Abstract
We characterized a new transposon, Tn5406 (5,467 bp), in a clinical isolate of Staphylococcus aureus (BM3327). It carries a variant of vgaA, which encodes a putative ABC protein conferring resistance to streptogramin A but not to mixtures of streptogramins A and B. It also carries three putative genes, the products of which exhibit significant similarities (61 to 73% amino acid identity) to the three transposases of the staphylococcal transposon Tn554. Like Tn554, Tn5406 failed to generate target repeats. In BM3327, the single copy of Tn5406 was inserted into the chromosomal att554 site, which is the preferential insertion site of Tn554. In three other independent S. aureus clinical isolates, Tn5406 was either present as a single plasmid copy (BM3318), as two chromosomal copies (BM3252), or both in the chromosome and on a plasmid (BM3385). The Tn5406-carrying plasmids also contain two other genes, vgaB and vatB. The insertion sites of Tn5406 in BM3252 were studied: one copy was in att554, and one copy was in the additional SCCmec element. Amplification experiments revealed circular forms of Tn5406, indicating that this transposon might be active. To our knowledge, a transposon conferring resistance to streptogramin A and related compounds has not been previously described.
Streptogramins (SGs) and related antibiotics are naturally produced by streptomycetes. They are classified as A and B compounds, according to their basic primary structures (9). The two classes of compounds bind different targets in the peptidyltransferase domain of the 50S ribosomal subunit and inhibit protein elongation at different steps (10). A and B compounds are bacteriostatic when used separately but act synergistically when combined, such that in some cases they are bactericidal, mainly against gram-positive bacteria. Natural mixtures, such as pristinamycin, synergistin, and mikamycin, are used in human medicine orally and topically. Quinupristin and dalfopristin (7), derivatives of streptogramin B (SGB) and A SGA, respectively, is an injectable semisynthetic mixture which has been available since 1999 for use in hospitals to treat infections due to gram-positive cocci that are resistant to other antibiotics. Virginiamycin, a related SG, was long used as a growth promoter in both Europe and the United States but was banned in Europe in 1999.
Staphylococcal resistance to synergistic mixtures of A and B compounds (pristinamycin MICs of >2 mg liter−1) is always associated with resistance to A compounds (pristinamycin IIA MICs of ≥8 mg liter−1) but not necessarily with resistance to B compounds (1, 12). Seven genes and a variant encoding resistance to A compounds have been isolated from staphylococcal and enterococcal plasmids. The genes vatA (6, 31), vatB (3, 31), vatC (4, 31), vatD (30, 31), and vatE (13, 31, 35) encode related proteins (50.4 to 60.1% identical amino acids [aa]) conferring resistance to SGA and similar compounds by acetylation of the drugs. The staphylococcal genes vgaA (5, 31) and vgaB (2, 31) encode related putative ATP-binding proteins (58.8% identical aa) that are probably involved in the active efflux of A compounds. The variant of vgaA (14), referred to here as “vgaAv,” was recently described in a clinical strain of Staphylococcus aureus (BM3327) and confers resistance to SGA but not to pristinamycin. Despite 83.2% identity with vgaA, vgaAv is distinguishable by its higher G+C content (35.6% instead of 29% for vgaA). In clinical isolates of S. aureus hybridizing with vgaAv, one to two copies of this gene are present on the chromosome and/or plasmids. Analysis of the sequences flanking vgaAv led to the identification of a new transposon, Tn5406 (5,467 bp) similar to Tn554 (11, 23, 24), also named Tn3853 (34). Tn554 (6,691 bp) contains three transposase genes (tnpA, tnpB, and tnpC), the gene ermA conferring inducible resistance to macrolide-lincosamide-SGB (MLSB) antibiotics, and the gene spc conferring spectinomycin resistance. Tn554 has no inverted terminal repeats: it fails to generate target repeats and transposes preferentially to a single chromosomal site, att554, with a frequency close to 100% (19, 23, 25, 27, 29). When att554 is occupied, deleted, or naturally absent, Tn554 can insert at a lower frequency into secondary insertion sites most of which are in the chromosome (att155 and att137) (11, 17, 23, 25, 32, 33). In some cases it inserts into penicillinase plasmids (pI524 and pI258) (23, 24). Five copies of Tn554 have been detected in the methicillin-resistant S. aureus strain N315, whose chromosome has been sequenced (21). One of the five copies is in the staphylococcal cassette chromosome mec (SCCmec) element (16, 21). Here we describe the sequence, distribution, and insertion sites of the transposon Tn5406 harboring vgaAv.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The relevant characteristics of the strains used are reported in Table 1. We used pUC18 as cloning vector. pIP1799 (14), pIP1692 (3), and pIP1705 (2) were used as probes to detect vgaAv, vatB, and vgaB, respectively.
TABLE 1.
