Abstract
Caspofungin inhibits the synthesis of 1, 3-β-d-glucan, an essential cell wall target in fungi. Genetic studies in the model yeast Saccharomyces cerevisiae have shown that mutations in FKS1 and FKS2 genes result in caspofungin resistance. However, direct demonstration of the role of gene overexpression in caspofungin resistance has been lacking. We transformed wild-type S. cerevisiae with an S. cerevisiae URA3-based GAL1 cDNA library and selected transformants in glucose synthetic complete plates lacking uracil (glucose SC minus uracil plates). We then moved the transformants to galactose SC minus uracil plates containing caspofungin (1 μg/ml) and looked for caspofungin-resistant colonies. We retested the candidates (true positives were sensitive on glucose caspofungin and resistant on galactose caspofungin media, respectively). We identified 16 caspofungin-resistant candidates. Restriction analysis and hybridization confirmed that 15 of the 16 clones were identical. We sequenced one of the cDNA clones and found that it contained the cDNA for SBE2. SBE2 has been described in S. cerevisiae to encode a Golgi protein involved in the transport of cell wall components (B. Santos and M. Snyder, Mol. Biol. Cell, 11:435-452, 2000). The SBE2 cDNA plasmid conferred again galactose-dependent caspofungin resistance when transformed back into the wild-type S. cerevisiae. Finally, the SBE2 deletion mutant was hypersensitive to caspofungin. In conclusion, overexpression of Sbe2p under the regulated control of the GAL1 promoter results in caspofungin resistance in S. cerevisiae. This transport pathway may provide insight into the tolerance or lack of sensitivity to caspofungin of some pathogenic fungi.
Caspofungin (CAS) is a novel echinocandin lipopeptide that selectively inhibits 1,3-β-d-glucan synthase, a fungus-specific enzyme that is critical for fungal cell wall biosynthesis (7, 12, 17). In vitro studies indicate that CAS has a broad-spectrum fungicidal activity against Candida species, including azole-resistant Candida albicans (7). Against Aspergillus species, CAS displays a complex pattern of growth inhibition in vitro that results in death of actively growing hyphal tips (13, 14; C. Douglas, J. Bowman, G. Abruzzo, A. Flattery, G. Gill, L. Kong, C. Leighton, J. Smith, V. Pikounis, K. Bartizal, M. Kurtz, and H. Rosen, Abstr. 40th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1683, 2000). CAS is effective in vivo against Candida species and Aspergillus fumigatus in mouse models of disseminated candidiasis and aspergillosis (7, 12, 13). CAS is currently approved for the treatment of refractory aspergillosis, and clinical testing is ongoing in patients with candidiasis. However, as in all classes of antifungals, there is a potential for the emergence of resistance to CAS following its expected extensive use in the future. A better understanding of the molecular responses of pathogenic fungi to CAS could enable physicians to make more-effective use of this promising, nontoxic antifungal agent.
Relatively little is known about the molecular mechanisms of CAS resistance in fungi. In the model yeast Saccharomyces cerevisiae, genetic studies have shown that glucan synthase is composed of at least two subunits: a putative catalytic subunit encoded by two related genes, FKS1 and FKS2, and a regulatory subunit, a GTP-bound protein encoded by RHO1 (12). Mutations in FKS1, FKS2, or RHO1 genes have been associated with altered susceptibility to glucan synthase inhibitors (12). However, a direct demonstration of the role of gene overexpression in CAS resistance has been lacking.
To that end, we looked for S. cerevisiae genes that would confer resistance to CAS when overexpressed. As a screening strategy, we used the regulated system of GAL1 cDNA overexpression in S. cerevisiae (11). More specifically, the GAL1 promoter is repressed when S. cerevisiae utilizes glucose as a carbon source and is derepressed, leading to overexpression, when growth is shifted to galactose as a sole carbon source (11). We identified that SBE2, a novel gene involved in cell wall formation, results in CAS resistance when overexpressed in S. cerevisiae.
(This work was presented in part at the 41st Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, Ill., 16 to 19 December 2001.)
MATERIALS AND METHODS
Strains.
Standard methods were used for making yeast growth media, for yeast manipulation, and for the molecular biology techniques (2, 8). The S. cerevisiae strain 10560-14C (MATa ura3-52 leu2::hisG his3::hisG) (10) was used for transformation. Dropout mix lacking uracil (adenine hemisulfate salt, l-arginine [HCl], l-aspartic acid, l-glutamic acid monosodium salt, l-histidine, l-leucine, l-lysine mono-HCl, l-methionine, l-phenylalanine, l-serine, l-threonine, l-tryptophan, l-tyrosine, l-valine [8]) was added to glucose yeast nutrient broth and galactose yeast nutrient broth media (synthetic complete medium lacking uracil [SC minus uracil medium]). CAS was obtained from Merck Research Laboratories (Rahway, N.J.), itraconazole was obtained from Janssen Pharmaceutical (Titusville, N.J.), and fluconazole was obtained from Pfizer (New York, N.Y.). Amphotericin B, cycloheximide, trifluoroperazine, and tunicamycin were purchased from Sigma Chemicals (St. Louis, Mo.).
Yeast transformation and screening for CAS-resistant colonies.
