Abstract
The penetration of ampicillin and ciprofloxacin through biofilms formed by Klebsiella pneumoniae was confirmed by transmission electron microscopic observation of antibiotic-affected cells at the distal edge of the biofilm. Because the bacteria nevertheless survived antibiotic treatment, some protective mechanism other than inadequate penetration must have been at work in the biofilm.
Bacteria that group together in biofilms are protected from killing by antibiotics. Researchers are still debating and investigating the mechanisms behind this protection (3-5). One of the critical questions is whether the antibiotic penetrates throughout the biofilm. In a previous report, we described the use of a primitive diffusion cell to measure the permeation of ampicillin and ciprofloxacin through biofilms of Klebsiella pneumoniae (1) Here we present evidence confirming penetration of these antibiotics as determined by transmission electron microscopy (TEM).
Colony biofilms of K. pneumoniae were scarcely affected by 24 h of treatment with concentrations of ampicillin or ciprofloxacin that quickly eradicated free-floating cells (1). For example, the log reduction in viable-cell numbers in a colony biofilm treated for 24 h with 1.8 μg of ciprofloxacin per ml was 1.07 ± 0.18. The same antibiotic treatment of a planktonic culture caused a log reduction of 4.14 ± 0.33 after only 4 h of exposure (1). In that same study, a simple diffusion cell was used to measure penetration of these antibiotics through colony biofilms sandwiched between two polycarbonate filter membranes. This method showed that ciprofloxacin penetrated readily and that ampicillin could penetrate through a biofilm formed by a β-lactamase-negative strain but did not penetrate through a biofilm formed by the β-lactamase-positive wild type. One criticism of this approach is that one cannot rule out the possibility that the antibiotic moved through gaps or channels in the biofilm without accessing all of the cells. Using TEM, it is possible to directly visualize spatial patterns of antibiotic action within the biofilm and thereby identify those regions of the biofilm which the antibiotic must have reached.
Biofilms of K. pneumoniae were developed on membranes resting on a glucose minimal agar medium. Polycarbonate membrane filters were sterilized by exposure to UV light, placed on an agar plate, and seeded with a 50-μl drop of an overnight culture of either K. pneumoniae Kp1, a β-lactamase-positive wild type, or Kp102, a β-lactamase-negative derivative. The plates were inverted and incubated at 37°C for 48 h. Each colony biofilm was transferred to a fresh plate every 8 to 10 h during this period. Further details of the colony biofilm growth procedure are described by Anderl et al. (1). After 48 h of development, the average number of bacteria per membrane was 9.24 ± 0.02 log10 CFU.
Colony biofilms were exposed to antibiotic by simply transferring the membrane and associated bacteria to an agar plate containing an antibiotic at the desired concentration. These plates were prepared from the same glucose minimal medium used to grow the biofilms and were amended with either 1.8 μg of ciprofloxacin per ml or 5,000 μg of ampicillin per ml. These antibiotic concentrations were 10 times the MIC measured with suspended bacteria. The exposure duration was 12 h at a temperature of 37°C.
Antibiotic-treated and untreated colony biofilms were examined by TEM. Six samples were prepared: a 48-h-old Kp1 biofilm not exposed to antibiotic (time zero control), 48-h-old Kp1 and Kp102 biofilms transferred to agar without antibiotic for an additional 12 h (12-h controls), a 48-h-old Kp1 biofilm transferred to agar containing 1.8 μg of ciprofloxacin per ml for 12 h, and 48-old-Kp1 and Kp102 biofilms transferred to agar containing 5,000 μg of ampicillin per ml for 12 h. These biofilms were fixed with 5% glutaraldehyde, stained with osmium tetroxide (1%), and washed. Specimens were then dehydrated in an ethanol series, which included a staining step with 1% uranyl acetate-1% phosphotungstic acid. The dehydrated samples were embedded in Spurr’s epoxy resin, which was polymerized for 14 h at 70°C. Thin sections were cut and examined using a Jeol JEM-100CX electron microscope.
