Nikaido. 2000. AcrD of Escherichia coli is an aminoglycoside efflux pump. J. Bacteriol. 182:1754-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.SaiSree, L., M. Reddy, and J. Gowrishankar. 2000. lon incompatibility associated with mutations causing SOS induction: null uvrD alleles induce an SOS response in Escherichia coli. J. Bacteriol. 182:3151-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sassanfar, M., and J. W. Roberts. 1990. Nature of the SOS-inducing signal in Escherichia coli: the involvement of DNA replication. J. Mol. Biol. 212:79-96. [DOI] [PubMed] [Google Scholar]
- 32.Schnarr, M., P. Oertel-Buchheit, M. Kazmaier, and M. Granger-Schnarr. 1991. DNA binding properties of the LexA repressor. Biochimie 73:423-431. [DOI] [PubMed] [Google Scholar]
- 33.Senior, K. 2000. FDA approves first drug in new class of antibiotics. Lancet 355:1523.. [DOI] [PubMed] [Google Scholar]
- 34.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Simic, D., B. Vukovic-Gacic, and J. Knezevic-Vukcevic. 1998. Detection of natural bioantimutagens and their mechanisms of action with bacterial assay system. Mutat. Res. 402:51-57. [DOI] [PubMed] [Google Scholar]
- 36.Sulavik, M. C., C. Houseweart, C. Cramer, N. Jiwani, N. Murgolo, J. Greene, B. DiDomenico, K. J. Shaw, G. H. Miller, R. Hare, and G. Shimer. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45:1126-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trias, J., and Z. Yuan. 1999. Mining bacterial cell wall biosynthesis with new tools: multitarget screens. Drug Resist. Updat. 2:358-362. [DOI] [PubMed] [Google Scholar]
- 38.Vaara, M. 1992. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 56:395-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Bogelen, R. A., and F. C. Neidhardt. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:5589-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Dyk, T. K., W. R. Majarian, K. B. Konstantinov, R. M. Young, P. S. Dhurjati, and R. A. LaRossa. 1994. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminescence gene fusions. Appl. Environ. Microbiol. 60:1414-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vollmer, A. C., S. Belkin, D. R. Smulski, T. K. Van Dyk, and R. A. LaRossa. 1997. Detection of DNA damage by use of Escherichia coli carrying recA′::lux, uvrA′::lux, or alkA′::lux reporter plasmids. Appl. Environ. Microbiol. 63:2566-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker, G. C. 1996. The SOS response of Escherichia coli, p. 1400-1416. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 43.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]