Abstract
The carbenicillin, gentamicin, kanamycin, streptomycin, spectinomycin, sulfonamide, and tobramycin resistance determinants found on Pseudomonas aeruginosa plasmid R151 have previously been shown to translocate to another plasmid, R388, and it was inferred that a transposon, Tn1404, carried the resistance determinants. Sequencing of the cassette array from the plasmid known as R388::Tn1404 revealed two known gene cassettes, oxa10 and aadB, and a previously unidentified cassette determining resistance to streptomycin and spectinomycin, here designated aadA10, in the order oxa10-aadB-aadA10. These cassettes replaced the dfrB2-orfA cassette array of R388, indicating that movement of the resistance determinants from R151 to R388 resulted from recombinational exchange between two class 1 integrons rather than transposition. The AadA10 protein is most closely related to AadA6 (85% identical) and AadA7 (80% identical). The aadA10 cassette found here has only a simple site containing a 7-bp spacer derived from attI1 in place of a 59-base element and is likely to represent a derivative of the complete cassette. IntI1-mediated deletion of the aadA10 cassette was not detected, indicating that this single simple site is either inactive or only weakly active.
Multiresistance plasmid R151 was originally identified in Pseudomonas aeruginosa strain POW 151, isolated in Chicago, Ill., in or prior to 1973 (5). R151 is an IncP-11 plasmid that confers resistance to carbenicillin, gentamicin, kanamycin, streptomycin, spectinomycin, sulfonamides, and tobramycin (5, 38). The carbenicillin resistance of R151 is due to the PSE-2 β-lactamase (38), since renamed OXA-10. Philippon et al. (38) used recombinant plasmids of unknown structure derived from R151 and broad-host-range plasmid pUZ8 (IncP-1; tetracycline resistant [22]) to move the R151 resistance markers into Escherichia coli. Trimethoprim and sulfonamide resistance plasmid R388 (IncW) (1, 14) was introduced as the target for transposition. Ampicillin-resistant transconjugants were obtained and were also resistant to gentamicin, kanamycin, streptomycin, spectinomycin, sulfonamides, and tobramycin but not to tetracycline. The resistance determinants were then found to be associated with an IncW plasmid, suggesting that they had been translocated to R388, and the unit carrying them was named Tn1404. However, although the plasmid was designated R388::Tn1404, it appeared to be the same size as R388 and did not confer the trimethoprim resistance of R388, implying that this determinant had been lost or inactivated.
The sequence of the oxa10 gene originating from R151 (GenBank accession no. J03427) (23) indicated that it was part of the first gene cassette in a class 1 integron (17, 23). Further sequencing (GenBank accession no. U37105.1) (40) revealed the beginning of an aadB cassette adjacent to the oxa10 cassette, accounting for the gentamicin-adenylating activity (5) and the gentamicin, kanamycin, and tobramycin resistances of strain POW 151. Class 1 integrons usually contain two conserved segments (CS) flanking the antibiotic resistance gene cassettes (45). The 5′-CS (20, 45) includes the intI1 gene, which encodes the IntI1 integrase (27); the attI1 recombination site (36); and the Pc promoter, which directs transcription of cassette genes (10, 45). The 3′-CS can vary in length but usually includes the sul1 sulfonamide resistance determinant (4, 18, 45). Gene cassettes can be inserted between the two CS of the integron by IntI1-catalyzed site-specific recombination between the 59-base element (59-be) of the cassette and the attI1 site of the integron (9), and IntI1 also catalyzes cassette excision (11, 12). As R388 also includes the class 1 integron In3, which contains the dfrB2 (trimethoprim resistance) and orfA (unknown function) cassettes (45), the resistance genes of R151 could have been acquired by R388 via cassette exchange, with the simultaneous loss of trimethoprim resistance.