Relevant characteristics of the strains used in this study
| Strain | Characteristics | Resistancea | MIC (μg ml−1)
|
MLSB and SGA resistance genesb | Tn5406 copy no. (location)c | Insertion site of Tn5406 | Detection of Tn5406 circular forms | Source or reference | |
|---|---|---|---|---|---|---|---|---|---|
| PIIA | PRI | ||||||||
| XL2-Blue | E. coli recipient (Stratagene) | No drug resistance marker | |||||||
| RN450 | S. aureus NCTC 8325-4 | No drug resistance marker | 1 | 0.250 | None | 28 | |||
| RN4220 | S. aureus NCTC 8325-4r− | No drug resistance marker | 2 | 0.06 | None | 18 | |||
| BM3252 | S. aureus clinical isolate: mecA | LIN SGA PEN MET TET MIN KAN NEO TOB GEN STR SUL CAD ARS MER ETB | 64 | 1 | vgaAv | 2 (C) | One copy in att554 site; one copy in SCCmec mobile element | No | 14 |
| BM3318 | S. aureus clinical isolate; mecA; Tn554 | MLSBc SGA PRI PEN MET TET MIN KAN NEO TOB GEN STR SPT SUL CAD ARS MER ETB ABR | 128 | 16 | ermA ermB vatB vgaB vgaAv | 1 (P) | NDd | Yes | 14 |
| IPF110 | BM3318 derivative, susceptible to SGA obtained after serial passages on drug-free medium; mecA; Tn554 | MLSBc PEN MET TET MIN KAN NEO TOB GEN STR SPT SUL CAD ARS MER ETB ABR | 1 | 0.12 | ermA ermB | This study | |||
| BM3327 | S. aureus clinical isolate | MLSBc SGA PRI PEN TET MIN KAN NEO TOB GEN STR SUL PEF | 128 | 8 | ermC vgaAv | 1 (C) | att554 site | Yes | 14 |
| BM3385 | S. aureus clinical isolate; pIP1156 | LIN SGA PRI PEN SUL TMP RIF CAD ARS ETB MER ABR | 128 | 4 | vatB vgaB vgaAv | 2 (C + P) | One copy (C) in att554 site; one copy (P): ND | No | 14 |
| BM12940 | BM3385 derivative, susceptible to SGA obtained after serial passages on drug-free medium | RIF CAD ARS MER | 2 | 0.06 | None | This study | |||
Abbreviations: ABR, acetyltrimethylammonium bromide; AMP, ampicillin; ARS, sodium arsenate; CAD, cadmium acetate; ETB, ethidium bromide; GEN, gentamicin; KAN, kanamycin; LIN, lincomycin; MER, mercuric nitrate; MET, methicillin; MLSBc, constitutive resistance to macrolides-lincosamides-SGB; MIN, minocycline; NEO, neomycin; PEN, penicillinase; PEF, pefloxacin; PRI, pristinamycin; PIIA, pristinamycin IIA; RIF, rifampin; SGA, streptogramin A; SPT, spectinomycin; STR, streptomycin; SUL, sulfonamide; TET, tetracycline; TOB, tobramycin; TMP, trimethoprim.
The strains were screened by PCR experiments for the following genetic elements: Tn554, ermA, ermB, ermC, mecA, vatB, vgaB, vgaAv, vgbA and vgbB.
Location: C, chromosome; P, plasmid.
ND, not done.
Media.
Staphylococci were grown in brain heart infusion (Difco Laboratories, Detroit, Mich.), and Escherichia coli was grown in Luria broth (Difco). Susceptibility to antibiotics was tested on Mueller-Hinton agar (Bio-Rad, Hercules, Calif.).
Susceptibility to antimicrobial drugs.
Susceptibility to antibiotics was determined by a disk diffusion assay with commercially available antibiotic disks (Bio-Rad) performed according to the recommendations of the French Society of Microbiology and with disks prepared in our laboratory as described previously (14). The MICs of pristinamycin IIA and pristinamycin (Rhône-Poulenc Rorer, Vitry, France) were determined with serial 1:2 dilutions of antibiotics in Mueller-Hinton agar (1).
DNA isolation and analysis.
Total cellular DNA was isolated from staphylococcal strains and purified by using the QIAamp tissue kit from Qiagen (Hilden, Germany). Plasmid DNA was extracted and purified from E. coli by using the QIAprep spin plasmid kit from Qiagen. Restriction endonucleases were obtained from Amersham-Pharmacia Biotech, Inc. (Piscataway, N.J.), and were used according to the manufacturer's instructions. DNA fragments of <500 bp were separated by electrophoresis in 4% NuSieve GTG agarose gels (FMC BioProducts, Rockland, Maine). SmaI digestion and pulsed-field gel electrophoresis were performed as described previously (14).
Cloning and DNA sequencing.
DNA restriction fragments were inserted into E. coli vectors by using the ligase of the Fast-Link ligation kit (Epicenter Technologies Corp., Madison, Wis.), and recombinant plasmids were introduced into competent E. coli XL-2 Blue cells (Stratagene, La Jolla, Calif.) by transformation according to the manufacturer's instructions.
An Applied Biosystems (Foster City, Calif.) automated 373A DNA sequencer was used for sequencing according to the protocol provided by the manufacturer.
Labeling of DNA probes, blotting, and hybridization.
Hybridization experiments were performed at 65°C as described previously (14)
PCR.
