Abstract
For the present report, a novel complex class 1 integron, In60, was characterized. Part of this integron includes the blaCTX-M-9 gene and its downstream nucleotide sequence, which shares 81% and 78% nucleotide identity with those of kluA-1 β-lactamase and orf3 of K. ascorbata, respectively. Furthermore, a new insertion sequence, IS3000, has been found in In60. PCR analysis indicates that integron In60 is present in 33 of 34 nonclonal enterobacterial isolates carrying the putative β-lactamase CTX-M-9.
Class 1 integrons are the most common integrons found in gram-negative clinical isolates (5). Members of this class of integrons contain a 5′-conserved segment (5′-CS) harboring the integrase gene (intI1); a promoter region, the attI1 site (within which the 59-be or attC elements of gene cassettes, some of which carry resistance genes, are recombined); and a 3′-conserved segment (3′-CS) that harbors a defective version of qacEΔ1 and a complete sul1 gene (5). Integrons have normally been reported in members of the Enterobacteriaceae family and other gram-negative and gram-positive bacteria. While most integrons of class 1 present the structure described above, three unusual class 1 integrons have been identified: In6, In7 (5, 6, 16), and the integron described for plasmid pSAL-1, from the Salmonella enterica serovar Enteritidis (18). These integrons contain the complete 5′-CS and two copies of part of the 3′-CS (Fig. 1). Gene cassettes are located between the 5′-CS and the first copy of the 3′-CS. Between the two copies of the 3′-CS lies a region of 2.1 kb that is nearly identical in In6, In7, and the integron from pSAL-1 and is followed by a variable region that contains resistance genes. These three integrons, In6, In7, and the integron from pSAL-1, have similar genetic organizations, suggesting that they have a common origin (16, 18).
FIG. 1.
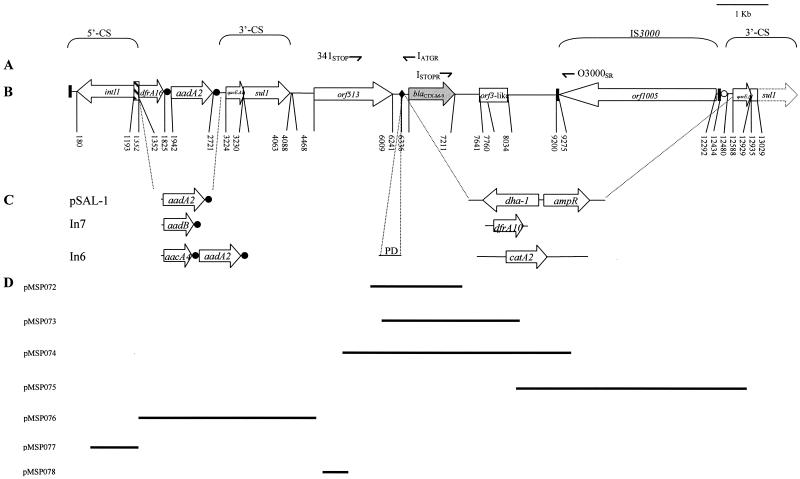
(A) Primers used for studying the environment of In60 (Table 1). (B) Schematic presentation of integron In60 carrying blaCTX-M-9 (accession number AF174129). Open arrows, ORFs; dotted arrow, the unsequenced fragment of the sul1 gene; open box, the region that shows high similarity to orf3 of K. ascorbata; gray arrow, the previously reported sequence (13); •, 59-be; ○, shorter 59-be; diagonally striped box, the recombination site attI1; ♦, hypothetical recombination region; solid boxes, IR. Relevant nucleotide translational start and stop codons of genes and ORFs are numbered. (C) Comparison of the variable regions of different sul1-type complex integrons. The partial duplication of In6 (PD) is also shown (16). (D) Recombinant plasmids (containing fragments of In60) obtained in this work.