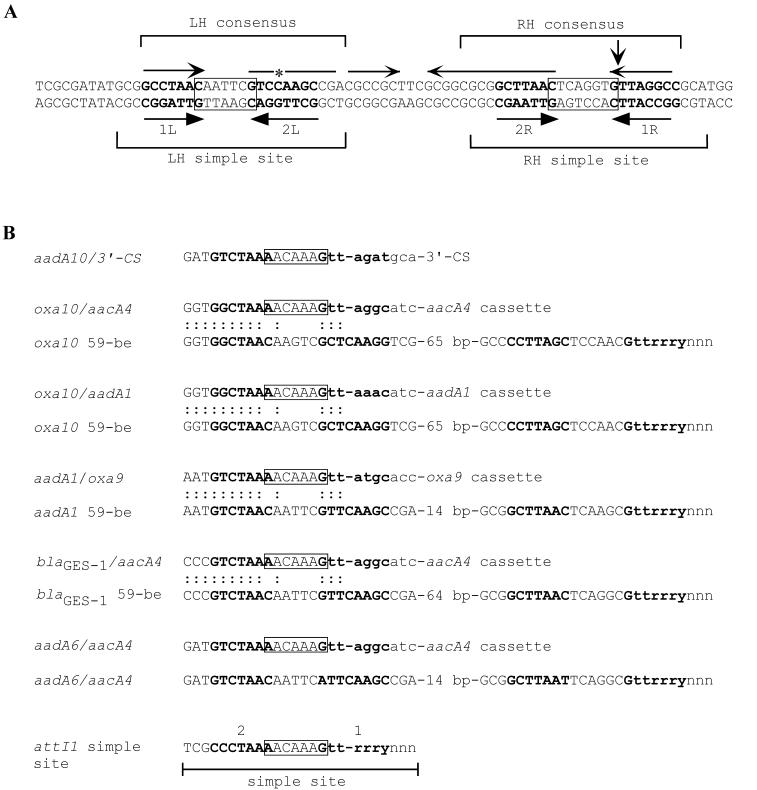
Gene cassettes are small, mobile genetic elements consisting of a single gene (or occasionally two genes) plus a recombination site called the 59-be (6, 12, 17, 40). At least 75 different antibiotic resistance gene cassettes have now been identified; and the genes that they contain confer resistance to aminoglycosides, β-lactams, chloramphenicol, trimethoprim, erythromycin, and rifampin (28, 40; S. R. Partridge and R. M. Hall, unpublished data). The 59-be found in different gene cassettes vary in length (55 to 141 bp) and sequence, but they all contain consensus regions of about 25 bp at each end (11, 17, 46) (Fig. 1A). The consensus regions are imperfect inverted repeats of one another and are separated by a region of variable length that is generally also an inverted repeat (17). Each consensus region contains a simple site (Fig. 1A) made up of a pair of inversely oriented core sites (GTTRRRY) separated by a 7- or 8-bp spacer that overlaps the core sites by 1 bp at each end (46). The extents of these simple sites are inferred from integrase binding data (13, 16). The core sites are designated 1L and 2L at the left-hand (LH) end and 2R and 1R at the right-hand (RH) end (46), and the recombination crossover occurs between the G and the first T in the 1R core site (46). Hence, in the linear, integrated form of a cassette the 1R site, except for the initial G residue, is situated at the start of the cassette. Occasionally, incomplete versions of known cassettes containing a single simple site in place of a 59-be have been found (7, 31, 33, 36, 39, 41). The central part of the 59-be has been lost or replaced by the spacer from the attI1 site of the integron (36). However, the effects of these changes on cassette mobility have not been investigated.
FIG. 1.
Recombination sites associated with gene cassettes. (A) Structure of 59-be. The 59-be from the circular form of the aadB cassette (GenBank accession no. L06418) is shown. Core sites, labeled as described by Stokes et al. (46), are in boldface type, with their relative orientations indicated by the arrows below the sequence. Spacer regions are boxed (note that one base at each end of the spacers is also part of the adjacent core site). Inverted repeat regions are indicated by the arrows immediately above the sequence, and an asterisk indicates the extra base present in 2L compared with the sequence of 2R. The extents of the LH and RH simple sites and consensus regions are also indicated. The vertical arrow shows the position of the recombination crossover point. (B) Simple sites with an attI1 spacer found in derivatives of gene cassettes. The simple site at the end of the R151-derived aadA10 cassette is compared with those of all known cassettes with single simple sites containing the attI1 spacer. The latter are aligned with the 59-be associated with the complete cassette, and colons indicate identity. The attI1 simple site, with the 7-bp core sites labeled as described by Partridge et al. (36) in boldface type and the 7-bp spacer boxed (note that A and G belong both to the spacer and the core site), is shown below. The sequence from the adjacent cassette or 3′ CS is indicated with lowercase letters. The sources of the sequences are as follows: oxa10 and aacA4, GenBank accession no. Z22590; oxa10, GenBank accession no. U37105; oxa10 and aadA1, GenBank accession no. AF205943; aadA1 and oxa9, GenBank accession no. M55547; aadA1, GenBank accession no. X12870; blaGES-1 and aacA4, GenBank accession no. AF156486; blaIBC-1 (equivalent to blaGES-1), reference 15; aadA6 and aacA4, GenBank accession no. AF453998; and aadA6, GenBank accession no. AF140629.
In the study described here, we have investigated the possibility that the loss of trimethoprim resistance observed on formation of R388::Tn1404 could be due to replacement of the R388 cassette array by the one that is present on pUZ8-R151. The sequence of the cassette array in R388::Tn1404 was determined, and a new aadA gene cassette was identified.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli DH5α [supE44 ΔlacU169 (φ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] was used to propagate plasmid DNA. R388::Tn1404 in E. coli strain C600 was kindly supplied by G. A. Jacoby and was confirmed to confer resistance to ampicillin, gentamicin, kanamycin, streptomycin, spectinomycin, and sulfamethoxazole. It was transformed into DH5α with selection for sulfamethoxazole and spectinomycin. Descriptions of the plasmids used in the study are provided in Table 1. The 5.3-kb BamHI fragment from R388::Tn1404 was cloned in pACYC184 (8) by standard procedures (42), and plasmids with the insert in opposite orientations were designated pRMH549 and pRMH857. pRMH549Δoxa10 and pRMH549ΔaadB were obtained by IntI1-mediated deletion of the aadB cassette or the oxa10 cassette, respectively, from pRMH857 to give the cassette orderes aadB-aadA10 and oxa10-aadA10. pRMH260 contains part of integron In6 from pSa, consisting of 567 bp of the 5′ CS, the aacA4-aadA2 cassette array, and 193 bp of the 3′ CS, in pACYC184 (10). A SalI-ClaI fragment containing this truncated integron was end filled and ligated to EcoRV-digested pACYC184, and a plasmid with the integron region in the orientation opposite that in pRMH260 was designated pRMH328. Bacteria were routinely cultured at 37°C in Luria-Bertani (LB) medium or on LB agar, with Mueller-Hinton agar being used to select for resistance to trimethoprim and sulfamethoxazole. The following antibiotics (Sigma) were added as appropriate at the indicated concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 25 μg ml−1; gentamicin, 2 or 8 μg ml−1; kanamycin, 15 μg ml−1; streptomycin, 25 μg ml−1; spectinomycin, 25 μg ml−1; trimethoprim, 25 μg ml−1; and sulfamethoxazole, 25 μg ml−1.