DNA was amplified by PCR by using the Ready-To-Go kit (Amersham), according to the manufacturer's instructions, in a Crocodile III apparatus (Appligène, Illkirch, France). The primers used are described in Table 2. Primer pairs 1 and 2, 3 and 4, 2 and 3, 5 and 6, mecA1 and mecA2, ermB1 and ermB2, and ermC1 and ermC2 were used for amplification by PCR at high stringency (initial cycle of 5 min at 95°C and 2 min at 55°C, followed by 35 cycles of 1 min at 72°C, 30 s at 95°C, and 1 min at 55°C, with a final extension step of 5 min at 72°C). All other amplifications by PCR used the same conditions, replacing the annealing temperature of 55°C by the following temperatures depending on the primers: 53°C with primers ermA1 and spc1 (to amplify internal region of Tn554), 51°C with primers ermA2 and 12 (to amplify the 3′ region of Tn554 inserted in att554), 47°C with primers 1 and 12, and 42°C with primers 15 and 16 (to amplify vgaAv).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′) | Target | Accession no. | Positions (nt) | Source or reference |
|---|---|---|---|---|---|
| 1 | GTTCGATTGTACATCCACG | att554 | AF186237 | 1156-1174 | 25 |
| 2 | ATCCACTACTATTTTTTGCAC | Tn554 tnpA-like | AF186237 | 1410-1390 | This study |
| 3 | GTGTAAAGGTTATTACATCGGC | Tn5406 3′ end | AF186237 | 6665-6687 | This study |
| 4 | CAATGAAAGTACTTCGTAGG | 3′ region from att554 | AF186237 | 6920-6901 | This study |
| 5 | GATGAACAATTTGAAGTAGTTGAACC | Tn554 tnpA-like | AF186237 | 1441-1466 | This study |
| 6 | GATACTCATATTGAGTGGG | Tn554 tnpB-like | AF186237 | 3084-3066 | This study |
| 12 | CCCGCTTCTACAAGACTGG | att554 | AF186237 | 6842-6824 | 25 |
| 15 | ATGAAAATATTGTTAGAGG | vgaAv | AF186237 | 5065-5083 | This study |
| 16 | AGACATGTCCTTAAAGTGATTC | Tn5406 3′ region | AF186237 | 6659-6638 | This study |
| mecA1 | GGGATCATAGCGTCATTATTC | mecA | Y14051 | 3851-3871 | 36 |
| mecA2 | TATCGTCAACGATTGTGACACG | mecA | Y14051 | 4384-4363 | 36 |
| ermA1 | TATCTTATCGTTGAGAAGGGATT | ermA | K02987 | 4913-4891 | 22, 24 |
| ermA2 | CTACACTTGGCTTAGGATGAAA | ermA | K02987 | 4775-4796 | 22, 24 |
| spc1 | GTTGTCCCTTGGCAATATCCTC | spc | K02987 | 3928-3949 | 24 |
| ermB1 | CTATCTGATTGTTGAAGAAGGATT | ermB | U35228 | 366-389 | 22 |
| ermB2 | GTTTACTCTTGGTTTAGGATGAAA | ermB | U35228 | 484-507 | 22 |
| ermC1 | CTTGTTGATCACGATAATTTCC | ermC | M17990 | 214-235 | 22 |
| ermC2 | ATCTTTTAGCAAACCCGTATTC | ermC | M17990 | 382-403 | 22 |
Nucleotide sequence accession number.
The nucleotide sequences of Tn5406 in the att554 site of S. aureus BM3327 have been submitted to GenBank under accession no. AF186237. The 5′- and 3′-flanking regions of Tn5406 in the SCCmec type element of S. aureus BM3252 are registered under accession no. AF411128 and AF411129, respectively.
RESULTS
Characterization of Tn5406 isolated from the chromosome of BM3327.
We recently characterized vgaAv (accession no. AF186237, nucleotides [nt] 5065 to 6639) (14). The two overlapping restriction fragments carrying the 5′ and 3′ parts of the sequenced copy of vgaAv, i.e., a 1.3-kb HindIII fragment and a 7-kb EcoRI fragment, respectively, were separately ligated into pUC18 cleaved by the same enzymes. The resulting recombinant plasmids, pIP1805 and pIP1809, respectively, were used to sequence the chromosomal regions adjacent to vgaAv (accession no. AF186237, nt 1 to 7089, including vgaAv). The three putative genes upstream from vgaAv exhibited significant similarities with the genes involved in the transposition of Tn554: tnpA, tnpB, and tnpC (70, 71.5, and 65% identities, respectively) (23, 24). The amino acid sequences of the predicted translation products of Tn5406 transposases were 71, 73, and 61% identical to those of Tn554, respectively (Fig. 1). Significant similarities with the 5′ and 3′ ends of Tn554 (accession no. K02987) (66.7 and 69.7% identity, respectively) were also observed between nt 1251 and 1383 and between nt 6642 and 6717 (accession no. AF186237). In contrast, the similarity between the region from the end of the Tn554-tnpC-like gene to the start of vgaAv (318 nt), as well as that of Tn554 between the end of tnpC and the start of spc (152 nt), is restricted to a region of 27 nt only (66.7% identity).
