Abstract
Mutations in the quinolone resistance-determining region of the gyrA gene from 138 ciprofloxacin-resistant (MIC, ≥4 μg/ml) and 24 ciprofloxacin-susceptible (MIC, ≤1 μg/ml) clinical Campylobacter jejuni isolates were subjected to single-strand conformation polymorphism analysis and sequencing. All of the isolates could be assigned to three genotypic clusters based on silent mutations. All resistant isolates had a point mutation at codon 86.
Fluoroquinolones are the antimicrobials most commonly used for treatment of adults with Campylobacter infections. During the 1990s, however, resistance to this antimicrobial group has increased worldwide among Campylobacter spp. (3, 8, 9, 12-15, 18, 19, 21, 22). The primary target of the fluoroquinolones in Campylobacter jejuni has been shown to be DNA gyrase, a type II topoisomerase that is an essential enzyme for DNA replication (2, 20). DNA gyrase is composed of two A subunits and two B subunits encoded by the gyrA and gyrB genes, respectively. A point mutation in the quinolone resistance-determining region (QRDR) of the gyrA gene at codon 86 (ACA to ATA), substituting isoleucine for threonine, is the most common cause of class-wide fluoroquinolone resistance among C. jejuni isolates (17, 24, 27). There have also been anecdotal reports of mutations leading to additional amino acid changes, as well as silent nucleotide mutations (1, 24, 27).
The purpose of the present study was to explore the molecular epidemiology of the QRDR of gyrA among C. jejuni isolates. To this end, we analyzed the QRDR (codons 69 to 120) of gyrA from 138 ciprofloxacin-resistant and 24 ciprofloxacin-susceptible clinical C. jejuni isolates by single-strand conformation polymorphism (SSCP) analysis and DNA sequencing.
A total of 162 clinical C. jejuni isolates collected in Finland between 1995 and 2000 were included in this study. All isolates were from stool samples, and the majority (n = 158) were collected from Finnish travelers returning from 22 different countries. Four isolates were from Finnish patients without any travel history. One hundred thirty-eight of the isolates were resistant to ciprofloxacin (MIC, ≥4 μg/ml), and 24 were ciprofloxacin susceptible (MIC, ≤1 μg/ml). The ciprofloxacin MICs were 8 μg/ml for 10 of the ciprofloxacin-resistant isolates and ≥16 μg/ml for 128 of the ciprofloxacin-resistant isolates. The resistant isolates were from travelers to 20 different countries, the majority being from travelers to Spain (34%) and Thailand (21%). The susceptible isolates were from travelers to 12 countries, the number of isolates varying between one and three from each country.
Chromosomal DNA from each strain was prepared by boiling for 10 min and proteinase K (Promega, Madison, Wis.) digestion for 30 min. DNA was amplified by PCR for the gyrA gene as described by Wang et al. (24). One microliter of [α-32P]dCTP (activity, 10 μCi/μl) was added to each PCR mixture for SSCP. SSCP was run in accordance with the manufacturer's instructions on nondenaturing MDE Gel (BioWhittaker Molecular Applications, Rockland, Maine) at 4°C. Autoradiograms were incubated overnight. The QRDR of the gyrA gene was sequenced in both directions with PCR primers from one or more representative isolates belonging to a different SSCP pattern (Table 1). Sequencing of a nonradioactive PCR product was performed with an ABI Prism BigDye Terminator Kit (Applied Biosystems, Foster City, Calif.). All of the gyrA sequences of the different SSCP patterns were confirmed by PCR and sequencing with two additional oligonucleotide primers, Cj-gyrA-393 (5′-CTTTGCCTGACGCAAGAG-3′) and Cj-gyrA-759r (5′-TCGCTTTCTGAACCATCA-3′). One representative isolate of every different SSCP pattern was confirmed to be C. jejuni by 16S rRNA gene sequencing (6). C. jejuni RH3583 was used as a control strain.
TABLE 1.