The S. cerevisiae strain 10560-14C was transformed with a URA3-based cDNA library under the control of the GAL1 promoter cloned into the centromeric plasmid pRS 316 (15), and Ura+ transformants were selected in glucose SC minus uracil plates. The transformants were then pooled and spread (concentration, approximately 100 colonies/plate) to galactose SC minus uracil CAS (1 μg/ml) plates. CAS-resistant colonies were identified after 48 h of incubation at 30°C. Previous pilot experiments determined that the 10560-14C strain transformed by the URA3 centromeric plasmid pRS 316 (control) fails to grow on galactose SC minus uracil CAS (1 μg/ml) plates. Resistant candidates were retested by streaking them on to glucose SC minus uracil CAS (1 μg/ml) plates and galactose SC minus uracil CAS (1 μg/ml) plates. True CAS-resistant colonies were plasmid mediated (CAS sensitive and CAS resistant on glucose and galactose medium, respectively).
CAS sensitivity testing.
Drug sensitivity tests were performed in the Ura+ transformants of the 10560-14C strain and in the Y270, Y1942, Y1943, and Y1944 strains (kindly provided by M Snyder, Yale University [18]). Four different methods were performed to measure sensitivity to CAS. Three independent experiments, each performed in triplicate at different time points, were performed per each susceptibility assay. For the first assay, the growth of each yeast strain streaked out to form single colonies was examined (after an incubation for 48 h at 30°C) on galactose SC minus uracil and glucose SC minus uracil agar plates containing various concentrations of CAS. Second, we used a disk diffusion assay, in which yeast growth was examined by plating approximately 105 yeast cells in late logarithmic growth phase on galactose SC minus uracil and glucose SC minus uracil agar plates, respectively. CAS (or the other inhibitors) was placed on a 0.25-in.-diameter paper disk (Schleicher and Schuell, Keene, N.H.) in a final volume of 5 μl. The radius of the zone of growth inhibition was measured after 48 h of growth at 30°C. Third, we measured the MIC of CAS for the Ura+ transformant CAS-resistant candidate strains recovered from our screen by using the broth microdilution method (NCCLS document M27-A [16]). In order to maintain the selection and to evaluate the relationship between CAS resistance and the carbon source (glucose versus galactose), the selective liquid media glucose SC minus uracil and galactose SC minus uracil were used, instead of the standard RPMI 1640. Both the control and the SBE2 cDNA-containing strains (each taken from a single colony grown on glucose SC minus uracil plates) were pregrown in both glucose SC minus uracil and galactose SC minus uracil liquid media. A standardized suspension of yeast cells (1 × 106 to 5 × 106) was inoculated onto glucose SC minus uracil CAS (concentrations from 0.06 to 32 μg/ml) and galactose SC minus uracil CAS (concentrations from 0.06 to 32 μg/ml) in 96-well microtiter plates. The MIC was determined after 48 h of growth at 30°C. Fourth, we assayed the growth kinetics of the control and the SBE2cDNA-containing strain, each taken from a single colony grown on glucose SC minus uracil plates and pregrown (overnight incubation at 30°C with constant shaking) in both glucose SC minus uracil and galactose SC minus uracil liquid media. Cells were diluted to a starting inoculum of 3 × 106 (optical density [OD] = 0.01) and inoculated onto 25 ml of liquid glucose SC minus uracil with or without CAS (concentration of 1.0 μg/ml) and 25 ml of liquid galactose SC minus uracil with or without CAS (concentration of 1.0 μg/ml). Cultures were grown at 30°C with constant shaking, and the OD was measured at 2-, 4-, 6-, 8-, 10-, and 24-h intervals.
Microscopy. (i) Differential interference contrast (DIC) microscopy.
Yeast cells were grown for 48 h on galactose SC minus uracil and glucose SC minus uracil agar plates in the presence or absence of CAS (0.25 μg/ml). Images were collected using a Nikon Microphot-SA microscope and a Hamamatsu model C2400 camera and transferred to Adobe Photoshop (version 4.0). Several fields containing 500 to 1,000 cells were examined and photographed.
(ii) Electron microscopy.
Yeast cells were grown for 48 h on galactose SC minus uracil and glucose SC minus uracil agar plates in the presence or absence of CAS (0.5 μg/ml). Preparation of samples for transmission electron microscopy was carried out as previously described (9). Yeast cells were fixed by the addition of glutaraldehyde (Electron Microscopy Sciences, Fort Washington, Pa.) to growth medium to a final concentration of 5%. After incubation for 3 h at room temperature, cells were concentrated by centrifugation at 10,000 × g for 10 min at room temperature and then washed two times with 0.9% NaCl. Samples were suspended in 4% KMnO4 in 0.1 M Na-cacodylate, pH 7.4 (Electron Microscopy Sciences) and incubated at 4°C for 1 h. After two washes with 0.9% NaCl, samples were suspended in 2% uranyl acetate and incubated for 1 h at room temperature. Samples were washed three times, dehydrated through a graded series of ethanol solutions, infiltrated with propylene oxide for 10 min, and then embedded in Epon-812 (Tousimis Research Co., Rockville, Md.). Ultra thin sections were stained for 5 min with 1% lead citrate before viewing with a transmission electron microscope model JEM-1010 (JEOL USA, Peabody, Mass.). Several fields containing 50 to 100 cells were examined, and representative portions were recorded. The cells exhibiting visible defects such as irregularities and aberrations in the electron-dense outer layer of the cell wall or thickening or deformation of the lighter middle layer of the cell wall were scored as having abnormal changes in cell wall. Approximately 500 cells were viewed per result point.