TABLE 1.
Plasmids used in the study
| Plasmid | Description | Relevant phenotypea | Reference |
|---|---|---|---|
| R388 | 33-kb IncW plasmid containing In3; cassette order, dfrB2-orfA | Sur Tpr Tra+ | 1 |
| R388::Tn1404 | “Tn1404” transposed into R388 | Apr Cbr Gmr Kmr Smr Spr Sur | 38 |
| pSU2056 | 1.2-kb BamHI-RsaI fragment from Tn21 inserted into pUC9 | Apr IntI1+ | 27 |
| pRMH260 | 2.3 kb of In6 from pSa in pACYC184; cassette order, aacA4-aadA2 | Cmr Kmr Smr Spr | 10 |
| pRMH328 | 2.3 kb of In6 from pSa in pACYC184 in opposite orientation to pRMH260; cassette order, aacA4-aadA2 | Cmr Kmr Smr Spr | This work |
| pRMH549 | 5.3-kb BamHI fragment of R388::Tn1404 in pACYC184 | Apr Cbr Cmr Gmr Smr Spr Sur | This work |
| pRMH857 | 5.3-kb BamHI fragment of R388::Tn1404 in pACYC184 | Apr Cbr Cmr Gmr Smr Spr Sur | This work |
| pRMH549Δoxa10 | IntI1-mediated deletion of oxa10 cassette from pRMH549; cassette order, aadB-aadA10 | Cmr Gmr Smr Spr Sur | This work |
| pRMH549ΔaadB | IntI1-mediated deletion of aadB cassette from pRMH549; cassette order, oxa10-aadA10 | Apr Cbr Cmr Smr Spr Sur | This work |
Ap, ampicillin; Cb, carbenicillin; Cm, chloramphenicol; Km, kanamycin; Sm, streptomycin; Sp, spectinomycin; Su, sulfonamide; Tc, tetracycline; Tp, trimethoprim.
DNA isolation and restriction mapping.
Plasmid DNA for restriction analysis and cloning was isolated by an alkaline lysis method (2). Restriction enzymes were used in accordance with the manufacturers' instructions. Fragments were separated by electrophoresis on 1% (wt/vol) agarose gels and visualized by staining with ethidium bromide. EcoRI-digested bacteriophage SPP1 DNA (Geneworks) and HindIII-digested bacteriophage λ DNA (Progen) were used as size markers. Plasmid DNA for sequencing was purified with Wizard maxiprep (Promega) or Jetstar midi prep (Genomed) kits.
DNA sequencing and analysis.
The DNA sequence on at least one strand of the insert in pRMH857 was determined. The sequences on both strands of the entire aadA10 gene cassette and of regions where there were differences from standard or prototype sequences were determined. The sequences of the integron boundaries were obtained by sequencing R388::Tn1404 directly. Automated sequencing was performed by the sequencing facility at the Department of Biological Sciences, Macquarie University, Sydney, New South Wales, Australia, on an ABI-PRISM 377 sequencer with the Big Dye system. DNA sequences were assembled by using MacVector (version 6.5) software and AssemblyLIGN software (Oxford Molecular). GenBank searches were performed by using the BLASTN and FastA programs available through WebANGIS (Australian National Genomic Information Service). Programs in the Genetics Computer Group Wisconsin package (version 8.1.0) were used via WAG (WebANGIS GCG) to align and analyze the DNA and protein sequences.
Cassette deletion experiments.
Strains for deletion experiments were constructed by introducing each of pRMH857, pRMH549ΔaadB, pRMH549Δoxa10, or pRMH328 into DH5α containing plasmid pSU2056, which contains the intI1 gene under the control of the lac promoter (27). Cassette deletion experiments were carried out as described by Collis and Hall (12), except that prior to DNA isolation the strains were grown in LB medium containing only chloramphenicol and ampicillin. The DNA recovered was used to transform DH5α by electroporation with selection on plates containing chloramphenicol, and the resultant colonies were patched onto plates containing chloramphenicol alone and chloramphenicol plus gentamicin, ampicillin, or spectinomycin, as appropriate. The patches were scored for resistance, and plasmid DNA prepared from selected patches was digested with restriction enzymes to confirm that a cassette had been lost.
Nucleotide sequence accession number.
The sequence with GenBank accession no. U37105.1 has been extended to include the sequence of the whole cassette array in the R388-R151 hybrid plasmid and is now GenBank accession no. U37105.2.
RESULTS
Movement of resistance determinants.