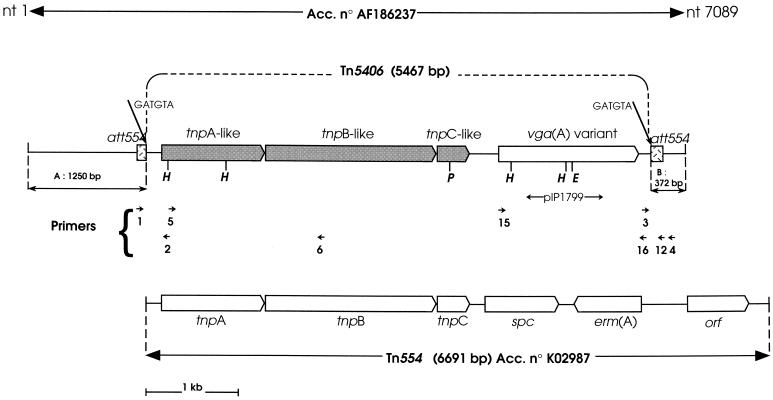
FIG. 1.
Schematic representation of Tn554 and the sequenced copy of Tn5406 from the chromosome of S. aureus BM3327. The position and orientation on Tn5406 of primers described in Table 2 are indicated by arrows. The vgaAv intragenic fragment inserted in pIP1799 used as a probe is indicated by an arrow. Abbreviations: H, HindIII; P, PstI; E, EcoRI. Regions (indicated by double-headed arows): A:1,250 bp, region adjacent to the 5′ part of Tn5406 and exhibiting 99.9% identity with chromosomal DNA of S. aureus N315 (accession no. AP003134, nt 192697 to 191446); B:372 bp, region adjacent to the 3′ part of Tn5406 and exhibiting 99.2% identity with chromosomal DNA of S. aureus N315 (accession no. AP003134, nt 184733 to 184362).
The region extending from nt 1251 to 6717 (accession no. AF186237) and including vgaAv and the three transposase genes is flanked by sequences corresponding to att554 (accession no. K02985) (23, 29), the preferential insertion site of Tn554 in the chromosome of S. aureus strain RN450. Indeed, from nt 1135 to 1250 (accession no. AF186237), the sequence is identical to that of RN450 att554 from nt 1 to 116 (accession no. K02985): both end with GATGTA, which is the 6-bp central core of att554. Moreover, from nt 6717 to 6851 (accession no. AF186237), the sequence is almost identical (one additional nucleotide in position 6843) to that of the RN450 att554 site from nt 117 to 249. These data suggested that vgaAv of BM3327 was within a Tn554-like transposon extending from nt 1251 to 6717. This 5,467-bp putative transposon was named Tn5406 (Fig. 1).
Analysis of the regions flanking Tn5406 in strain BM3327.
We sequenced 1,250 nt 5′ to Tn5406 and 372 nt 3′ to Tn5406 in BM3327 (Fig. 1). The 1,250 nt 5′ to Tn5406 (nt 1 to 1250) exhibited 99.9% identity with the S. aureus N315 genome from nt 192697 to 191446 (accession no. AP003134) (21). From nt 32 to 712 there is a putative gene whose translation product is a hydrophobic protein of 227 aa. A gene encoding a putative and closely related protein (98.7% aa identity) similar to type IV prepilin peptidase was detected at the same position relative to the 5′ end of Tn554 in the chromosome of N315 (accession no. AP003134, nt 192689 to 191982). The sequenced region of 372 nt 3′ to Tn5406 (nt 6718 to 7089) (Fig. 1) exhibited 99.2% identity with the 3′ part of the copy of Tn554 in the att554 site of the S. aureus N315 genome extending from nt 184733 to 184362 (accession no. AP003134) (21).
In the chromosome of BM3327, as well as in N315 (21) and NCTC8325 (http://www.genome.ou.edu/staph.html), the att554 sequence is within a putative 669-bp gene (Fig. 2). The translation product of this gene is similar (53.1% aa identity) to the Bacillus subtilis protein Ysxa (accession no. Z99118, nt 65971 to 66666) (20) believed to be involved in DNA repair. In the chromosomes of BM3327 and N315, this putative gene is interrupted at the same site by either Tn5406 or Tn554, respectively. A frameshift mutation is present in the gene of BM3327 interrupted by Tn5406.
FIG. 2.
Alignments of the nucleotide sequences of att155 (33) and those flanking Tn554 and Tn5406 in the chromosomes of the following S. aureus strains: BM3327 (accession no. AF186237, nt 713 to 1250, and nt 6718 to 6851), N315 (accession no. AP003134, nt 191453 to 191985 and nt 184362 to 184503), BM3252 (accession no. AF411128, nt 844 to 1146, and accession no. AF411129, nt 124 to 265), and 85/2082 (accession no. AB037671, nt 57652 to 58803 and nt 49780 to 50963). The sequences from these latter strains are delimited by the start and stop codons (reported in boldface) of the genes interrupted by the transposons. Symbols: #, nucleotides common to all the aligned sequences; *, nucleotides common to BM3327 and N315; +, nucleotides common to BM3252, 85/2082, and att155.