In vitro glucan synthase activity.
Crude membrane fractions were prepared according to previously described methods (5). Briefly, standardized inocula (106 cells) of yeast taken from a single colony were grown in 250 ml of liquid galactose SC minus uracil and glucose SC minus uracil media at 30°C until cultures reached an OD of 1.5. The cells were collected by centrifugation at 5,000 × g for 5 min at 4°C. The supernatant was decanted, and cells were washed twice with 25 ml of breakage buffer (0.1 M HEPES [pH 7], 1 mM EDTA, and 1 mM dithiothreitol). Twenty-five grams of acid-washed glass beads (diameter, 0.2 μm; Sigma) were added, and the cell suspension was shaken vigorously for 3 to 5 min until 80% breakage was achieved. Cellular debris was then spun down at 5,000 × g for 5 min. The supernatant from the low-speed spin was spun at 50,000 × g for 1 h at 4°C to obtain a crude membrane pellet. The crude membrane pellet was suspended in 2.5 ml of breakage buffer plus 25% glycerol. Using a homogenizer (no. 77272 [2 ml]; Pyrex, Birmingham, England), membranes were homogenized using 10 strokes. Protein concentration was determined using the Bradford assay. A concentration of 1 to 10 mg/ml was typically obtained. The membranes were then frozen in 100-μl aliquots in liquid nitrogen and stored at −80°C. The whole procedure was conducted at 4°C in a cold room. To measure 1,3-β-d-glucan synthase activity, we modified the procedure previously reported by Douglas et al. (5). We used 80 μl of reaction mixture and 25 μg (20 μl) of membrane per reaction. The reaction mixture consisted of 0.1 M Tris-HCl (pH 7), 1 mM EDTA, 60 mM NaCl, 25 mM NaF, 0.4 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 0.2% (wt/vol) bovine serum albumin, 40 U of α-amylase, 3 μM GTP-γ-S, UPD-d-[6-3H]glucose (80,000 dpm/nmol), and 0.4 mM UPD-glucose. We exposed the membranes to six different concentrations of CAS: 0.0, 0.0001, 0.001, 0.01, 0.1, and 1 μg/ml. The reactions were performed at room temperature for 1 h, during which the reaction was linear. The reactions were agitated every 15 min. After 1 h, each reaction was terminated with the addition of 80 μl of ice-cold 20% trichloroacetic acid. The reaction was then filtered through glass fiber filters (#31 grade; Schleicher and Schuell), washed with distilled water and dried for 1 h. Each filter was then placed in a scintillation tube with 4 ml of aquasol-2 (universal LSC cocktail Dupont). The radioactivity was determined using an LS 6500 scintillation counter (Beckman Coulter, Fullerton, Calif.). Enzyme activity was determined in triplicate for three independent assays. Specific activity was expressed as nanomoles of radiolabeled product per hour per mg of protein.
Statistical analysis.
Data comprised three independent experiments with three replicates within each experiment. All comparisons were considered statistically significant for P values of 0.05 or less. No adjustments were made for multiple comparisons. Statistical analyses were made using the SAS software system.
RESULTS
Identification of a novel gene that when overexpressed gives resistance to CAS.
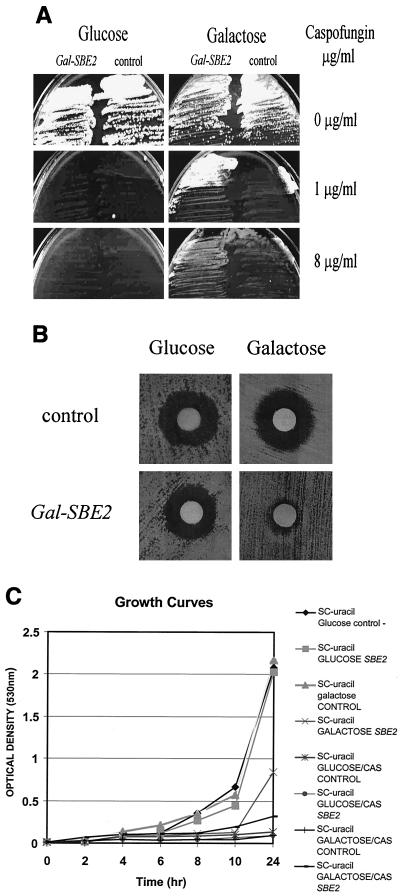
A total of 16 galactose-dependent, CAS-resistant isolates out of approximately 3 × 105 Ura+ transformants were identified. We cloned and analyzed the cDNAs from all 16 isolates. Restriction analysis and hybridization confirmed that 15 of the 16 clones were identical. We sequenced one of the 15 cDNA clones (GAL1 primer: 690-5′-TGGATAACCACTTTAACT-3′-707) and found that it contained a truncated SBE2 cDNA (amino acids 106 to 824). Sbe2p is a novel protein required for cell wall formation (18). The clone was fully sequenced and found to contain an SBE2 cDNA with no point mutations. It was retransformed into S. cerevisiae strain 10560-14C. The transformants were, as were the initial 10560-14C GAL1 SBE2 cDNA clones, resistant to CAS (32-fold more resistant than the control strain by the NCCLS M27-A microdilution method) in a galactose-dependent manner (Table 1; Fig. 1). However, some degree of leakiness of the GAL1 promoter in the glucose media was appreciated by the microdilution method (Table 1). Finally, the growth rate of the strain containing the GAL1 cDNA clone was identical to that of the control strain in glucose SC minus uracil liquid medium, but it was lower in the galactose SC minus uracil liquid medium (Fig. 1C). However, the strain containing the GAL1 cDNA clone grew better than the control strain in galactose SC minus uracil CAS (1 μg/ml) liquid medium, while the growth was almost identical to that of the control strain in glucose SC minus uracil CAS (1 μg/ml) liquid medium (Fig. 1C).