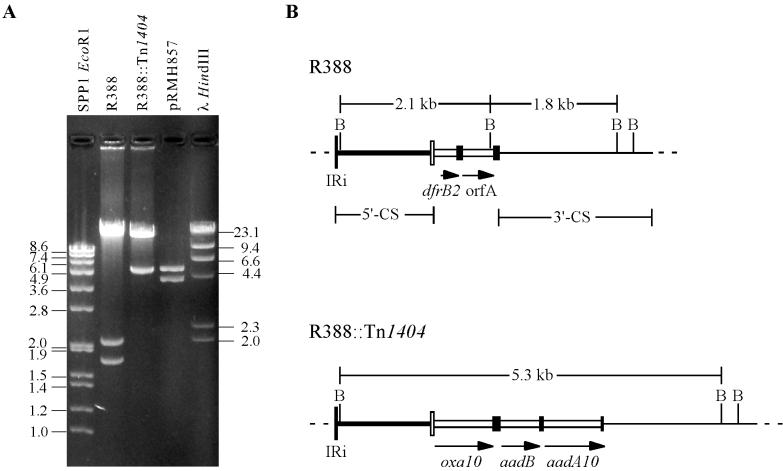
The possibility that an exchange of gene cassettes was responsible for the loss in R388 of the trimethoprim resistance accompanying the acquisition of R151 resistance markers (38) was investigated. Integron In3 in R388 contains characteristic BamHI sites present in the 5′-CS and the 3′-CS of class 1 integrons plus an additional BamHI site in the orfA cassette (Fig. 2). As no other BamHI sites are present in R388 (1), BamHI digestion gives a large fragment, comprising the bulk of the plasmid, plus fragments of 2.1 and 1.8 kb derived from integron In3 (Fig. 2). In BamHI digests of R388::Tn1404, the 2.1- and 1.8-kb fragments of R388 were replaced by a single band of 5.3 kb (Fig. 2). This indicates that only a single integron is present in R388::Tn1404 and that replacement of the R388 cassette array (dfrB2-orfA) had taken place. This was confirmed by determining the sequences at the boundaries of the integron in R388::Tn1404, which were found to correspond exactly to those seen between In3 and the plasmid backbone in R388 (4, 34), indicating that the new cassettes lie within the boundaries of In3. We suggest that R388::Tn1404 should in future be described as an R388-R151 hybrid.
FIG. 2.
Structures of R388 and R388::Tn1404. (A) BamHI digests of R388, R388::Tn1404, and pRM857. Size markers are SPP1 digested with EcoRI and bacteriophage λ digested with HindIII, and the sizes of the fragments (in kilobases) are indicated. (B) Maps of integron In3 in R388 and the cassette array in R388::Tn1404. The 5′-CS (thick line) is bounded at one end by IRi and ends with the attI1 site (narrow open box). Gene cassettes are shown as open boxes and an adjacent filled box representing the 59-be or simple site. The 3′-CS region present is represented by a thin line, and flanking regions are shown as dashed lines. BamHI sites (labeled B) and the sizes of the fragments expected on BamHI digestion are shown.
Cassette array in R388-R151.
The 5.3-kb BamHI fragment of the R388-R151 hybrid plasmid containing most of the integron (Fig. 2) was cloned in pACYC184 (8) to give pRMH857, and the insert was sequenced (GenBank accession no. U37105.2). The 5′-CS has the strong version of the Pc promoter (TTGACA-17 bp-TAAACT), which is also found in In3 (34, 45), and neither the three G residues in In2 that create the P2 promoter nor the 19-bp duplication of attI1 sequence in In4 (35) was present. As expected, the first cassette was oxa10 (carbenicillin resistance), and the sequence was identical to that of the sequence with GenBank accession no. U37105.1. The adjacent aadB gene cassette (gentamicin, kanamycin, and tobramycin resistance) was found to be identical to the prototype version of this cassette (GenBank accession no. L06418) (6). The third cassette was a novel aadA cassette, designated aadA10, which confers resistance to streptomycin and spectinomycin.
aadA10 gene cassette.
The aadA10 cassette of R388-R151 is 822 bp in length, and in place of the typical 59-be configuration of two simple sites separated by a central region (46), a single simple site is present. The simple site that would be found in the circularized version of this aadA10 cassette appears to consist of the first 6 bp of the 1L core site separated from the last 6 bp of 1R (found at the beginning of the linear cassette) by a 7-bp region that corresponds to the spacer of the attI1 simple site (Fig. 1B). Other gene cassettes with similar 1L-attI1 spacer-1R simple sites are known, and for all of them complete cassettes with a 59-be have been identified (Fig. 1B) (15, 36). It is therefore possible that a longer version of the aadA10 cassette with a complete 59-be will be discovered in the future.
The sequence of the aadA10 cassette is most closely related to those of the aadA6 (88% identical) and aadA7 (83% identical) cassettes (Fig. 3). There are two possible initiation codons for the aadA10 gene: an ATG at positions 10 to 12 that is also found in all of the other aadA cassettes and a GTG at positions 22 to 24 that is also found in the aadA7 cassette (Fig. 3). A GTG is also present in this position in the aadA1 and the aadA2 cassettes, and a potential ribosome binding site (GAGG) is present upstream of this GTG, which appears to be the major initiation codon for at least aadA2 (3). In the aadA7 and aadA10 cassettes a potential ribosome binding site (GAG) is present in the equivalent position, and it is possible that the GTG codon is also the main initiation codon for these genes. The aadA10 reading frame extends through the part of the simple site sequence (GTCTAAAACAAAG) found at the end of the cassette and into the adjacent 3′-CS (the sequence with lowercase letters in Fig. 3). An in-frame TAA codon in the 3′-CS would terminate translation in this particular configuration, yielding a 6-amino-acid terminal extension (the sequence with lowercase letters in Fig. 4). The terminal extension would differ if the cassette was in another position. In the aadA6 cassette, which has a complete 59-be, the reading frame ends within the 59-be, whereas in the aadA7 cassette, deletion of a single base has brought a TGA codon preceding the start of the 59-be into frame (Fig. 3).
