Abstract
Yersinia pestis, the cause of bubonic plague, blocks feeding by its vector, the flea. Recent evidence indicates that blockage is mediated by an in vivo biofilm. Y. pestis and the closely related Yersinia pseudotuberculosis also make biofilms on the cuticle of the nematode Caenorhabditis elegans, which block this laboratory animal's feeding. Random screening of Y. pseudotuberculosis transposon insertion mutants with a C. elegans biofilm assay identified gmhA as a gene required for normal biofilms. gmhA encodes phosphoheptose isomerase, an enzyme required for synthesis of heptose, a conserved component of lipopolysaccharide and lipooligosaccharide. A Y. pestis gmhA mutant was constructed and was severely defective for C. elegans biofilm formation and for flea blockage but only moderately defective in an in vitro biofilm assay. These results validate use of the C. elegans biofilm system to identify genes and pathways involved in Y. pestis flea blockage.
Yersinia pestis, the causative agent of bubonic plague, is transmitted to mammals primarily by the bites of infected fleas. The bacteria heavily colonize the insect's proventriculus, a valve-like organ connecting the esophagus to the midgut, and thereby block the passage of food. As fleas starve they bite repeatedly in search of blood meals, thus spreading Y. pestis to new hosts.
Y. pestis cells in the blocked proventriculus are surrounded by extracellular material that is positive for polysaccharide in histological tests (12). Y. pestis lacking the hmsHFRS operon, which encodes predicted polysaccharide biosynthetic proteins, fails to block fleas (10). Y. pestis flea blockage therefore appears to depend on creation of an in vivo biofilm, i.e., a population of bacteria in a self-synthesized matrix containing exopolysaccharide (EPS).
Y. pestis and its recent ancestor, Yersinia pseudotuberculosis (1), make biofilms on the head of the model nematode Caenorhabditis elegans that cover the mouth and block the worm from feeding on bacteria (4, 13). Like flea blockage, this process requires hmsHFRS, suggesting that the nematode can be used as a flea surrogate to study plague transmission (4). Lectin staining confirms that the matrix on nematodes contains polysaccharide (21).
The outer membranes of gram-negative bacteria contain either lipopolysaccharide (LPS) or lipooligosaccharide (LOS). Y. pseudotuberculosis makes a typical LPS, which comprises three covalently linked domains. Lipid A is innermost; core oligosaccharide, a branched, nonrepeating unit, is linked to lipid A; and O antigen, a branched polysaccharide containing many repeated units, is linked to core. Y. pestis makes only LOS, which contains lipid A and core oligosaccharide but lacks O antigen (17).
Core oligosaccharide is divided into inner and outer domains. Inner core contains 3-deoxy-d-manno-octulosonic acid, of which one residue is linked to lipid A, and l-glycero-d-manno heptose (heptose), of which one residue is attached to 3-deoxy-d-manno-octulosonic acid. This arrangement is common to enteric bacteria (18), including both Y. pestis and Y. pseudotuberculosis (24). Heptose is the attachment point for the more variable outer-core sugars, which in turn are the attachment sites for O antigen in organisms that make complete LPS.