TABLE 1.
MICs of CASa
| Medium and strain | Median MIC (μg/ml) of CAS |
|---|---|
| Glucose SC minus uracil caspofungin | |
| GAL1 URA3 vector control | 0.5 |
| GAL1 URA3 truncated SBE2 cDNA | 2 |
| GAL1 URA3 full-length SBE2 cDNA | 2 |
| Galactose SC minus uracil caspofungin | |
| GAL1 URA3 vector control | 0.5 |
| GAL1 URA3 truncated SBE2 cDNA | 16 |
| GAL1 URA3 full-length SBE2 cDNA | 16 |
The (median) MIC of CAS for the strain transformed by the GAL1URA3 centromeric plasmid pRS 316 (control) and the same strain transformed by the same vector containing the SBE2 cDNA (truncated or full length) on glucose and galactose SC minus uracil CAS liquid (NCCLS M-27A microdilution method).
FIG. 1.
GAL1-SBE2 cDNA confers galactose-dependent resistance to CAS. (A) Growth inhibition responses of the control 10560-14C strain transformed by the GAL1 URA3 centromeric plasmid pRS 316 (control-right) and the same vector containing the SBE2 cDNA (left) on glucose and galactose SC minus uracil agar plates in the presence or absence of CAS. (B) Disk diffusion assays of the 10560-14C strain transformed by the GAL1 URA3 centromeric plasmid pRS 316 (top panels) and the same vector containing the SBE2 cDNA (bottom panels) on glucose and galactose SC minus uracil agar plates. The disks contained 5 μg of CAS. (C) Growth curves of the 10560-14C strain transformed by the GAL1 URA3 centromeric plasmid pRS 316 (control) and the same vector containing the SBE2 cDNA on glucose and galactose SC minus uracil agar liquid medium in the presence or absence of CAS (1 μg/ml).
To obtain the full-length SBE2 cDNA, we amplified by PCR the missing 5′ coding region of SBE2 from wild-type S. cerevisiae strain 10560-14C genomic DNA (primers SBE2BHI, 5′-GATCGGATCCTTTTCCCAAAGAGTTGAT-3′, and SBE2SalI, 5′-AATGTTCCAAATTAACGAGCC-3′). The resulting 900-bp PCR product was ligated into the truncated SBE2 cDNA initially isolated following digestion with BamHI/SalI and gel purification. The full-length GAL1 SBE2 cDNA was then transformed into the S. cerevisiae strain 10560-14C. These transformants containing the full-length GAL1 SBE2 cDNA were, as the transformants containing the original truncated SBE2 cDNA clone, equally resistant to CAS in a galactose-dependent manner (Table1). Finally, we sequenced the clone that was not SBE2 and found it to contain the GZF3 cDNA, a GATA factor involved in nitrogen-regulated transcription in yeast (19).
Galactose-dependent overexpression of SBE2 attenuates the abnormal cell wall ultrastructure and growth defects caused by CAS.
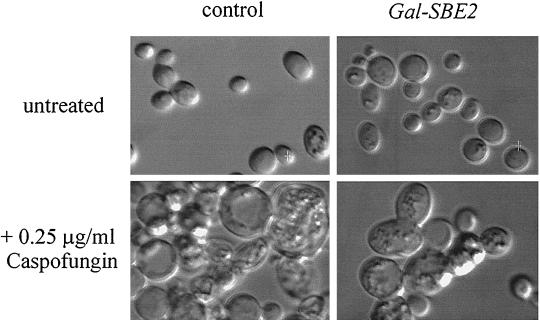
We analyzed by DIC microscopy the morphology of both the 10560-14C strain transformed by the GAL1 URA3 vector (control) and the same strain transformed by the same vector containing the GAL1 SBE2 cDNA growing on agar plates for 48 h in the following media: glucose SC minus uracil, galactose SC minus uracil, glucose SC minus uracil CAS (0.25 μg/ml), and galactose SC minus uracil CAS (0.25 μg/ml). We found that only the strain containing the GAL1 SBE2 cDNA, and only when grown in galactose, had an attenuation of the CAS-induced cell wall abnormalities (Fig. 2).
FIG. 2.
Galactose-dependent expression of SBE2 cDNA reduces cellular swelling and vacuolization in the presence of CAS. Control (left) and GAL1-SBE2 (right) S. cerevisiae strains were grown on galactose agar plates for 48 h at 30°C in the absence (top) or presence (bottom) of CAS. DIC microscopy and image analysis were performed as described in Materials and Methods.