Cell sizes were determined by analysis of TEM images of control and antibiotic-exposed biofilm samples printed at a ×15,000 magnification. Three locations in each biofilm sample were analyzed: near the air interface, in the middle of the colony biofilm, and near the membrane. The major and minor axis lengths of individual cells were recorded for a minimum of 20 cells at each location, and the average cell cross-sectional area was calculated along with associated standard deviations. To eliminate bias in the selection of cells for analysis, all of the cells in a TEM print were individually numbered and then 20 numbers were randomly chosen for measurement. A two-sample t test was used to test for statistically significant differences between the means of cell lengths in different samples.
In colony biofilms that were not exposed to antibiotic (time zero and 12-h controls), bacteria were evenly distributed throughout the colony. These bacteria exhibited a typical short-rod morphology that did not change much with location in the biofilm (Fig. 1 and 2; Table 1). Bacterial cells in untreated controls were slightly longer when they were near the membrane (mean length, 1.39 μm) than when they were near the air interface (mean length, 1.05 μm).
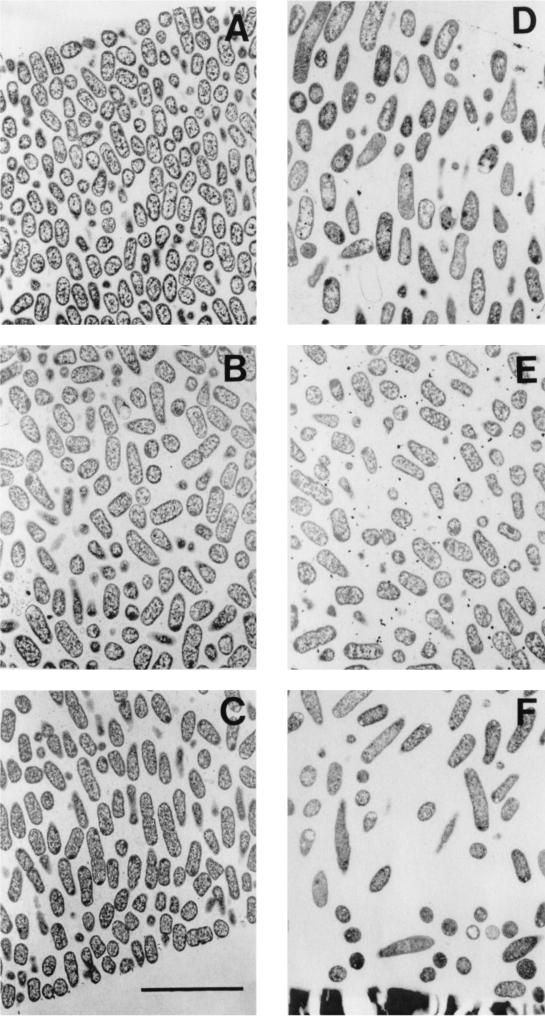
FIG. 1.
Transmission electron micrographs of β-lactamase-positive K. pneumoniae colony biofilms from the untreated 12-h control (A to C) and exposed to 1.8 μg of ciprofloxacin per ml for 12 h (D to F). (A and D) Spots near the air interface, which is just visible in the upper left (A) and right (D) corners; (B and E) locations near the middle of the biofilm; (C and F) regions near the membrane. The membrane is visible in the bottom of panel F. The membrane detached from the specimen shown in panel C; the former location of the membrane was along the bottom right-hand corner of the panel. Scale bar, 5 μm.