Testing for circular forms of Tn5406 in the cellular DNA of clinical isolates.
Murphy (23) proposed a transposition model, including excision and circularization steps, preceding integration in the new target. To check whether this model can be proposed for Tn5406, we looked for the presence of cicular forms with primers 2 and 3 (Table 2). Amplification of the cellular DNA of strains BM3318 and BM3327 (Table 1) by PCR with these primers gave a 213-bp amplicon. This demonstrates the presence of circular forms of Tn5406, suggesting that it might be functional according to the model proposed for Tn554. We were not able to detect these forms in strains BM3252 and BM3385.
Investigation for presence of Tn5406 in clinical isolates carrying vgaAv.
We have already shown (14) that a vgaAv-probe (pIP1799) hybridizes with (i) a single chromosomal SmaI fragment in BM3327; (ii) two chromosomal SmaI fragments in BM3252; (iii) a single plasmid SmaI fragment containing two other SGA resistance genes, vatB and vgaB, in BM3318; and (iv) both a chromosomal SmaI fragment and a large vatB-vgaB plasmid not cleaved by SmaI in BM3385. We used PCR, hybridization, and sequencing to determine whether these various copies of vgaAv are carried by Tn5406.
Each of the four wild-type strains tested (Table 1) gave two to four EcoRI fragments and two or three HindIII fragments hybridizing with the vgaAv probe, including the 0.57-kb HindIII intragenic fragment (results not shown). Since the intragenic probe used (from pIP1799) contained a HindIII site (14), these data suggested that each strain contained no more than one or two copies of vgaAv.
PCR with primer pairs 1-2 and 3-4 (Table 2) amplified fragments of 255 and 256 bp, respectively, from BM3327, BM3252, and BM3385. Thus, each of these strains contained a copy of Tn5406 inserted in att554. These results were confirmed by hybridization experiments with an “att554 probe” consisting of a 220-bp fragment amplified from within the att554 of RN4220 with the primer pair 1-12 (Table 2): each of the three strains contained two att554-hybridizing HindIII fragments (a 1.3-kb fragment cohybridizing with vgaAv and a 3.2-kb fragment) (results not shown).
The finding that BM3318 appears to contain a circular form of Tn5406 suggests that the single plasmid copy of vgaAv is carried by this transposon. Amplification with the primer pairs spc1-ermA1 and ermA2-12 (Table 2) and the detection of two HindIII fragments hybridizing with the att554 probe but not with vgaAv probe indicated that the single chromosomal att554 site in BM3318 was occupied by a copy of Tn554.
In BM3385, the 3′ part of the plasmid-borne vgaAv is carried by a 1.1-kb HindIII fragment (results not shown). An indistinguishable fragment is present in BM3318 (results not shown). The 5′ part of this vgaAv copy in BM3385 is on an EcoRI fragment of ∼20 kb giving no hybridization signal with the att554 probe. However, this EcoRI fragment hybridized with a “Tn5406 transposase A-B probe” (a 1,640-bp amplicon obtained with primers 5 and 6 [Table 2]) (results not shown). Thus, the plasmid copy of vgaAv in BM3385 is, like the chromosomal copy, carried by Tn5406.
HindIII and EcoRI digests of BM3252 and BM3327 DNAs were hybridized with the vgaAv probe (results not shown). BM3252 gave three HindIII fragments hybridizing with vgaAv probe, two of which were also present in BM3327: HindIII fragments of 0.57 kb (internal to vgaAv) and of 1.3 kb. The third HindIII fragment (3 kb), unique to BM3252, was suspected to contain a vgaAv not on the copy of Tn5406 inserted in att554. It was inserted into pUC18. The ends of this fragment were sequenced: it indeed carried part of a vgaAv gene, including 123 nt identical to the 3′ end of BM3327 Tn5406 (accession no. AF186237, nt 6594 to 6717). Two of the four EcoRI fragments (7 and 7.5 kb) of BM3252 hybridized with both the vgaAv probe and the probe obtained with primers 1 and 2 (Table 2). The 7.5-kb EcoRI fragment of BM3252 was not found in BM3327. Therefore, it was suspected to carry the 5′ part of the copy of vgaAv, which is not in the Tn5406 copy at the att554 site. This fragment was inserted into pUC18, and its ends were sequenced: it was mapped to the 5′ part of the transposon. It contained 134 nt identical to the 5′ end of BM3327 Tn5406 (accession no. AF186237, nt 1251 to 1385). These data suggested that both copies of vgaAv in BM3252 were carried by Tn5406-related transposons.
Sequence analysis of the regions flanking the chromosomal copy of Tn5406 in BM3252 not inserted in the att554 site.