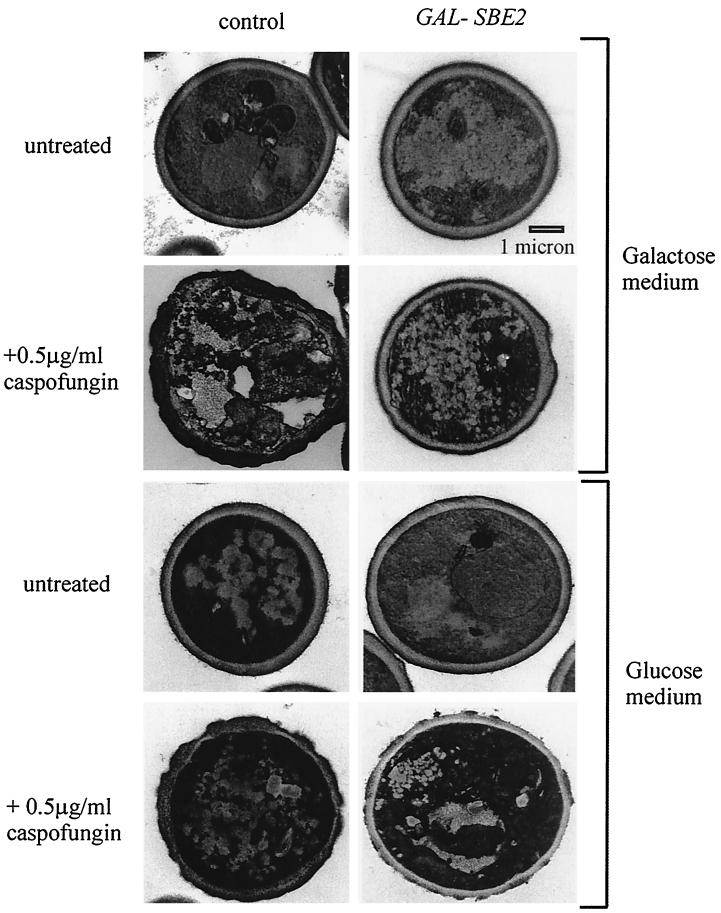
We also analyzed by electron microscopy the cell wall ultrastructure of these strains (control and GAL1 SBE2 cDNA-containing strain) growing for 48 h in the following media (agar plates): glucose SC minus uracil, galactose SC minus uracil, glucose SC minus uracil CAS (0.5 μg/ml), and galactose SC minus uracil CAS (0.5 μg/ml). We found that only the strain containing the GAL1 SBE2 cDNA, and only when grown in the presence of galactose, had reduced amounts of the CAS-induced cell wall abnormalities (Fig. 3; Table 2). However, even in the Sbe2p overexpressing cells, defects in the integrity of the cell wall, although milder, were seen, such as irregularities in the electron-dense outer layer or uneven thickness of the lighter middle layer (Fig. 3).
FIG. 3.
Galactose-dependent expression of SBE2 cDNA reduces CAS-induced cell wall damage. Control (left) and GAL1-SBE2 (right) S. cerevisiae strains were grown on galactose (top panel) and glucose (lower panel) minimal medium agar plates for 48 h at 30°C in the absence (top) or presence (bottom) of CAS. Transmission electron microscopy was performed as described in Materials and Methods.
TABLE 2.
Overexpression of SBE2 attenuates the cell wall abnormalities that are induced by CASa
| Medium | % of cells with abnormal cell wall
|
|
|---|---|---|
| SBE2 GAL1 | Vector control | |
| Glucose SC minus uracil | 1 | 1 |
| Galactose SC minus uracil | 4 | 3 |
| Glucose SC minus uracil CAS (0.5 μg/ml) | 35 | 32 |
| Galactose SC minus uracil CAS (0.5 μg/ml) | 7 | 38 |
S. cerevisiae cells in electron microscopy sections (approximately 500 cells viewed per result point) were examined, and the proportion that exhibited visible cell wall defects such as irregularities and aberrations in the electron-dense outer layer of the cell wall or thickening or deformation of the lighter middle layer were scored as having abnormal changes in the cell wall.
Galactose-dependent overexpression of Sbe2p results in an increase of glucan synthase activity.