In 1996, an Escherichia coli strain (785-D) carrying plasmid pMSP071 containing the extended-spectrum β-lactamase CTX-M-9 was isolated in our laboratory (13). Upstream of the blaCTX-M-9 gene, a sequence having homology with the orf513 present in sul1-type integrons (In6 and In7) was detected (accession number AF174129). This fact suggested that the surrounding region of blaCTX-M-9 is related to a complex integron. Then, several PCR fragments obtained from pMSP071 with primers corresponding to internal regions of In7 and the blaCTX-M-9 gene (Table 1; Fig. 1) were cloned in pGEM-T plasmid (9) and sequenced according to the dideoxy method (4) using fluorescent primers and the Automatic Laser Fluorescent DNA Sequencer (Amersham Pharmacia Biotech).
TABLE 1.
Primer sequences used for PCR analysis of In60
| Primer | Nucleotide sequence (5′ to 3′) | Position in published sequence (bp) | Related feature | Accession numbera |
|---|---|---|---|---|
| 5′-CS | GGC ATC CAA GCA GCA AG | 1190-1206 | Conserved sequence of 5′-CS integrons | M73819 |
| 341IR | GTA ACC GTT TGT TTG AGT GGG | 3627-3607 | Internal sequence of orf513 | L06418 |
| 5′-CSR | CTT GCT GCT TGG ATG CC | 1206-1190 | Reverse and complementary sequence of 5′-CS integrons | M73819 |
| IR | TGT CAT TTT CAG AAG ACG ACT GCA C | 214-238 | Belonging to the IR sequence present in class 1 and 2 integrons | X72585 |
| 785up | GGC TGT TTT CAC TCT CG | 8962-8978 | Internal sequence of In60 | AF174129 |
| sul1 | TGA AGG TTC GAC AGC AC | 1463-1447 | Internal sequence of sul1 | X12869 |
| 341A | CGC CCA CTC AAA CAA ACG | 3605-3622 | Internal sequence of orf513 | L06418 |
| IATG | GTG ACA AAG AGA GTG CAA CGG | 6339-6359 | Reverse and complementary sequence corresponding to blaCTX-M-9 translational starting codon region | AF174129 |
| ISTOP | ATG ATT CTC GCC GCT GAA GCC | 7195-7175 | Sequence corresponding to blaCTX-M-9 translational stop codon region | AF174129 |
| IATGR | CCG TTG CAC TCT CTT TGT CAC | 6359-6339 | Reverse and complementary sequence of blaCTX-M-9 translational starting codon region | AF174129 |
| 341STOP | ACA TTA GTC GGC CAG CGG | 5453-5470 | Internal sequence of orf513 | AF174129 |
| O3000SR | GGA GCT TAT GCG CTC AAT CG | 9323-9304 | Reverse complementary sequence of orf1005 translational stop codon region | AF174129 |
| ISTOPR | GGC TTC AGC GGC GAG AAT CAT | 7175-7195 | Sequence of blaCTX-M-9 translational stop codon region | AF174129 |
| 341B | GAG GCT TTG GTG TAA CCG | 4073-4056 | Internal sequence of orf513 | L06418 |
Accession numbers are for the EMBL/GenBank database.
The nucleotide sequences of these overlapping PCR fragments confirmed that the blaCTX-M-9 gene is inserted into a complex sul1-type integron, which has been designated as integron 60 (In60).
Characterization of the 5′-CS to the first 3′-CS region.
The first 1,332 nucleotides of In60 are identical to those of the other class 1 integrons, including the intI1 gene and the attI1 site (5). Two different gene cassettes were detected between the 5′-CS and the first 3′-CS. The first one contains a point mutant derivative of the dfrA16 gene encoding a dihydrofolate reductase (accession number AJ131405). The sequence of this gene cassette was almost identical to the previously described sequence, except at the position of nucleotide 281 (numbered with respect to its position in the gene cassette; bp 1 and 2, TT; last base, G) (nucleotide 1613 in In60), which changed from nucleotide G to T. The second gene cassette contained an aadA2 gene encoding an aminoglycoside adenylyltransferase. The nucleotide sequence of aadA2 was nearly identical to that previously found in both the In6 carried in plasmid pSa (accession number L06822) (1) and another sul1-type integron contained in pSAL-1 (accession number AJ237702) (18). The only discrepancies were at nucleotides 398 and 495 (with respect to position on the gene cassette [nucleotides 2318 and 2415 in In60]), which changed from nucleotide A to G with a consequent amino acid change from Asp to Gly and from A to G with no amino acid change, respectively. Nevertheless, there was some difference between the imperfect inverted repeats (IRs) of its 59-be and those of aadA2 from pSa and pSAL-1. All of these nucleotide discrepancies were verified in two independent PCR-cloned fragments. Following the 59-be of aadA2, the 3′-CS conserved region of class 1 integrons (positions 1 to 1313 of the 3′-CS), including the qacEΔ1 and sul1 genes, was detected in In60 (Fig. 1) (from nucleotide 2775 to 4087 in In60).