We sequenced 1,146 nt 5′ and 1,185 nt 3′ to the chromosomal copy of Tn5406 in BM3252 that was not inserted into att554. These flanking regions were more similar to the 240-nt att155 insertion site of Tn554 in methicillin-resistant S. aureus and S. epidermidis clinical strains isolated in 1988 (33) (81.7% identity on a 229-nt overlap) than to att554 (58.1%). Nevertheless, this Tn5406 insertion site in BM3252 was clearly neither att554 nor att155: the central hexanucleotide core sequence GATATA, differed by a single nucleotide from those (GATGTA) of att155 and att554. The sequence of the region 5′ to Tn5406 in BM3252 from nt 1 to 1146 (accession no. AF411128) is at least 87.6% identical to the 5′-flanking regions of Tn554 inserted in the type III SCCmec of S. aureus strains 85/3907 (accession no. AB047089, nt 68 to 1213) (16) and 85/2082 (accession no. AB037671, nt 57652 to 58803) (16). The sequence of the region 3′ to Tn5406 (accession no. AF411129, nt 124 to 1308) is highly similar (97.5% nt identity) to the 3′-flanking region of Tn554 in the type III SCCmec of strain 85/2082 (accession no. AB037671, nt 49780 to 50963) (16). Moreover, the 128-nt 3′ to BM3252 Tn5406 are also identical to the right arm of the secondary Tn554 attachment site (accession no. M32312, nt 1 to 128) in the methicillin-resistant S. aureus isolate, ANS46. This strain is typical of endemic and epidemic methicillin-resistant S. aureus strains found in Australia and England (8).
The BM3252 Tn5406 chromosomal copy which is not in att554 appears to be inserted into a gene encoding a 148-aa putative protein. This gene is closely related to those of strains 85/3907 and 85/2082 (16), which are interrupted by Tn554 at the same relative position (Fig. 2). These genes are assumed to be involved in DNA repair because of the significant similarity of their products with the Bacillus sp. strain NEB-606 RadC (15).
Analysis of SGA-susceptible derivatives of S. aureus clinical isolates, BM3318 and BM3385.
BM3318 and BM3385 derivatives susceptible to SGA, IPF110 and BM12940, respectively (Table 1), were obtained after 20 passages of each strain on drug-free medium. The BM3318 derivative, IPF110, had lost resistance to SGA, vatB, vgaB, Tn5406, and also the single plasmid SmaI band (results not shown), suggesting the loss of the large SGA resistance plasmid. This loss was not associated with the loss of LIN resistance (ermB conferring constitutive MLSB resistance) or with that of penicillinase (BM3318 carries two penicillinase plasmids). The SGA-susceptible variant of BM3385, BM12940, whose SmaI pattern was identical to that of BM3385, had lost the antibiotic resistances conferred by pIP1156 and Tn5406 (resistance to SGA, lincomycin, trimethoprim, and penicillinase). With SmaI-digested DNA from strain BM12940, no hybridization was detected with vgaB, vatB, and vgaAv probes (results not shown). Conversely, with the same probes and SmaI-digested BM3385 DNA, hybridization was detected at the level of the wells and ∼670 kb (14). The att554 probe hybridized with a single HindIII fragment of 3.6 kb in the cellular DNA of BM12940 as in RN4220 (results not shown). Primer pair 1-12 (Table 2) amplified a 220-bp fragment from BM12940. Its sequence was identical to part of att554. This indicates that the chromosomal copy of Tn5406 had been precisely excised from BM3385.
DISCUSSION
The sequencing of regions adjacent to vgaAv in BM3327 allowed us to characterize a new transposon, Tn5406, that is similar to Tn554. We detected circular forms of Tn5406 in some clinical isolates, and it was able to excise precisely from its insertion site. There is thus strong evidence of its ability to transpose. Four wild-type S. aureus clinical strains were studied, and in all cases vgaAv was on Tn5406 (one or two copies per isolate). Thus, vgaAv may always be carried by Tn5406. This transposon is therefore presumably not uncommon among SGA-resistant staphylococcal strains, since vgaAv was detected in 25 of the 56 independent staphylococci belonging to five species investigated (14). All plasmid copies of Tn5406 were on large self transferable plasmids, carrying two other SGA resistance genes, vatB and vgaB, which are cotranscribed and functional in BM3385. These plasmids are unstable in the absence of selection with SGA. The advantage of Tn5406 being carried by these plasmids may be the maintenance of SGA resistance through its transposition to the multiple available chromosomal insertion sites. However, although Tn5406 is active in BM3318, it did not transpose to the chromosome of the SGA-susceptible derivative IPF110. This may have been because IPF110 was derived in the absence of selective pressure with SGA. In BM3252, which is susceptible to erythromycin and SGB, the two chromosomal copies of Tn5406 conferred resistance to SGA (pristinamycin IIA MIC of 64 μg ml−1) but not to pristinamycin (MIC of 1 μg ml−1). In contrast, BM3385, which carries vgaB and vatB in addition to two copies of Tn5406, is resistant not only to SGA (pristinamycin IIA MIC of 128 μg ml−1) but also to pristinamycin (MIC of 4 μg ml−1). It is not known which of the SGA resistance genes and Tn5406 copies is functional in any strain. The multiplicity of Tn554 copies in clinical isolates has been explained by mutational drift toward inactivation in the spc gene (32). Similarly, the presence of more than one copy of Tn5406 could possibly be a consequence of mutations inactivating some copies of vgaAv. Different copies of Tn5406 carried by individual strains should be cloned to test their expression.