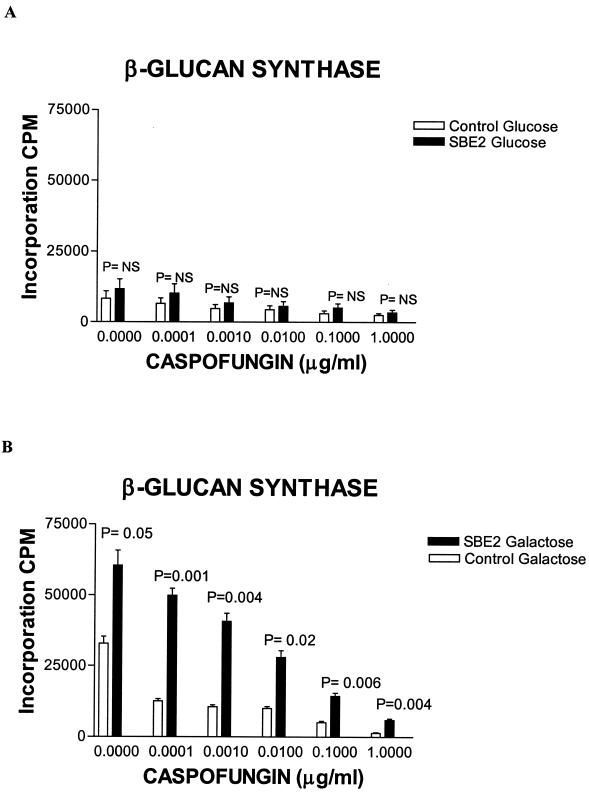
The glucan synthase activity was inhibited in a linear dose-dependent manner in both strains with increasing concentrations of CAS (Fig. 4). In the galactose SC minus uracil medium, both the baseline glucan synthase activity and its activity in the presence of increasing concentrations of CAS was much higher in the strain containing the SBE2 cDNA clone compared to control (Fig. 4B). A higher level of glucan synthase activity was seen in the strain containing the SBE2 cDNA in glucose SC minus uracil medium, again implying some degree of leakiness of the GAL1 promoter in glucose (Fig. 4A). A higher glucan synthase activity was observed when both the control and the strain containing the SBE2 cDNA were grown on galactose SC minus uracil, compared with the enzymatic activity of the corresponding strains measured in glucose SC minus uracil (Fig. 4).
FIG. 4.
Galactose-dependent expression of SBE2 cDNA increases the baseline glucan synthase activity and decreases its degree of inhibition by CAS. Control and GAL1-SBE2 S. cerevisiae strains were grown on glucose (A) and galactose (B) SC minus uracil liquid media. Membrane fractions were harvested, different concentrations of CAS were added, and glucan synthase activity was measured as described in Materials and Methods. The mean and standard deviation (error bars) of the three experiments, after first averaging the replicates in each experiment, are plotted. The statistical significance of the measurements of the control strain versus the SBE2-overexpressing strain is shown. Comparisons were performed, at each concentration separately, using two-sample t tests.
The specificity of the resistance or susceptibility to CAS in strains overexpressing or lacking of Sbe2p.
We used the disk diffusion assay to test the resistance phenotype of the control and the GAL1 SBE2 cDNA-containing strain in galactose SC minus uracil and glucose SC minus uracil plates against a variety of commonly used antifungals—polyenes (amphotericin B, 20 μg in disk) and triazoles (fluconazole, 20 μg in disk; itraconazole, 10 μg in disk)—as well as against a variety of other inhibitors such as protein synthesis (cycloheximide, 0.5 μg in disk), secretion (trifluoroperazine, 10 μg in disk), and glycosylation (tunicamycin, 20 μg in disk) inhibitors. Interestingly, the GAL1 SBE2 cDNA-overexpressing strain appeared to be more susceptible to itraconazole and fluconazole (Table 3).
TABLE 3.
Specificity of resistance to CAS in a strain overexpressing GAL1 SBE2
| Inhibitor (μg/disk) | Mean zone of inhibitiona (mm) ± SD seen on strain
|
|||
|---|---|---|---|---|
| Galactose SC minus uracil
|
Glucose SC minus uracil
|
|||
| GAL1 URA3 SBE2 cDNA (A) | GAL1 URA3 vector control (B) | GAL1 URA3 SBE2 cDNA (C) | GAL1 URA3 vector control (D) | |
| CAS (5) | 1.6 ± 0.02 | 3.8 ± 0.51 + | 3.6 ± 0.19 ++ | 3.0 ± 0.29 |
| Itraconazole (10) | 11.1 ± 0.9 | 7.2 ± 1.1 + | 9.1 ± 0.1 ++ | 5.0 ± 0.2 +++ |
| Fluconazole (20) | 8.6 ± 0.6 | 6.2 ± 0.3 + | 7.4 ± 0.3 ++ | 4.3 ± 0.0 +++ |
| Amphotericin B (20) | 3.8 ± 0.7 | 3.5 ± 0.2 | 3.0 ± 0.0 | 4.1 ± 0.1 |
| Cycloheximide (0.5) | 10.0 ± 1.1 | 11.2 ± 0.1 | 11.4 ± 0.7 | 9.9 ± 0.3 |
| Trifluoroperazine (10) | 1.7 ± 0.1 | 1.4 ± 0.3 | 1.0 ± 0.0 | 1.1 ± 0.2 |
| Tunicamycin (20) | 6.2 ± 0.2 | 5.3 ± 0.2 | 6.2 ± 0.2 | 7.0 ± 0.3 |
The measurements are based on triplicate samples from three independent experiments, each performed at a different time point. The means and standard deviations of the three experiments, after first averaging the replicates in each experiment, are indicated. Statistical comparisons for each column (as lettered parenthetically) were performed using one-way analysis of variance followed by pairwise comparisons between groups of interest. Symbols: +, P < 0.05 (comparison of A to B); ++, P < 0.05 (comparison of A to C); +++, P < 0.05 (comparison of C to D).