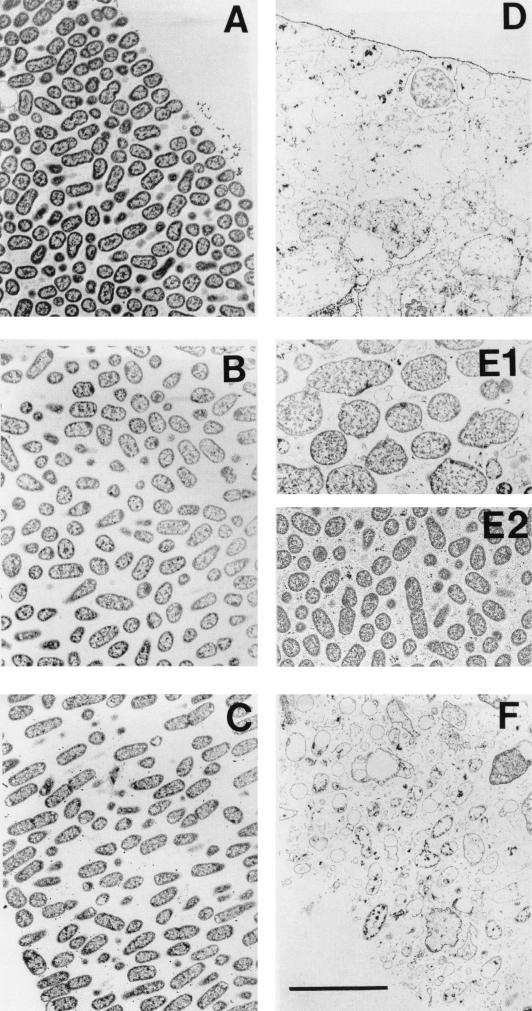
FIG. 2.
Transmission electron micrographs of β-lactamase-negative K. pneumoniae colony biofilms from the untreated 12-h control (A to C) and exposed to 5,000 μg of ampicillin per ml for 12 h (D to F). (A and D) Spot near the air interface; (B, E1, and E2) locations near the middle of the biofilm; (C and F) region near the membrane. The interior of the ampicillin-treated biofilm contained both zones of obvious antibiotic action (E1) and little antibiotic action (E2). Scale bar, 5 μm.
TABLE 1.
Mean major axis dimension of K. pneumoniae cells in TEM images
| Treatment (h) | β-Lactamase | Major axis length (μm) ± SD
|
||
|---|---|---|---|---|
| Air interface | Middle | Membrane | ||
| Control (0) | + | 1.10 ± 0.28 | 1.30 ± 0.41 | 1.52 ± 0.42 |
| Control (12) | + | 1.06 ± 0.28 | 1.11 ± 0.34 | 1.29 ± 0.34 |
| Control (12) | − | 0.98 ± 0.22 | 1.13 ± 0.31 | 1.36 ± 0.31 |
| Ciprofloxacin (12) | + | 1.53 ± 0.57 | 1.08 ± 0.30 | 1.44 ± 0.83 |
| Ampicillin (12) | + | 0.91 ± 0.30 | 1.10 ± 0.58 | 1.32 ± 0.43 |
| Ampicillin (12) | − | 3.23 ± 1.40 | 2.08 ± 0.61 | 2.90 ± 0.94 |
In ciprofloxacin-treated biofilm, antibiotic action was evident, especially near the air interface (Fig. 1). Cells near the air interface in this sample were longer than normal, and the cell density was less than in the corresponding control. The elongation of bacterial cells near the air interface in the ciprofloxacin-treated biofilm compared to lengths of cells of the 12-h untreated control was statistically significant (P < 10−3). No statistically significant cell elongation could be discerned in the middle of the biofilm (P = 0.60) or near the membrane (P = 0.25). Cell elongation is a well-known response to fluoroquinolone antibiotics, which prevent cell division by interfering with chromosome segregation. These results confirm the idea that the antibiotic was able to penetrate through the entire extent of the colony biofilm and act on bacteria at the distal edge. Antibiotic action was apparent along the full length of the air interface of the colony biofilm, showing that ciprofloxacin penetrated uniformly in this system, not just at isolated locations.