The G+C content of the transposons Tn554 (32.6%) and Tn5406 (34.9%), including the antibiotic resistance genes, is similar to that of the staphylococcal genome (32 to 36%) but slightly higher than that of the two putative ATP-binding proteins conferring resistance to SGA, i.e., vgaA (29%) and vgaB (27.2%). The evolutionary ancestor of vgaA and vgaB probably belongs to a genus other than staphylococcus. If vgaAv has the same ancestor, it was acquired by staphylococci much earlier. Tn554 and Tn5406 have very similar transposases, and both were found in S. aureus chromosomal insertion sites that were either identical (att554) or very similar (att155 in the SCCmec region). In both sites, the transposition of Tn554 or Tn5406 interrupts genes encoding putative DNA repair proteins (Ysxa-like or RadC-like) with 31.7% identical amino acids. This relatedness is significantly higher in the regions flanking Tn5406 and Tn554: 64.3% for the 28 aa upstream and 48.5% for the 33 aa downstream. The nucleotide sequences encoding these highly related flanking regions (84 nt upstream and 99 nt downstream), may thus serve as insertion sites for these two transposons.
All attempts to detect repeats in the regions flanking the transposase genes in Tn554 and Tn5406 failed. Detection of such repeats, which may be subject to site-specific breakage and joining, would make the transposase region available as a cassette to pick up different antibiotic resistance genes and thus to trigger their transfer.
Acknowledgments
We thank C. Tran for secretarial assistance. We are grateful to the Staphylococcus aureus Genome Sequencing Project and to B. A. Roe, Y. R. Tian, H. Jia, S. Li, S. Lin, S. Kenton, H. Lai, J. D. White, A. Dorman, F. Z. Najar, S. Clifton, V. Worrell, and J. Iandolo, with funding from the NIH and the Merck Genome Research Institute, for the use of NCTC 8325 DNA sequence data prior to publication.
J.H. received grants from the Fonds d'Etudes et de Recherche du Corps Médical des Hôpitaux de Paris, Institut Lilly (Bourse d'Etude en Infectiologie SPILF-Lilly), and from the Institut SmithKline Beecham.
REFERENCES
- 1.Allignet, J., S. Aubert, A. Morvan, and N. El Solh. 1996. Distribution of the genes encoding resistance to streptogramin A and related compounds among the staphylococcci resistant to these antibiotics. Antimicrob. Agents Chemother. 40:2523-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allignet, J., and N. El Solh. 1997. Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transporter conferring resistance to streptogramin A and related compounds. Gene 202:133-138. [DOI] [PubMed] [Google Scholar]
- 3.Allignet, J., and N. El Solh. 1995. Diversity among the gram-positive acetyltransferases inactivating streptogramin A and structurally related compounds, and characterization of a new staphylococcal determinant, vatB. Antimicrob. Agents Chemother. 39:2027-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allignet, J., N. Liassine, and N. El Solh. 1998. Characterization of a staphylococcal plasmid related to pUB110 and carrying two novel genes, vatC and vgbB, encoding resistance to streptogramins A and B and similar antibiotics. Antimicrob. Agents Chemother. 42:1794-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allignet, J., V. Loncle, and N. El Solh. 1992. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamycin A-like antibiotics. Gene 117:45-51. [DOI] [PubMed] [Google Scholar]
- 6.Allignet, J., V. Loncle, C. Simenel, M. Delepierre, and N. El Solh. 1993. Sequence of a staphylococcal gene, vat, encoding an acetyltransferase inactivating the A-type compounds of virginiamycin-like antibiotics. Gene 130:91-98. [DOI] [PubMed] [Google Scholar]
- 7.Barriere, J. C., N. Berthaud, D. Beyer, S. Dutka-Malen, J. M. Paris, and J. F. Desnottes. 1998. Recent developments in streptogramin research. Curr. Pharm. Design 4:155-180. [PubMed] [Google Scholar]
- 8.Chikramane, S. G., P. R. Matthews, W. C. Noble, P. R. Stewart, and D. T. Dubin. 1991. Tn554 inserts in methicillin-resistant Staphylococcus aureus from Australia and England: comparison with an American methicillin-resistant group. J. Gen. Microbiol. 137:1303-1311. [DOI] [PubMed] [Google Scholar]
- 9.Cocito, C. 1979. Antibiotics of the virginiamycin family, inhibitors which contain synergistic components. Microbiol. Rev. 43:145-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocito, C., M. Digambattista, E. Nyssen, and P. Vannuffel. 1997. Inhibition of protein synthesis by streptogramins and related antibiotics. J. Antimicrob. Chemother. 39:7-13. [DOI] [PubMed] [Google Scholar]
- 11.Dubin, D. T. 1990. Tn554 and related DNA: mapping and tracking the staphylococcal chromosome, p. 85-98. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 12.El Solh, N., and J. Allignet. 1998. Staphylococcal resistance to streptogramins and related antibiotics. Drug Resist. Update 1:169-175. [DOI] [PubMed] [Google Scholar]