Finally, the sbe2 (strain Y1942) and sbe22 deletion diploid mutants (strain Y1943) and the sbe2 sbe22 double mutant (strain Y1944) as well as the isogenic control strain Y270 (18) were tested by disk diffusion assay in SC plates and were found to be hypersensitive to CAS (Table 4). These mutants (sbe2, sbe22, and the sbe2 sbe22 double mutant as well as the isogenic wild-type control strain) were also tested by disk diffusion assay against the aforementioned antifungals (amphotericin B, itraconazole, fluconazole) and growth inhibitors (cycloheximide, trifluoroperazine, and tunicamycin) and had a comparable degree of susceptibility on galactose and glucose media, with the possible exception being the sbe2 deletion mutant, which appeared to be less susceptible to fluconazole (Table 4).
TABLE 4.
Specificity of sensitivity to caspofungin in the SBE deletion mutants
| Inhibitor (μg/disk) | Mean zone of inhibitiona (mm) ± SD seen on strain
|
|||
|---|---|---|---|---|
| Isogenic control (A) | Δ SBE2 (B) | Δ SBE22 (C) | Δ SBE2 Δ SBE22 (D) | |
| CAS (2.5) | 4.4 ± 0.4 | 5.7 ± 0.0 + | 6.0 ± 0.1 ++ | 8.7 ± 0.5 +++ |
| Itraconazole (10) | 3.3 ± 0.0 | 3.0 ± 0.0 + | 2.3 ± 0.0 ++ | 2.4 ± 0.1 +++ |
| Fluconazole (20) | 6.0 ± 0.0 | 3.4 ± 0.1 + | 9.1 ± 0.4 ++ | 6.3 ± 0.2 |
| Amphotericin B (20) | 1.3 ± 0.1 | 1.1 ± 0.0 | 1.0 ± 0.0 | 1.7 ± 0.1 |
| Cycloheximide (0.5) | 11.2 ± 0.2 | 6.2 ± 0.2 | 11.1 ± 0.2 | 10.9 ± 0.5 |
| Trifluoroperazine (10) | 2.0 ± 0.0 | 2.7 ± 0.1 | 2.0 ± 0.0 | 2.2 ± 0.1 |
| Tunicamycin (20) | 2.7 ± 0.2 | 3.3 ± 0.0 | 2.6 ± 0.1 | 2.0 ± 0.0 |
The measurements are based on triplicate samples from three independent experiments, each performed at a different time point. The means and standard deviations of the three experiments, after first averaging the replicates in each experiment, are indicated. Statistical comparisons for each column (as lettered parenthetically) were performed using one-way analysis of variance followed by pairwise comparisons between groups of interest. Symbols: +, P < 0.05 (comparison of A to B); ++, P < 0.05 (comparison of A to C); +++, P < 0.05 (comparison of A to D).
DISCUSSION
We found for the first time that overexpression of Sbe2p under the regulated control of the GAL1 promoter results in resistance to CAS in S. cerevisiae. This is a novel mechanism of resistance to echinocandins. Neither of the genes previously associated with resistance to CAS (FKS1 or FKS2) was identified in our screen. One explanation could be that Fks1p and Fks2p, which are components of the larger enzymatic complex of glucan synthase (12), would have to be overexpressed along with the other proteins of that active complex in order to confer resistance to CAS; such overexpression of a multicomponent complex cannot be done with our genetic screen. Our experimental conditions may have underestimated the degree of resistance to CAS conferred by the overexpression of GAL1 SBE2. Because FKS2 activity is upregulated by the absence of glucose (12), it is possible that overexpression of SBE2 under the control of another strong promoter that is not regulated by carbon source could result in further resistance to CAS.
On the other hand, the strain overexpressing the SBE2 cDNA grew slower than the control in the presence of galactose (Fig. 1C). Therefore, the relative CAS resistance of that strain could be attributed to the fact that CAS acts more effectively against rapidly growing cells (14; Douglas et al, 40th ICAAC) and would therefore be relatively less effective against this slow-growing strain. However, if Sbe2p were providing protection to CAS only by slowing growth in general, one would probably have recovered several different cDNAs in our screen, since overexpression of many genes would be expected to result in slowed growth. In contrast, we found only two genes, with the SBE2 being found in the vast majority (15 of 16) of isolates.
SBE2 has been described in S. cerevisiae to encode a Golgi protein involved in the transport of cell wall components (18). SBE2 is 43% similar to the highly related gene SBE22 (18). The sbe2 sbe22 double mutant exhibits severe cell wall and polarity defects (18), implying that these two genes have overlapping function, possibly as a result of gene duplication. However, we did not find SBE22 in our screen. Interestingly, SBE2 but not SBE22 was found to suppress, when overexpressed in high copy number, the lethality of the chs5 spa2 mutant (18). This could suggest a degree of divergence of their structural features, physiologic role, and regulation. For example, the potential coiled-coil region (amino acids 510 to560) is conserved in Sbe2p but not Sbe22p (18).
Even though there is clear evidence that SBE2 is involved in maintaining cell wall integrity, the exact mechanism of protection from the glucan synthase inhibitors such as CAS is unclear. In view of the fact that we found that both baseline activity and the degree of inhibition of glucan synthase were higher in the strain overexpressing Sbe2p, it is possible that the aforementioned protein might be involved in the export or assembly of some regulatory subunits of the glucan synthase enzyme complex. Alternatively, a nonspecific protective mechanism, independent of the glucan synthase—such as the increased transport of other cell wall constituents compensating for impairment of cell wall synthesis and assembly caused by CAS—may be responsible for the resistance to CAS. In view of the recent insight regarding the regulation by sphingolipid biosynthetic pathway of FKS activity in S. cerevisiae (1) and of cell wall formation in Schizosaccharomyces pombe (6), it is tempting to speculate that the secretory machinery (of which Sbe2p is a part) that transports cell wall components could somehow be involved in those interactions. Further work that will examine the genetic interactions between sbe2 and those mutants known to affect cell wall biosynthesis as well as study of the transcriptional regulation of SBE2 in the presence or absence of CAS will further clarify the role of this gene in echinocandin resistance in yeast.