In ampicillin-treated β-lactamase-negative biofilm, antibiotic action was evident in much of the biofilm (Fig. 2). Bloated cells, lysed cells, and cell debris were particularly abundant near the air interface and membrane, while some areas of the middle of the colony appeared unaffected. The enlargement of bacteria in the ampicillin-treated biofilm in comparison to the lengths of untreated control bacteria (Table 1) was statistically significant at all three biofilm locations (P < 10−3). One explanation for these enlarged bacterial cells is that they are bacteria whose cell walls were destroyed by the antibiotic but whose integrity was stabilized by the extracellular matrix material of the biofilm. These data confirm that ampicillin was able to penetrate through the entire extent of the β-lactamase-negative colony biofilm and act on bacteria at the far edge. In biofilms formed by a β-lactamase-positive strain, there was no evidence of antibiotic action at any location in the biofilm (Table 1). This finding suggests that the hydrolysis of ampicillin by β-lactamase was sufficiently rapid to deplete the antibiotic from the agar before it could affect bacterial viability.
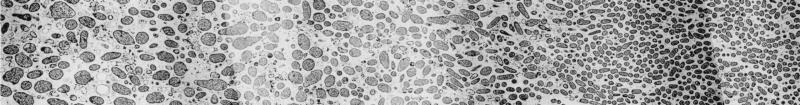
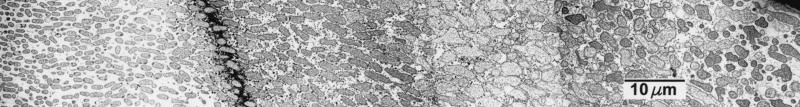
These results corroborate by an independent technique the effective antibiotic penetration reported by Anderl et al. (1). Both this work and the Anderl et al. study point to a critical role of bacterial physiology in modulating antibiotic susceptibility. In particular, it seems likely that bacteria in the interior regions of the colony are in a slow- or nongrowing state in which they are protected from killing (2, 6). We speculate that bacteria near the membrane grow by fermentation but that bacteria near the air interface grow aerobically on waste products of their fermenting neighbors. Bacteria in the center of the colony biofilm are likely starved for both the fermentable carbon source and for oxygen. This pattern of growth may explain the pattern of killing, particularly evident for ampicillin, in which bacteria at either edge of the biofilm were more strongly affected than bacteria in the colony interior (Fig. 3).
FIG. 3.
Spatial variation in ampicillin action in a β-lactamase-negative K. pneumoniae colony biofilm visualized by TEM. The biofilm was exposed to 5,000 μg of ampicillin per ml for 12 h. These two panels are montages along a single transect. The air interface was at the right and the membrane at the left. The gap between the two panels contained a bar of the copper grid supporting the section. This gap was about 50 μm.
We conclude that K. pneumoniae cells in biofilms are protected from killing by ciprofloxacin and ampicillin but that this protection is not due to inadequate penetration of the antibiotic into the bacterial aggregate except in the case of ampicillin acting on a β-lactamase-positive biofilm. In ongoing work, we are exploring the possibility that reduced rates of bacterial growth in the biofilm are responsible for biofilm resistance.
Acknowledgments
This work was supported through cooperative agreement EEC-8907039 between the National Science Foundation and Montana State University and by the industrial partners of the Center for Biofilm Engineering.
REFERENCES
- 1.Anderl, J. N., M. J. Franklin, and P. S. Stewart. 2000. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 44:1818-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, M. R. W., D. G. Allison, and P. Gilbert. 1988. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J. Antimicrob. Chemother. 22:777-783. [DOI] [PubMed] [Google Scholar]
- 3.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mah, T.-F. C., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 5.Stewart, P. S., and J. W. Costerton.2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 6.Xu, K. D., G. A. McFeters, and P. S. Stewart. 2000. Biofilm resistance to antimicrobial agents. Microbiology 146:547-549. [DOI] [PubMed] [Google Scholar]