The internal In60 common region.
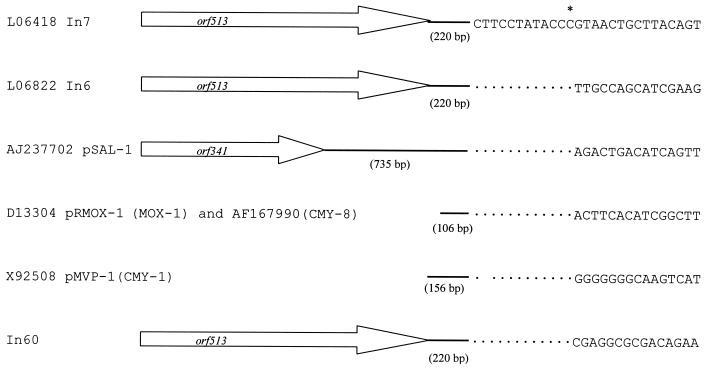
Downstream of the first sul1 gene, a region common to the three complex sul1-type integrons is found (from nucleotide 4088 to 6241 in In60). This region includes orf513, previously called orf341 which is present in In6 and In7 (accession numbers L06822 and L06418, respectively). The first 320 residues of orf513 are identical to those of orf341 from pSAL-1; however, by insertion of a G in position 960 of the pSAL-1 orf341 nucleotide sequence (with respect to its hypothetical translational starting codon), a longer open reading frame (ORF) corresponding to a 513-amino acid protein which exhibits 100% identity with our orf513 may be generated, suggesting that orf341 is a truncated derivative of orf513.
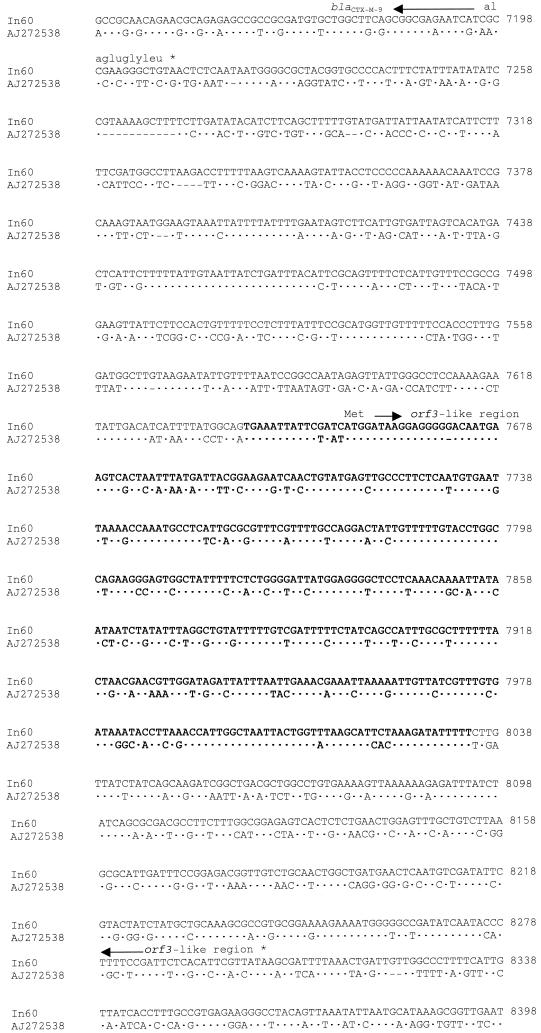
Orf513 is 65% identical to OrfA of E. coli 10507 (accession number AAG16658) (2). It has been proposed (16-18) that orf341 encodes a DNA recombinase, the preferential site of which is downstream of its encoding region and is the site at which insertion of different resistance genes into the complex structure of the integron takes place (Fig. 2). This hypothetical preferential site is also present 232 bp downstream of the orf513 stop codon in In60 (nucleotide 6241). At a location 99 nucleotides downstream of this boundary, the gene encoding the β-lactamase CTX-M-9 is found. A potential promoter, which shows similarity to the E. coli consensus promoter, was identified just upstream of the above-mentioned hypothetical recombination site which, as has been proposed for dfrA10 of In7, could be the promoter of blaCTX-M-9 (11). It has also been proposed that the kluA-1 or kluA-2 gene from Kluyvera ascorbata or kluC-1 from Kluyvera cryocrescens (3, 10) could be an ancestor of some plasmid-encoded CTX-M type β-lactamases, including CTX-M-9, because all share between 79.4% and 80.4% nucleotide identity rates with CTX-M-9. It is noteworthy that 429 bp downstream of the stop codon of blaCTX-M-9, a region of 394 bp (from nucleotide 7641 to 8034 of In60), showing a 78% identity with orf3 from K. ascorbata (accession number AJ272538), was observed (Fig. 3). The orf3 (645 bp long and encoding a putative protein of 215 amino acids) has been described as being close to the chromosomal gene responsible for production of the class A β-lactamase KluA-1 of