FIG. 3.
Alignment of sequences at the ends of the aadA6, aadA7, and aadA10 cassettes. Colons indicate identity between aligned sequences. Possible initiation codons (marked by the letter M) and stop codons are indicated by letters in larger type. Potential ribosome binding sites are underlined. Core sites in the 59-be are in boldface type and are labeled as described in the legend to Fig. 1. The aadA6 and aadA7 sequences shown end at the cassette boundary, and lowercase letters in the aadA10 sequence lie outside the cassette boundary and represent the 3′-CS sequence. Bases corresponding to the attI1 spacer are boxed. The sources of the sequences are as follows: aadA6, GenBank accession no. AF140629; aadA7, GenBank accession no. AF224733.
FIG. 4.
Alignment of cassette-encoded AadA proteins. The sequences are identified on the left by the numeral in the protein name, and more closely related sequences are grouped. The first ATG after the beginning of the cassette has been used as the start codon, although AadA1, AadA2, AadA7, and AadA9 also have a second potential start codon (GTG) which corresponds to the valine at position 5. Residues identical in seven or all eight (boldface type) sequences are indicated in the consensus sequence shown below. Residues that are also conserved in the Aad proteins from the sequences with GenBank accession nos. X68089, X02588, M69221, AF288536, AF408195, AL392149, and AE007309 are boxed. The protein sequences are translations of DNA sequences in AadA1 (GenBank accession no. X12870), AadA2 (GenBank accession no. X68227), AadA4 (GenBank accession no. Z50802.3), AadA5 (GenBank accession no. AF137361), AadA6 (GenBank accession no. AF140629), AadA7 (GenBank accession no. AF224733), and AadA9 (GenBank accession no. AX135967 or AJ420072). The AadA3 (GenBank accession no. AF047479) and AadA8 (GenBank accession no. AF326210) proteins have not been included in the analysis, as they can be considered hybrids of AadA1 and AadA2.
The aadA10 product, AadA10, is an aminoglycoside (3")(9)-adenylytransferase [AAD(3")(9)], as expected from adenylation studies (26). It is most closely related to AadA6 (85% identical; GenBank accession no. AF140629) (32) and AadA7 (80% identical; GenBank accession no. AF224733) (29). AadA10 is also related to all of the other cassette-encoded AadA proteins (59 to 75% identical), and Fig. 4 shows the alignment of the amino acid sequences of these proteins, which fall into groups of more closely related sequences. AadA6, AadA7, and AadA10 are 80 to 85% identical to each other, AadA1 and AadA2 are 86% identical, and AadA4 and AadA5 are 95% identical, while AadA9 is less than 62% identical to any of the other proteins. One hundred twelve amino acids are completely conserved in all of the cassette-encoded AadA proteins (sequences in boldface type in Fig. 4), and at least some of these conserved residues are likely to be important in enzyme function.
Database searches with the sequence of the AadA10 protein also identified several related proteins that are not encoded by a cassette. One of these, from a streptomycin-resistant mutant of Corynebacterium acetoacidophilum (GenBank accession no. AJ278607) (J. Deb and G. Karan, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., 2000), is 74% identical to that of AadA10 and differs by only 1 amino acid (V31M in Fig. 4) from AadA2 if the GTG codon is used as the start codon for the aadA2 gene (position 5 in Fig. 4). The nucleotide sequences of the aadA2 gene and the C. acetoacidophilum aadA gene are also 95% identical over the length of the reading frame, with the last 437 of the total 779 nucleotides being 100% identical. However, this similarity drops away completely on either side of the reading frame, and no vestige of a 59-be is detectable. This high degree of identity suggests that the C. acetoacidophilum aadA gene could represent the direct precursor of the one in the aadA2 cassette.
Other Aad proteins that are not cassette associated are less closely related to AadA10 (29 to 45% identical; 51 to 62% similar). These include the Salmonella enterica serovar choleraesuis AAD(3")(9) protein (GenBank accession no. X68089), which confers resistance to both streptomycin and spectinomycin (25); the AAD(9) spectinomycin adenylyltransferases from Tn554 of Staphylococcus aureus (GenBank accession no. X02588) (30), Enterococcus faecalis (GenBank accession no. M69221) (24), and Legionella longbeachae (GenBank accession no. AF288536); and a number of other proteins (GenBank accession nos. AF408195, AL392149, and AE007309) that are predicted to be aminoglycoside-modifying enzymes on the basis of the similarities of their sequences to that of the S. aureus AAD(9) protein. The amino acids that are conserved among all of these proteins are boxed in Fig. 4.
Activity of the aadA10 simple site.