SSCP patterns and QRDRs of gyrA of 138 ciprofloxacin-resistant and 24 ciprofloxacin-susceptible C. jejuni isolates
| SSCP pattern | No. (%) of isolates analyzeda
|
Ciprofloxacin MIC range (μg/ml) | Nucleic acid codons and corresponding amino acids of C. jejuni QRDR of gyrAb
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP-S | CIP-R | Codon | Amino acid | Codon | Amino acid | Codon | Amino acid | Codon | Amino acid | Codon | Amino acid | Codon | Amino acid | Codon | Amino acid | Codon | Amino acid | ||
| ICIP-S | 7 (29) | 0.064-0.5 | GGT | Gly-78 | CAC | His-81 | ACA | Thr-86 | GAT | Asp-90 | CCA | Pro-104 | GGC | Gly-110 | AGT | Ser-119 | GCC | Ala-120 | |
| ICIP-R1 | 21 (15) | 16->64 | --- | - | --- | - | -T- | Ile-86 | --- | - | --- | - | --- | - | --- | - | --- | - | |
| ICIP-R2 | 1 (0.7) | >64 | --- | - | --- | - | -T- | Ile-86 | A-- | Asn-90 | --- | - | --- | - | --- | - | --- | - | |
| ICIP-R3 | 1 (0.7) | 16 | --- | - | --- | - | -T- | Ile-86 | --- | - | T-- | Ser-104 | --- | - | --- | - | --- | - | |
| IICIP-S | 2 (8) | 0.25-0.5 | --C | - | --- | - | --- | - | --- | - | --- | - | --- | - | --C | - | --- | - | |
| IIICIP-S | 15 (63) | 0.125-1 | --- | - | --T | - | --- | - | --- | - | --- | - | --- | - | --C | - | --T | - | |
| IIICIP-R1 | 113 (82) | 8->64 | --- | - | --T | - | -T- | Ile-86 | --- | - | --- | - | --- | - | --C | - | --T | - | |
| IIICIP-R2 | 2 (1.4) | 32 | --- | - | --T | - | -T- | Ile-86 | --- | - | --- | - | --T | - | --C | - | --T | - | |
Number of isolates sequenced per SSCP pattern: ICIP-S, 4; ICIP-R1, 2; ICIP-R2, 1; ICIP-R3, 1; IICIP-S, 2; IIICIP-S, 9; IIICIP-R1, 12; IIICIP-R2, 2. CIP-S, ciprofloxacin susceptible (MIC, ≤1 μg/ml); CIP-R, ciprofloxacin resistant (MIC, ≥4 μg/ml).
-, no change.
On the basis of the SSCP patterns of the QRDR of gyrA, all 138 ciprofloxacin-resistant C. jejuni isolates could be distinguished from the 24 susceptible isolates. Eight different SSCP patterns were found: five among the ciprofloxacin-resistant isolates and three among the ciprofloxacin-susceptible isolates. These patterns were designated as follows. Roman numerals I to III were used to define the three susceptible SSCP patterns. The subscript CIP-S indicates ciprofloxacin susceptibility, and the subscript CIP-R indicates ciprofloxacin resistance. The numerals after the subscript CIP-R define resistant variants within each cluster.
The most common susceptible SSCP pattern was IIICIP-S, with 15 (63%) isolates, followed by patterns ICIP-S and IICIP-S, with 7 (29%) and 2 (8%) isolates, respectively (Fig. 1). The most common resistant SSCP pattern, IIICIP-R1, was exhibited by 113 (82%) resistant isolates. Pattern ICIP-R1 was exhibited by 21 (15%) resistant isolates, pattern IIICIP-R2 was exhibited by 2 (1.4%) resistant isolates, and patterns ICIP-R2 and ICIP-R3 were exhibited by 1 resistant isolate each.
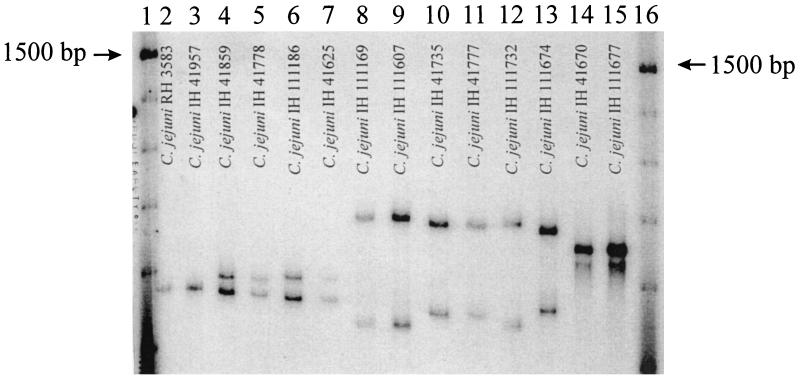
FIG. 1.