- 13.Haroche, J., J. Allignet, S. Aubert, A. E. van den Bogaard, and N. El Solh. 2000. satG, conferring resistance to streptogramin A, is widely distributed in Enterococcus faecium strains but not in staphylococci. Antimicrob. Agents Chemother. 44:190-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haroche, J., J. Allignet, C. Buchrieser, and N. El Solh. 2000. Characterization of a variant of vga(A) conferring resistance to streptogramin A and related compounds. Antimicrob. Agents Chemother. 44:2271-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh, P.-C., J.-P. Xiao, D. O'Loane, and S.-Y. Xu. 2000. Cloning, expression, and purification of a thermostable nonhomodimeric restriction enzyme, BslI. J. Bacteriol. 182:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and H. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreiswirth, B. N., G. R. Kravitz, P. M. Schlievert, and R. P. Novick. 1986. Nosocomial transmission of a strain of Staphylococcus aureus causing toxic shock syndrome. Ann. Intern. Med. 105:704-707. [DOI] [PubMed] [Google Scholar]
- 18.Kreiswirth, B. N., S. Lofdahl, M. J. Bethey, M. O'Reilly, P. M. Shlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock exotoxin structural gene is not detectably transmitted by a prophage. Nature 306:709-712. [DOI] [PubMed] [Google Scholar]
- 19.Krolewski, J. J., E. Murphy, R. P. Novick, and M. G. Rush. 1981. Site specificity of the chromosomal insertion of Staphylococccus aureus transposon Tn554. J. Mol. Biol. 152:19-33. [DOI] [PubMed] [Google Scholar]
- 20.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessières, A. Bolotin, S. Borchert, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 21.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, et al. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 22.Martineau, F., F. J. Picard, N. Lansac, C. Ménard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 2000. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy, E. 1990. Properties of the site-specific transposable element Tn554, p. 123-135. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 24.Murphy, E., L. Huwyler, and M. do Carmo de Freire Bastos. 1985. Transposon Tn554: complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants. EMBO J. 4:3357-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy, E., and S. Lofdahl. 1984. Transposition of Tn554 does not generate a target duplication. Nature 307:292-294. [DOI] [PubMed] [Google Scholar]
- 26.Murphy, E., S. Phillips, I. Edelman, and R. P. Novick. 1981. Tn554: isolation and characterization of plasmid insertions. Plasmid 5:292-305. [DOI] [PubMed] [Google Scholar]
- 27.Murphy, E., E. Reinheimer, and L. Huwyler. 1991. Mutational analysis of att554, the target of the site-specific transposon Tn554. Plasmid 26:20-29. [DOI] [PubMed] [Google Scholar]
- 28.Novick, R. P. 1990. The Staphylococcus as a molecular genetic system, p. 1-37. In R. P. Novick (ed.), Molecular biology of staphylococci. VCH Publishers, New York, N.Y.
- 29.Phillips, S., and R. P. Novick. 1979. Tn554-a site-specific repressor-controlled transposon in Staphylococcus aureus. Nature 278:476-478. [DOI] [PubMed] [Google Scholar]
- 30.Rende-Fournier, R., R. Leclercq, M. Galimand, J. Duval, and P. Courvalin. 1993. Identification of the satA gene encoding a streptogramin A acetyltransferase in Enterococcus faecium BM4145. Antimicrob. Agents Chemother. 37:2119-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. Bogo-Jensen, J. Rood, and H. Seppala. 1999. Nomemclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 42:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thakker-Varia, S., W. D. Jenssen, L. Moon-McDermott, M. P. Weinstein, and D. T. Dubin. 1987. Molecular epidemiology of macrolide-lincosamide-streptogramin B resistance in Staphylococcus aureus and coagulase-negative staphylococci. Antimicrob. Agents Chemother. 31:735-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tillotson, L. E., W. D. Jenssen, L. Moon-McDermott, and D. T. Dubin. 1989. Characterization of a novel insertion of the macrolides-lincosamides-steptogramin B resistance transposon Tn554 in methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 33:541-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Townsend, D. E., S. Bolton, N. Ashdown, D. I. Annear, and W. B. Grubb. 1986. Conjugative staphylococcal plasmids carrying hitch-hiking transposons similar to Tn554: intra- and interspecies dissemination of erythromycin resistance. Aust. J. Exp. Biol. Med. 64:367-379. [DOI] [PubMed] [Google Scholar]
- 35.Werner, G., and W. Witte. 1999. Characterization of a new enterococcal gene, satG, encoding a putative acetyltransferase conferring resistance to streptogramin A compounds. Antimicrob. Agents Chemother. 7:1813-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, S., H. de Lencastre, and A. Tomasz. 1998. Genetic organization of the mecA region in methicillin-susceptible and methicillin-resistant strains of Staphylococcus sciuri. J. Bacteriol. 180:236-242. [DOI] [PMC free article] [PubMed] [Google Scholar]