A typical active 59-be consists of two simple sites separated by a central region (46), and deletion of either the LH or the RH simple site of the aadA1 59-be has been shown to reduce the activity of this recombination site to undetectable levels in conduction assays (27). Removal of 1L from the aadB 59-be was also found to dramatically decrease the activity of this 59-be in conduction assays (19). On the basis of these findings, it seemed unlikely that the single simple site configuration found in the aadA10 cassette of R388-R151 would function efficiently in recombination. To determine whether this simple site is functional, IntI1-mediated excision of cassettes from pRMH857 was followed by assaying for loss of the associated antibiotic resistance determinants (Table 2). Deletion of the aadA10 cassette was not detected in over 600 colonies screened, while aadB, the central cassette in the array, was frequently excised and the first cassette, oxa10, was occasionally lost. Loss of the aadA10 cassette from derivatives of pRMH857 containing only two cassettes (aadB-aadA10 or oxa10-aadA10) was not detected in over 1,000 colonies screened, and the first cassette was lost infrequently. When another array containing two complete gene cassettes, aacA4-aadA2, was examined, the first cassette, aacA4, was again lost at very low levels, but the aadA2 cassette in the second position was frequently excised, which is in contrast to the lack of excision of the incomplete aadA10 cassette from the equivalent position. Together, these results indicate that the simple site present in the aadA10 cassette is not functional at a significant level, at least for IntI1-mediated excision reactions.
TABLE 2.
IntI1-mediated excision of gene cassettes
| Plasmid | Cassette array | % Colonies with cassettes deleted
|
No. of colonies screened | |||
|---|---|---|---|---|---|---|
| 1 | 2 | 1 and 2 | 3 | |||
| pRMH549 | oxa10-aadB-aadA10 | 1.1 | 25.5 | 1.1 | <0.2 | 663 |
| pRMH549Δoxa10 | aadB-aadA10 | 0.2 | <0.1 | <0.1 | NAa | 1,069 |
| pRMH549ΔaadB | oxa10-aadA10 | <0.1 | <0.1 | <0.1 | NA | 1,034 |
| pRMH328 | aacA4-aadA2 | 0.3 | 30.4 | 0.5 | NA | 382 |
NA, not applicable.
DISCUSSION
We have shown that the resistance genes present on R151-pUZ8 were translocated to R388 by exchange of the gene cassette region between two integrons rather than by transposition. A similar exchange of cassette arrays was recently found to account for the movement of resistance markers from the chromosome of the P. aeruginosa Dalgleish strain to R388 via a pUZ8 intermediate, forming the plasmid known as R388::Tn1405 (34). Although movement of resistance markers from R151-pUZ8 to R388 was recA independent in the original study (38), the investigators suggested that the gene exchange may have occurred by homologous recombination between identical regions present in both plasmids, i.e., the 5′ CS and the 3′ CS. An alternate explanation is that IntI1-mediated site-specific recombination may have been involved. If this was the route, two site-specific recombination events must have occurred, one at each extremity of the two cassette arrays.
However, the findings reported here do not preclude the existence of Tn1404 as a transposon. Philippon et al. (38) also used pUB5573, a sulfonamide-sensitive PstI deletion derivative of R388 that has lost part of the orfA cassette and part of the 3′ CS region, instead of R388 as the recipient for R151-pUZ8 resistance markers. The putative pUB5573::Tn1404 transconjugants arose in a rec+ strain but were about 9.6 kb larger than pUB5573, retained trimethoprim resistance, and had acquired sulfonamide resistance together with the other resistance markers (26, 38). This suggests that a transposable unit carrying all of the resistance determinants of R151 may have translocated to pUB5573 in this case. The size of this unit is similar to those of class 1 integrons such as In5 (9 kb), In2 (11 kb), and In4 (8 kb), which all carry the sul1 gene (4, 35), and it is possible that Tn1404 corresponds to a class 1 integron. Although most class 1 integrons lack a complete set of transposition genes and are unable to transpose themselves (4, 35, 37), if they have intact terminal inverted repeats they may be moved in trans by appropriate transposition proteins that happen to be present in the same cell. Indeed, integron In33, which corresponds to both Tn2521 and Tn1405, can move by transposition catalyzed by transposition proteins supplied in trans (44). Thus, it is possible that movement of an integron from R151-pUZ8 to pUB5573 created pUB5573::Tn1404. However, this event could have occurred only if the R151-pUZ8 recombinant carries the appropriate genes for transposition functions. In this regard, it would be interesting to look at the structures of pUB5573::Tn1404 and of the integron in the original plasmid R151 to determine if transposition genes are present.
The aadA10 cassette found here may not represent the complete cassette, as a single simple site was found in place of a 59-be. IntI1-mediated excision of this incomplete aadA10 cassette was not detected, and this is consistent with previous observations that deletion of all or part of either the LH or the RH simple site from a 59-be reduces the activity of the 59-be to undetectable levels (19, 27). Similar observations have been made in the case of the attI1 recombination site, which consists of one simple site plus two additional, but directly oriented, integrase binding domains (13). In cointegration assays, the attI1 simple site alone is active at low levels in reactions with a complete attI1 site but is not an efficient recombination site in reactions with the 59-be (21, 36). Further experiments are needed to determine if single simple sites derived from the 59-be, such as the one found in the aadA10 cassette, are able to participate in integration reactions.