Autoradiogram of SSCP patterns of the QRDR of gyrA of C. jejuni isolates. SSCP patterns are designated as explained in the text and Table 1. Lanes: 1 and 16, 100-bp DNA ladder (Gibco BRL, Life Technologies, Inc., Gaithersburg, Md.) labeled with [γ-32P]ATP by using T4 polynucleotide kinase; 2 and 3, ciprofloxacin-susceptible isolates showing pattern IIICIP-S; 4 and 5, ciprofloxacin-resistant isolates showing pattern IIICIP-R1; 6 and 7, ciprofloxacin-resistant isolates showing pattern III CIP-R2; 8 and 9, ciprofloxacin-susceptible isolates showing pattern ICIP-S; 10 and 11, ciprofloxacin-resistant isolates showing pattern ICIP-R1; 12, ciprofloxacin-resistant isolate showing pattern ICIP-R2; 13, ciprofloxacin-resistant isolate showing pattern ICIP-R3; 14 and 15, ciprofloxacin-susceptible isolates showing pattern IICIP-S.
We sequenced the gyrA QRDRs of 1 to 12 representative C. jejuni isolates of all eight defined SSCP patterns (Table 1). Compared to ciprofloxacin-susceptible C. jejuni UA580 (24) and C. jejuni NCTC 11168 (11), all of the resistant isolates sequenced had a point mutation at codon 86 (ACA to ATA), substituting isoleucine for threonine. Two resistant isolates harbored one additional mutation leading to an amino acid change each. In addition, silent mutations were identified among isolates with SSCP patterns IIICIP-R1 and IIICIP-R2 (Table 1).
Among members of the ciprofloxacin-susceptible C. jejuni population, the QRDRs of gyrA from the representative isolates (C. jejuni IH 111169 and C. jejuni IH 111607; Fig. 1 and Table 1) showing SSCP pattern ICIP-S were identical to those of C. jejuni UA580 (24) and C. jejuni NCTC 11168 (11). None of the susceptible C. jejuni isolates had mutations leading to amino acid changes. However, silent nucleic acid mutations were observed. The QRDR of gyrA from the representative isolates (C. jejuni IH 41670 and C. jejuni IH 111677; Fig. 1 and Table 1) showing SSCP pattern IICIP-S and from those showing SSCP pattern IIICIP-S (C. jejuni RH 3583 and C. jejuni IH 41957; Fig. 1 and Table 1) had mutations in two and three codons, respectively.
Genetic variation of the QRDR of gyrA in C. jejuni was demonstrated here by the finding of eight SSCP patterns among our whole study population and, in particular, by identification of three defined SSCP patterns among members of the ciprofloxacin-susceptible population. Considering the relatively small number of susceptible isolates studied, it seems likely that additional variants of C. jejuni do exist. This polymorphism is surprising because bacterial topoisomerase genes are generally highly conserved even across the borders of the bacterial genera (4, 5, 7, 10, 16, 23, 24). However, the present findings are in line with the results of the C. jejuni genome sequencing project reported by Parkhill et al. (11), who found hypervariability in the sequence of C. jejuni NCTC 11168. They suspected that C. jejuni might be lacking in DNA repair functions. They also assumed that it may not be possible to produce a single definitive sequence for the C. jejuni genome (11).
Since fluoroquinolones are currently recommended as the first-choice empirical therapy for adult patients with suspected bacterial gastroenteritis, recognition of C. jejuni as the causative agent of a patient's disease is important, as is its susceptibility to antimicrobial agents. Many studies have shown that the Thr-86-Ile mutation in the QRDR of gyrA is the most common mechanism causing ciprofloxacin resistance in C. jejuni (17, 24, 27). Consequently, several molecular techniques have been developed to detect this particular mutation (25-27). If specific primers or probes are used, the nucleic acid variation on the areas described in this report or in previous reports (1, 24, 27) might confuse the primer-target interaction and cause false results. This should be considered in clinical laboratories if traditional susceptibility testing methods are replaced with modern PCR- or hybridization-based methods to detect the mutation at codon 86 (Thr to Ile).
A total of 130 ciprofloxacin-resistant and 23 ciprofloxacin-susceptible C. jejuni isolates from patients with an identified travel history were evaluated for their SSCP pattern profiles (Table 2). Of the five different resistant genotypic variants, two SSCP patterns were dominant while the other three were exhibited by merely one or two resistant isolates. The main pattern, defined as IIICIP-R1, which was presented by 113 (82%) resistant isolates, has also prevailed in earlier studies (27). This pattern seems to have spread all over the world, whereas pattern ICIP-R1 was found mainly in Europe. Unexpectedly, pattern ICIP-S, which is identical to the C. jejuni wild-type sequence described by Wang et al. (24), was not our leading susceptible variant. Pattern IIICIP-S exceeded this wild-type pattern in frequency, with a percentages of 63 versus 29%.
TABLE 2.