K. ascorbata. Furthermore, the sequence between the blaCTX-M-9 gene and the orf3-like region is also quite extensively preserved (Fig. 3). Identification of this orf3-like downstream sequence of blaCTX-M-9 gives an additional clue to the origin of at least some CTX-M β-lactamase genes, suggesting that this region could come from a Kluyvera relative strain. Comparison of this orf3-like sequence with orf3 revealed one possible nucleotide insertion at the 13th nucleotide from the translational starting point of the former, at which nucleotide a reading frame change originates (Fig. 3). Following this orf3-like region, a new insertion sequence, designated IS3000, is found (extending from nucleotide 9200 to 12434 of In60). This IS3000 is composed of an ORF 3,015 bp long (orf1005) that is oriented in the opposite direction to the other ORFs (Fig. 1) and is flanked by two imperfect 83-bp IRs (Fig. 4). The deduced amino acid sequence of orf1005 had 28% amino acid identity with the amino acid sequence of the Tn5501 transposase of Pseudomonas putida (8).
FIG. 2.
Comparison of regions to which the proposed DNA recombinase preferential site of In60 and of other sul1-type complex integrons and/or plasmids belong (18). The β-lactamase gene detected downstream of these regions is presented in brackets. Solid lines and dots indicate nucleotide identities, and the asterisk represents the point break where the nucleotide sequence changes. In6 contains a direct tandem duplication of the last 392 bases of the common region (16).
FIG. 3.
Comparison of the region between blaCTX-M-9 and orf1005 of the In60 sequence and the region between the kluA-1 and orf3 genes of Kluyvera ascorbata (AJ272538). The relevant amino acid and translational stop codons (∗) are indicated. Dashes and dots indicate gaps and identical nucleotides, respectively. Nucleotide positions in In60 are shown. Regions sharing 78% identity between both sequences are in bold.
FIG. 4.
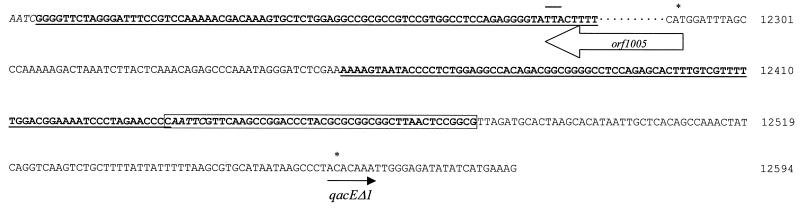
Sequence of the imperfect 83-bp IR (in bold and underlined) flanking orf1005. Imperfect direct duplications found at both ends of IS3000 are indicated in italics. The remnant sequence of the aadA2 59-be is boldface and boxed. The translational starting points of both orf1005 and qacEΔ1 are marked with asterisks, and the stop codon of orf1005 is overlined. Nucleotide positions in In60 are shown.