Finally, we found that the overexpression of Sbe2p appears to result in a paradoxical relative sensitivity to azoles. If Sbe2p is increasing the shuttling of vesicles containing cell wall membrane-related proteins, there may be subtle differences in cell membrane composition, leading to increased sensitivity to azoles. A feedback regulation of wall biosynthesis and remodeling following azole-induced ergosterol depleting conditions is suggested in S. cerevisiae and C. albicans (3, 4, 10)
The implications of our finding could have significant clinical relevance. This transport pathway may provide insight into the tolerance or lack of sensitivity to CAS seen in some pathogenic fungi (e.g., Cryptococcus and molds) and it needs to be further explored for diagnosis and therapy. Future work is needed to evaluate whether a constitutive hyperactivity of this novel pathway could explain the fundamental differences between yeasts and molds in regards to the type and degree of activity of echinocandins.
Acknowledgments
We thank C. Douglas from the Merck laboratories and R. E. Lewis from the University of Houston for useful discussions. We thank M. Snyder for kindly providing the strains Y270, Y1942, Y1943, and Y1944 and J. Thornby from the Houston VA Medical Center for his assistance in statistical analysis.
This work was supported by the Cancer Center (Core) grant CA16672 from The University of Texas M. D. Anderson Cancer Center and an educational grant from Merck and Company, Inc., to D.P.K.
REFERENCES
- 1.Abe, M., I. Nishida, M. Minemura, H. Qadota, Y. Seyama, T. Watanabe, and Y Ohya. 2001. Yeast 1,3-β-glucan synthase activity is inhibited by phytosphingosine localized to the endoplasmic reticulum. J. Biol. Chem. 276:26923-26930. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1998. Current protocols in molecular biology. Greene Publishing Associates/Wiley Inter Science, New York, N.Y..
- 3.Backer, M. D., T. Ilyina, X.-J. Ma, S. Vandoninck, W. Luyten, and H. Vanden Bossche. 2001. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob. Agents Chemother. 45:1660-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bammert, G. F., and J. M. Fostel. 2000. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob. Agents Chemother. 44:1225-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douglas, C. M., J. A. Marrinan, and M. B. Kurtz. 1994. A. Saccharomyces cerevisiae mutant with echinocandin-resistant 1, 3-β-d glucan synthase. J. Bacteriol. 176:5686-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feoktistova, A., P. Magneti, C. Abeijon, P. Perez, R. L. Lester, R. C. Dickson, and K. L. Gould. 2001. Coordination between fission yeast and glucan formation and growth requires a sphingolipase activity. Genetics 158:1397-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Georgopapadakou, N. H. 2001. Update on antifungals targeted to the cell wall: focus on beta-1, 3-glucan synthase inhibitors. Expert Opin. Investig. Drugs 10:269-280. [DOI] [PubMed] [Google Scholar]
- 8.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 9.Kaiser, C., and R. Schekman. 1990. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61:723-733. [DOI] [PubMed] [Google Scholar]
- 10.Kontoyiannis, D. P. 1999. Genetic analysis of azole resistance by transposon mutagenesis in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 43:2731-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kontoyiannis, D. P. 1999. Overexpression of Erg11p by the regulatable GAL1 promoter confers fluconazole resistance in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 43:2798-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurtz, M. B., and C. M. Douglas. 1997. Lipopeptide inhibitors of fungal glucan synthase. J. Med. Vet. Mycol. 35:79-86. [DOI] [PubMed] [Google Scholar]
- 13.Kurtz, M. B., E. M. Bernard, F. F. Edwards, et al. 1995. Aerosol and parenteral pneumocandins are effective in a rat model of pulmonary aspergillosis. Antimicrob. Agents Chemother. 39:1784-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurtz, M. B., I. B. Heath, J. Marrinan, S. Dreikorn, J. Onishi, and C. Douglas. 1994. Morphological effects of lipopeptides against Aspergillus fumigatus correlate with activities against (1,3)-beta-d-glucan synthase. Antimicrob. Agents Chemother. 38:1480-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, H., J. Krizek, and A. Bretscher. 1992. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics 132:665-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing for yeasts. Approved standard M-27A. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 17.Orlean, P. 1997. Biogenesis of yeast wall and surface components, p. 229-360. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cold Springs Harbor Laboratory Press, Plainview, N.Y.
- 18.Santos, B., and M. Snyder. 2000. Sbe2p and Sbe22p, two homologous golgi proteins involved in cell wall formation. Mol. Biol. Cell 11:435-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soussi-Boudekou, S., S Vissers, J. C. Jauniaux and B Andre. 1997. GZF3p, a fourth GATA factor involved in nitrogen-regulated transcription in Saccharomyces cerevisiae. Mol. Microbiol. 23:1157-1168. [DOI] [PubMed] [Google Scholar]