Geographical distribution of the SSCP patterns of gyrA of 130 ciprofloxacin-resistant and 23 ciprofloxacin-susceptible C. jejuni isolates
| Continent (no. of countries) | No. of isolatesa
|
No. of isolates with SSCP pattern:b
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CIP-S | CIP-R | ICIP-S | ICIP-R1 | ICIP-R2 | ICIP-R3 | IICIP-S | IIICIP-S | IIICIP-R1 | IIICIP-R2 | |
| Asia (10) | 7 | 59 | 1 | 3 | 1 | 6 | 54 | 1 | ||
| Africa (3) | 4 | 5 | 4 | 5 | ||||||
| Central America (1) | 0 | 1 | 1 | |||||||
| Europe (9) | 12 | 65 | 6 | 16 | 1 | 2 | 4 | 48 | ||
CIP-S, ciprofloxacin susceptible (MIC, ≤1 mg/ml); CIP-R, ciprofloxacin-resistant (MIC, ≥4 mg/ml).
SSCP patterns are designated as explained in the text and Table 1.
In conclusion, consistent with earlier findings, all of the resistant C. jejuni isolates sequenced here had the Thr-86-Ile mutation in the QRDR of gyrA. Only this mutation distinguished the two main resistant genotypic variants from the two main susceptible variants, suggesting a common origin. Considering that three different SSCP patterns were identified among the relatively small number of susceptible isolates studied, it seems likely that additional variants of C. jejuni exist. Polymorphism of the QRDR of gyrA should be considered when any PCR- or hybridization-based method is used to detect the Thr-86-Ile mutation in gyrA as an indication of C. jejuni fluoroquinolone resistance.
Acknowledgments
We are indebted to Tero Mustalahti for performing sequencing reactions and to Liisa Immonen, Minna Lamppu, Satu Linko, Tiina Muuronen, Erkki Nieminen, and Tuula Randell for technical assistance.
This work was supported by a grant from the Maud Kuistila Memorial Foundation and a special government (EVO) grant from Turku University Central Hospital (both to A.H.).
REFERENCES
- 1.Charvalos, E., E. Peteinaki, I. Spyridaki, S. Manetas, and Y. Tselentis. 1996. Detection of ciprofloxacin resistance mutations in Campylobacter jejuni gyrA by nonradioisotopic single-strand conformation polymorphism and direct DNA sequencing. J. Clin. Lab. Anal. 10:129-133. [DOI] [PubMed] [Google Scholar]
- 2.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaudreau, C., and H. Gilbert. 1998. Antimicrobial resistance of clinical strains of Campylobacter jejuni subsp. jejuni isolated from 1985 to 1997 in Quebec, Canada. Antimicrob. Agents Chemother. 42:2106-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griggs, D. J., K. Gensberg, and L. J. Piddock. 1996. Mutations in gyrA gene of quinolone-resistant Salmonella serotypes isolated from humans and animals. Antimicrob. Agents Chemother. 40:1009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hakanen, A., P. Kotilainen, J. Jalava, A. Siitonen, and P. Huovinen. 1999. Detection of decreased fluoroquinolone susceptibility in salmonellas and validation of nalidixic acid screening test. J. Clin. Microbiol. 37:3572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jalava, J., and E. Eerola. 1999. Phylogenetic analysis of Fusobacterium alocis and Fusobacterium sulci based on 16S rRNA gene sequences: proposal of Filifactor alocis (Cato, Moore and Moore) comb. nov. and Eubacterium sulci (Cato, Moore and Moore) comb. nov. Int. J. Syst. Bacteriol. 49:1375-1379. [DOI] [PubMed] [Google Scholar]
- 7.Kasai, H., K. Watanabe, E. Gasteiger, A. Bairoch, K. Isono, S. Yamamoto, and S. Harayama. 1998. Construction of the gyrB database for the identification and classification of bacteria. Genome Inform. Ser. Workshop Genome Inform. 9:13-21. [PubMed] [Google Scholar]
- 8.Li, C. C., C. H. Chiu, J. L. Wu, Y. C. Huang, and T. Y. Lin. 1998. Antimicrobial susceptibilities of Campylobacter jejuni and coli by using E-test in Taiwan. Scand. J. Infect. Dis. 30:39-42. [DOI] [PubMed] [Google Scholar]
- 9.Moore, J. E., M. Crowe, N. Heaney, and E. Crothers. 2001. Antibiotic resistance in Campylobacter spp. isolated from human faeces (1980-2000) and foods (1997-2000) in Northern Ireland: an update. J. Antimicrob. Chemother. 48:455-457. [DOI] [PubMed] [Google Scholar]