Orf1005 presents the DD(35)E motif, which is highly conserved in the C-terminal end of transposases (7). However, we were unable to detect IS3000 transposition, despite repeated attempts to do so by using killer-trap vectors according to a previously reported strategy (15). Therefore, the transposition ability of this putative insertion sequence remains to be determined. Moreover, no 59-be was detected at the end of either blaCTX-M-9, the orf3-like region, or orf1005.
Close to the downstream IS3000 IR, there is a 45-bp region which exhibits total identity with the last 45 nucleotides of the 59-be of the preceding aadA2 cassette. Immediately after this region, the second 3′-CS (only nucleotides 1 to 550 were sequenced), which includes the qacEΔ1 and sul1 genes, was found (nucleotides 12480 to 13029 in In60) (Fig. 4).
Despite the fact that different PCR analyses were carried out using primers belonging to the characteristic 25-bp IR present in integrons of class 1 (Table 1) (5), the presence of this IR at the 3′ end of In60 was not detected. Nevertheless, the IR primer allowed us to identify the 5′ sequence of In60.
Presence of In60 in other strains carrying putative β-lactamase CTX-M-9.
A total of 34 enterobacteria (30 E. coli and four S. enterica strains) from different and independent sources carrying putative β-lactamase CTX-M-9 (according to susceptibility pattern, isoelectric point, and PCR analysis) (13, 14) were isolated during the years 1996 to 1999 in our hospital. The 30 E. coli strains did not correspond to a single epidemic strain. Several groups can be established according to their biotypes, as determined by the method proposed by Richard (12) (data not shown). The four S. enterica strains correspond to the serovar Virchow (14). Analysis of the surrounding regions of blaCTX-M-9 by PCR using the 341STOP-IATGR and ISTOPR-O3000SR (Table 1) set of primers showed that 29 of 30 strains had the expected 907- and 2,149-bp regions between blaCTX-M-9 and both orf513 and orf1005, respectively. In the remaining E. coli strain, the 907-bp PCR band (containing orf513) was also detected, but a 600-bp band was obtained instead of the 2,149-bp band (accession number AY092058). The nucleotide sequence of this 600-bp fragment showed a deletion of 1,549 bp, which starts 449 bp downstream of the blaCTX-M-9 stop codon and ends 67 bp before the orf1005 translational stop codon and includes part of an IR (from nucleotide 7660 to 9208 in In60) (Fig. 1).
Comparative analysis of In60 with other complex sul1-type integrons.
In addition to In60, other complex integrons (In6, In7, and the integron in the pSAL-1 plasmid), as cited above, have been identified. In the 5′-CS, In60 and the pSAL-1 integron have a weak P1 promoter and a second strong promoter, P2, which was created by insertion of three G residues between preexisting −10 and −35 motifs to extend the spacing to 17 bp (5). The In6 and In7 integrons did not have these nucleotide insertions.
Between the 5′-CS and the first 3′-CS region, the complex integrons pSAL-l, In6, and In60 present an aadA2 cassette. Two of these integrons (In6 and In60) each have an unrelated additional cassette (aacA4 and dfrA16, respectively), and In7 contains the aadB cassette. In both 3′-CS regions, important differences are detectable between In60 and other complex integrons known. Thus, in In60 there is a 6.4-kb DNA fragment which includes blaCTX-M-9, the orf3-like region, and orf1005, while in In6, In7, and pSAL-1, there are a catA2 gene contained in a 2.4-kb fragment, a dfrA10 gene carried in a 0.7-kb fragment, and ampC and ampR genes contained in a 2.5-kb fragment, respectively (Fig. 1). Furthermore, and in contrast to what happens in In6, In7, and pSAL-1, the second 3′-CS of In60 has not lost any part of its 5′ end.
In conclusion, In60 is the fourth complex sul1-type integron described so far and the first one reported to contain two transposase-like regions (Orf513 and Orf1005). The knowledge concerning the new complex sul1-type integrons will be useful to clarify the origin and evolution of these types of integrons.
The accession number of the In60 is AF174129, which represents an update of the previously deposited sequence, AF174129 (containing blaCTX-M-9), and includes AF373103 (containing the 5′-CS and 3′-CS regions) and AF373104 (containing the region between the two 3′-CS regions).
Acknowledgments
We are grateful to P. Nordmann for helpful advice and comments.