The loss of the central region of the 59-be appears to have effectively fused the aadA10 cassette to the 3′-CS, with the consequence of fixing it as part of the class 1 integron backbone. There are four further examples in which the final gene cassette in an array is likely to be fused to the 3′-CS as a result of the replacement of a complete 59-be by a simple site (33, 39, 41). However, the structures of these simple sites differ from that of the one found in the R388-R151 aadA10 cassette (Fig. 5). The 1L and 1R sites of the original 59-be are present, but they are separated either by the RH spacer found in the complete 59-be or by the LH spacer. Thus, a 59-be can be converted to a single simple site in more than one way. We have previously suggested how simple sites with the attI1 spacer could have been created (36), and the role of the integron-encoded integrase in these processes deserves investigation.
FIG. 5.
Simple sites containing spacers derived from 59-be. The sequences at the ends of the aadA1 and aadA2 cassettes with the complete 59-be and derivatives with simple sites are aligned. Colons indicate identity. Core sites are in boldface type and are labeled as described in the legend to Fig. 1, and spacers are boxed. The sequence from the adjacent 3′-CS or gene cassette is indicated with lowercase letters. The stop codons of the aadA1 and aadA2 genes are marked by asterisks. The sources of the sequences are as follows: aadA1, GenBank accession no. X12870; aadA2, GenBank accession no. X68227; aadA1 and 3′ CS, GenBank accession no. AJ278514; aadA2 and 3′ CS with RH spacer, GenBank accession no. AF156486; aadA2 and 3′ CS with LH spacer, GenBank accession no. AF227505; blaVIM-2 and 3′ CS, GenBank accession no. AF302086; and blaVIM-2, GenBank accession no. AF191564.
The other cassette derivatives in which the central part of the 59-be is replaced by the attI1 spacer sequence have all been fused to a second cassette (Fig. 1B). Although these partial 59-be are presumably also nonfunctional, in these cases the first cassette is likely to retain mobility by moving together with the downstream cassette as a single two-gene unit. Conversion of the remaining 59-be in a fused cassette to a simple site could potentially extend such cassette fusions to include three or more genes. The genes situated in a cassette array usually constitute an operon, as they are transcribed together from the Pc promoter within the integron (10). Furthermore, as the numbers, orders, and identities of the cassettes in the array can be varied by adding or removing cassettes, a vast number of different operons can potentially be created. Conversion of the 59-be within a cassette array to simple sites would reduce the flexibility of an array of functional gene cassettes by fixing the cassette genes in a certain order. This cassette fusion process could therefore have consequences in terms of creating stable operons from the more flexible operons that the cassette arrays within integrons represent. It may also have played a role in the creation of the 3′-CS of class 1 integrons.
It should be noted that the putative Tn1404 transposon of R151 is not the same as the transposon recently designated Tn1404 (GenBank accession nos. AF157797 to AF1577801) by Schnabel and Jones (43).
Acknowledgments
S.R.P. was supported by a grant from the Australian National Health and Medical Research Council.
REFERENCES
- 1.Avila, P., and F. de la Cruz. 1988. Physical and genetic map of the IncW plasmid R388. Plasmid 20:155-157. [DOI] [PubMed] [Google Scholar]
- 2.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bito, A., and M. Susani. 1994. Revised analysis of aadA2 gene of plasmid pSa. Antimicrob. Agents Chemother. 38:1172-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, H. J., H. W. Stokes, and R. M. Hall. 1996. The integrons In0, In2, and In5 are defective transposon derivatives. J. Bacteriol. 178:4429-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryan, L. E., M. S. Shahrabadi, and H. M. van den Elzen. 1974. Gentamicin resistance in Pseudomonas aeruginosa: R-factor-mediated resistance. Antimicrob. Agents Chemother. 6:191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cameron, F. H., D. J. Groot Obbink, V. A. Ackerman, and R. M. Hall. 1986. Nucleotide sequence of the AAD(2") aminoglycoside adenyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhrfII in R388. Nucleic Acids Res. 14:8625-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centron, D., and P. H. Roy. 2002. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob. Agents Chemother. 46:1402-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, A. Y. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collis, C. M., G. Grammaticopoulos, J. Briton, H. W. Stokes, and R. M. Hall. 1993. Site-specific insertion of gene cassettes into integrons. Mol. Microbiol. 9:41-52. [DOI] [PubMed] [Google Scholar]
- 10.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collis, C. M., and R. M. Hall. 1992. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol. Microbiol. 6:2875-2885. [DOI] [PubMed] [Google Scholar]
- 12.Collis, C. M., and R. M. Hall. 1992. Site-specific deletion and rearrangement of integron insert genes catalyzed by integron DNA integrase. J. Bacteriol. 174:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collis, C. M., M.-J. Kim, H. W. Stokes, and R. M. Hall. 1998. Binding of the purified integron DNA integrase IntI1 to integron- and cassette-associated recombination sites. Mol. Microbiol. 29:477-490. [DOI] [PubMed] [Google Scholar]
- 14.Datta, N., and R. W. Hedges. 1972. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J. Gen. Microbiol. 72:349-355. [DOI] [PubMed] [Google Scholar]