- 10.Moore, R. A., B. Beckthold, S. Wong, A. Kureishi, and L. E. Bryan. 1995. Nucleotide sequence of the gyrA gene and characterization of ciprofloxacin-resistant mutants of Helicobacter pylori. Antimicrob. Agents Chemother. 39:107-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and, B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 12.Pigrau, C., R. Bartolome, B. Almirante, A.-M. Planes, J. Gavalda, and A. Pahissa. 1997. Bacteremia due to Campylobacter species: clinical findings and antimicrobial susceptibility patterns. Clin. Infect. Dis. 25:1414-1420. [DOI] [PubMed] [Google Scholar]
- 13.Rautelin, H., O.-V. Renkonen, and T. U. Kosunen. 1991. Emergence of fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli in subjects from Finland. Antimicrob. Agents Chemother. 35:2065-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reina, J., N. Borrell, and A. Serra. 1992. Emergence of resistance to erythromycin and fluoroquinolones in thermotolerant Campylobacter strains isolated from feces 1987-1991. Eur. J. Clin. Microbiol. Infect. Dis. 11:1163-1166. [DOI] [PubMed] [Google Scholar]
- 15.Reina, J., M. J. Ros, and A. Serra. 1994. Susceptibilities to 10 antimicrobial agents of 1,220 Campylobacter strains isolated from 1987 to 1993 from feces of pediatric patients. Antimicrob. Agents Chemother. 38:2917-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reyna, F., M. Huesca, V. Gonzalez, and L. Y. Fuchs. 1995. Salmonella typhimurium gyrA mutations associated with fluoroquinolone resistance. Antimicrob. Agents Chemother. 39:1621-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz, J., P. Goni, F. Marco, F. Gallardo, B. Mirelis, T. Jimenez De Anta, and J. Vila. 1998. Increased resistance to quinolones in Campylobacter jejuni: a genetic analysis of gyrA gene mutations in quinolone-resistant clinical isolates. Microbiol. Immunol. 42:223-226. [DOI] [PubMed] [Google Scholar]
- 18.Sáenz, Y., M. Zarazaga, M. Lantero, M. J. Gastañares, F. Baquero, and C. Torres. 2000. Antibiotic resistance in Campylobacter strains isolated from animals, foods, and humans in Spain in 1997-1998. Antimicrob. Agents Chemother. 44:267-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sánchez, R., V. Fernández-Baca, M. D. Díaz, P. Muñoz, M. Rodríguez-Créixems, and E. Bouza. 1994. Evolution of susceptibilities of Campylobacter spp. to quinolones and macrolides. Antimicrob. Agents Chemother. 38:1879-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segreti, J., T. D. Gootz, L. J. Goodman, G. W. Parkhurst, J. P. Quinn, B. A. Martin, and G. M. Trenholme. 1992. High-level quinolone resistance in clinical isolates of Campylobacter jejuni. J. Infect. Dis. 165:667-670. [DOI] [PubMed] [Google Scholar]
- 21.Sjögren, E., G.-B. Lindblom, and B. Kaijser. 1997. Norfloxacin resistance in Campylobacter jejuni and Campylobacter coli isolates from Swedish patients. J. Antimicrob. Chemother. 40:257-261. [DOI] [PubMed] [Google Scholar]
- 22.Smith, K. E., J. M. Besser, C. W. Hedberg, F. T. Leano, J. B. Bender, J. H. Wicklund, B. P. Johnson, K. A. Moore, M. T. Osterholm, and I. Team. 1999. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992-1998. N. Engl. J. Med. 340:1525-1532. [DOI] [PubMed] [Google Scholar]
- 23.Taylor, D. E., and A. S.-S. Chau. 1997. Cloning and nucleotide sequence of the gyrA gene from Campylobacter fetus subsp. fetus ATCC 27374 and characterization of ciprofloxacin-resistant laboratory and clinical isolates. Antimicrob. Agents Chemother. 41:665-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, Y., W. M. Huang, and D. E. Taylor. 1993. Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob. Agents Chemother. 37:457-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westin, L., C. Miller, D. Vollmer, D. Canter, R. Radtkey, M. Nerenberg, and J. P. O'Connell. 2001. Antimicrobial resistance and bacterial identification utilizing a microelectronic chip array. J. Clin. Microbiol. 39:1097-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson, D. L., S. R. Abner, T. C. Newman, L. S. Mansfield, and J. E. Linz. 2000. Identification of ciprofloxacin-resistant Campylobacter jejuni by use of a fluorogenic PCR assay. J. Clin. Microbiol. 38:3971-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zirnstein, G., Y. Li, B. Swaminathan, and F. Angulo. 1999. Ciprofloxacin resistance in Campylobacter jejuni isolates: detection of gyrA resistance mutations by mismatch amplification mutation assay PCR and DNA sequence analysis. J. Clin. Microbiol. 37:3276-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]