This work was supported partially by grants FIS 97/0623, FIS 98/1522, and 2001SGR-206. We thank the Fundación M Francisca de Roviralta for financial support.
REFERENCES
- 1.Bito, A., and M. Susani. 1994. Revised analysis of aadA2 gene of plasmid pSa. Antimicrob. Agents Chemother. 38:1172-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cloeckaert, A., S. Baucheron, G. Flaujac, S. Schwarz, C. Kehrenberg, J. L. Martel, and E. Chaslus-Dancla. 2000. Plasmid-mediated florfenicol resistance encoded by the floR gene in Escherichia coli isolated from cattle. Antimicrob. Agents Chemother. 44:2858-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decousser, J. W., L. Poirel, and P. Nordmann. 2001. Characterization of a chromosomally encoded extended-spectrum class A β-lactamase from Kluyvera cryocrescens. Antimicrob. Agents Chemother. 45:3595-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandez de Henestrosa, A. R., E. Rivera, A. Tapias, and J. Barbé. 1998. Identification of the Rhodobacter sphaeroides SOS box. Mol. Microbiol. 28:991-1003. [DOI] [PubMed] [Google Scholar]
- 5.Hall, R. M., and C. M. Collis. 1998. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updates 1:109-119. [DOI] [PubMed] [Google Scholar]
- 6.Hall, R. M., and H. W. Stokes. 1990. The structure of a partial duplication in the integron of plasmid pDG100. Plasmid 23:76-79. [DOI] [PubMed] [Google Scholar]
- 7.Haren, L., B. Ton-Hoang, and M. Chandler. 1999. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 53:245-281. [DOI] [PubMed] [Google Scholar]
- 8.Lauf, U., C. Muller, and H. Herrmann. 1998. The transposable elements resident on the plasmids of Pseudomonas putida strain H, Tn5501 and Tn5502, are cryptic transposons of the Tn3 family. Mol. Gen. Genet. 259:674-678. [DOI] [PubMed] [Google Scholar]
- 9.Miller, J. M. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Oliver, A., J. C. Pérez-Díaz, T. M. Coque, F. Baquero, and R. Cantón. 2001. Nucleotide sequence and characterization of a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-10) isolated in Spain. Antimicrob. Agents Chemother. 45:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons, Y., R. M. Hall, and H. W. Stokes. 1991. A new trimethoprim resistance gene, dhfrX, in the In7 integron of plasmid pDG0100. Antimicrob. Agents Chemother. 35:2436-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richard, C. 1981. Une méthode simple de marquage épidémiologique: la biotypie, application a Enterobacter cloacae et Escherichia coli. Bull. Assoc. Anciens Élèves Inst. Pasteur 87:14-21. [Google Scholar]
- 13.Sabaté, M., R. Tarragó, F. Navarro, E. Miró, C. Vergés, J. Barbé, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simarro, E., F. Navarro, J. Ruiz, E. Miró, J. Gómez, and B. Mirelis. 2000. Salmonella enterica serovar Virchow with CTX-M-like β-lactamase in Spain. J. Clin. Microbiol. 38:4676-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon, R., B. Hötte, B. Klauke, and B. Kosier. 1991. Isolation and characterization of insertion sequence elements from gram-negative bacteria by using new broad-host-range, positive selection vectors. J. Bacteriol. 173:1502-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stokes, H. W., C. Tomaras, Y. Parsons, and R. M. Hall. 1993. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDG0100 have a common origin. Plasmid 30:39-50. [DOI] [PubMed] [Google Scholar]
- 17.Valentine, C. R., M. J. Heinrich, S. L. Chissoe, and A. Roe. 1994. DNA sequence of direct repeats of the sulI gene of plasmid pSa. Plasmid 32:222-227. [DOI] [PubMed] [Google Scholar]
- 18.Verdet, C., G. Arlet, G. Barnaud, P. H. Lagrange, and A. Phillipon. 2000. A novel integron in Salmonella enterica serovar Enteritidis, carrying the blaDHA-1 gene and its regulator ampR, originated from Morganella morganii. Antimicrob. Agents Chemother. 44:222-225. [DOI] [PMC free article] [PubMed] [Google Scholar]