- 15.Giakkoupi, P., L. S. Tzouvelekis, A. Tsakris, V. Loukova, D. Sofianou, and E. Tzelepi. 2000. IBC-1, a novel integron-associated class A β-lactamase with extended-spectrum properties produced by an Enterobacter cloacae clinical strain. Antimicrob. Agents Chemother. 44:2247-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gravel, A., B. Fournier, and P. H. Roy. 1998. DNA complexes obtained with integron integrase IntI at the attI1 site. Nucleic Acids Res. 26:4347-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, R. M., D. E. Brookes, and H. W. Stokes. 1991. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol. Microbiol. 5:1941-1959. [DOI] [PubMed] [Google Scholar]
- 18.Hall, R. M., H. J. Brown, D. E. Brookes, and H. W. Stokes. 1994. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J. Bacteriol. 176:6286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall, R. M., C. M. Collis, M.-J. Kim, S. R. Partridge, G. D. Recchia, and H. W. Stokes. 1999. Mobile gene cassettes and integrons in evolution. Ann. N. Y. Acad. Sci. 870:68-80. [DOI] [PubMed] [Google Scholar]
- 20.Hall, R. M., and C. Vockler. 1987. The region of the IncN plasmid R46 coding for resistance to β-lactam antibiotics, streptomycin/spectinomycin and sulphonamides is closely related to antibiotic resistance segments found in IncW plasmids and in Tn21-like transposons. Nucleic Acids Res. 15:7491-7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansson, K., O. Sköld, and L. Sundström. 1997. Non-palindromic attI sites of integrons are capable of site-specific recombination with one another and with secondary sites. Mol. Microbiol. 26:441-453. [DOI] [PubMed] [Google Scholar]
- 22.Hedges, R. W., and M. Matthew. 1979. Acquisition by Escherichia coli of plasmid-borne β-lactamases normally confined to Pseudomonas spp. Plasmid 2:269-278. [DOI] [PubMed] [Google Scholar]
- 23.Huovinen, P., S. Huovinen, and G. A. Jacoby. 1988. The sequence of PSE-2 β-lactamase. Antimicrob. Agents Chemother. 32:134-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LeBlanc, D. J., L. N. Lee, and J. M. Inamine. 1991. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob. Agents Chemother. 35:1804-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leung, K. Y., S. R. Ruschkowski, and B. B. Finlay. 1992. Isolation and characterization of the aadA aminoglycoside-resistance gene from Salmonella choleraesuis. Mol. Microbiol. 6:2453-2460. [DOI] [PubMed] [Google Scholar]
- 26.Lévesque, R. C., and G. A. Jacoby. 1988. Molecular structure and interrelationships of multiresistance β-lactamase transposons. Plasmid 19:21-29. [DOI] [PubMed] [Google Scholar]
- 27.Martinez, E., and F. de la Cruz. 1990. Genetic elements involved in Tn21 site-specific integration, a novel mechanism for the dissemination of antibiotic resistance genes. EMBO J. 9:1275-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazel, D., and J. Davies. 1999. Antibiotic resistance in microbes. Cell. Mol. Life Sci. 56:742-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 2000. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 44:1568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy, E. 1985. Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3")(9). Mol. Gen. Genet. 200:33-39. [DOI] [PubMed] [Google Scholar]
- 31.Naas, T., Y. Mikami, T. Imai, L. Poirel, and P. Nordmann. 2001. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183:235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naas, T., L. Poirel, and P. Nordmann. 1999. Molecular characterisation of In51, a class 1 integron containing a novel aminoglycoside adenylyltransferase gene cassette, aadA6, in Pseudomonas aeruginosa. Biochim. Biophys. Acta 1489:445-451. [DOI] [PubMed] [Google Scholar]
- 33.Pallecchi, L., M. L. Riccio, J. Docquier, R. Fontana, and G. M. Rossolini. 2001. Molecular heterogeneity of blaVIM-2-containing integrons from Pseudomonas aeruginosa plasmids encoding the VIM-2 metallo-β-lactamase. FEMS Microbiol. Lett. 195:145-150. [DOI] [PubMed] [Google Scholar]
- 34.Partridge, S. R., H. J. Brown, and R. M. Hall. 2002. Characterization and movement of the class 1 integron known as Tn2521 and Tn1405. Antimicrob. Agents Chemother. 46:1288-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Partridge, S. R., G. D. Recchia, C. Scaramuzzi, C. M. Collis, H. W. Stokes, and R. M. Hall. 2000. Definition of the attI1 site of class 1 integrons. Microbiology 146:2855-2864. [DOI] [PubMed] [Google Scholar]
- 37.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Philippon, A. M., G. C. Paul, and G. A. Jacoby. 1983. Properties of PSE-2 β-lactamase and genetic basis for its production in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 24:362-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poirel, L., I. le Thomas, T. Nass, A. Karim, and P. Nordmann. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 41.Riccio, M. L., L. Pallecchi, R. Fontana, and G. M. Rossolini. 2001. In70 of plasmid pAX22, a blaVIM-1-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrob. Agents Chemother. 45:1249-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Schnabel, E. L., and A. L. Jones. 1999. Distribution of tetracycline resistance genes and transposons among phylloplane bacteria in Michigan apple orchards. Appl. Environ. Microbiol. 65:4898-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinclair, M. I., and B. W. Holloway. 1982. A chromosomally located transposon in Pseudomonas aeruginosa. J. Bacteriol. 151:569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3:1669-1683. [DOI] [PubMed] [Google Scholar]
- 46.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile genetic elements. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]