Phosphoheptose isomerase, encoded by gmhA (previously named lpcA), catalyzes conversion of sedoheptulose 7-phosphate to d-glycero-d-mannoheptose 7-phosphate, the first step in the heptose biosynthesis pathway (3). Escherichia coli strains with mutations in gmhA do not make heptose, and their LPS is truncated (3) because the attachment point for more distal moieties is absent. We show that Y. pestis and Y. pseudotuberculosis gmhA mutants have truncated LOS and LPS and makes small, aberrant biofilms on C. elegans. The Y. pestis gmhA mutant is almost completely defective in blocking the flea proventriculus.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and plasmids are described in Table 1. Bacteria were grown on L agar plates or in LB broth for genetic manipulation and strain construction. The antibiotics used and their concentrations for both Yersinia spp. and E. coli were ampicillin (100 to 200 μg/ml), chloramphenicol (10 μg/ml), kanamycin (30 μg/ml), and streptomycin (100 μg/ml).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and/or description | Reference or source |
|---|---|---|
| E. coli | ||
| DH10B | Cloning strain | Invitrogen |
| HB101 | Cloning strain | 19 |
| OP50 | Standard C. elegans food | 25 |
| SM10λpir | Conjugation donor | 20 |
| Y. pestis | ||
| KIM6+ | Wild type | 16 |
| CDY113 | Δpgm | 4 |
| CDY140 | ΔgmhA::cat | This study |
| CDY145 | ΔgmhA::cat/pCBD32 | This study |
| CDY146 | ΔgmhA::cat/pCR2.1-TOPO | This study |
| Y. pseudotuberculosis | ||
| YPIII | Wild type | 8 |
| CDY103 | hmsT::TnphoA | 4 |
| CDY139 | ΔgmhA::cat | This study |
| CDY143 | ΔgmhA::cat/pCBD32 | This study |
| CDY144 | ΔgmhA::cat/pCR2.1-TOPO | This study |
| JM427 | ddhC::mini-Tn5 | 14 |
| JM496 | wzx::mini-Tn5 | 14 |
| JM543 | gmd::mini-Tn5 | 14 |
| Plasmids | ||
| pCR2.1-TOPO | Cloning vector for PCR products | Invitrogen |
| pCBD32 | gmhA in pCR2.1-TOPO | This study |
| pCBD41 | gmhA allelic exchange plasmid | This study |
| pCVD442 | Allelic exchange vector | 6 |
| pRT733 | Suicide plasmid for TnphoA delivery | 22 |
| pRY109 | Source of cat gene | 26 |
For biofilm production, bacteria were grown overnight at 26°C in LB with shaking. Aliquots of 100 μl were pipetted onto 6-cm-diameter nematode growth medium (NGM) plates (25) that contained 2% agar. Lawns were grown 16 to 24 h at room temperature before addition of nematodes. For lawns containing mixtures of wild-type and hms mutant strains, each strain was grown overnight in LB, the optical density at 600 nm determined, and the cell densities adjusted with LB to make them equivalent. Mixtures at various ratios were made and immediately used to inoculate NGM plates.
Biofilm formation on nematodes.
Gravid C. elegans adult hermaphrodites (wild-type strain N2) were placed on bacterial lawns, allowed to lay eggs for 2 to 4 h, and then removed. The plates were incubated at 20 to 22°C for 2 days, and the developmental stage of the broods was scored. In every experiment a parallel assay was conducted with either E. coli OP50, the standard C. elegans food source, or E. coli HB101, a laboratory cloning strain that also supports nematode growth. Triplicate samples were used in each experiment.
Transposon mutagenesis of Y. pseudotuberculosis.
JM403, a spontaneous streptomycin-resistant variant of Y. pseudotuberculosis YPIII (gift of J. Mecsas), has no defect in nematode growth inhibition (data not shown). Plasmid pRT733, which contains transposon TnphoA and which does not replicate in Y. pseudotuberculosis, was delivered into JM403 by conjugation from E. coli strain SM10λpir essentially as described previously (22). Y. pseudotuberculosis containing the transposon was selected on L agar with kanamycin and the E. coli donors counterselected with streptomycin. Colonies were tested individually in the nematode growth assay, and mutants were kept either if they formed no biofilm on nematodes or if they formed a biofilm but the animals nevertheless grew more rapidly than on JM403. Although the alkaline phosphatase encoded by TnphoA can be used to identify genes whose products may be exported, screening was without regard to export phenotypes.
Transposon insertion mapping.
Chromosomal DNA was digested with BamHI and SalI, neither of which cuts in the kanamycin resistance gene of TnphoA. Fragments were ligated into double-digested pUC18 and transformed into E. coli with selection for kanamycin resistance. The sequence of Y. pseudotuberculosis DNA flanking the transposon was determined using a primer that hybridizes to the 5′ end of TnphoA (Table 2).
TABLE 2.
PCR and sequencing primers
| Primer | Sequencea | Purpose |
|---|---|---|
| TNPHOA5 | 5′-TACTTGTGTATAAGAGTCAG | Sequence out from TnphoA 5′ end |
| GMHF | 5′-GCTTGGATCCCATAATGAAGCTCCTGAGATGTAG | PCR, complete gmhA locus |
| GMHR | 5′-AGTGGGTCGACACAGAAGATTGAGGTGATCAAC | PCR, complete gmhA locus |
| GMHUPF | 5′-GTTCTAGAGCATCAGAACCAGCTTCC | PCR, upstream flanking DNA |
| GMHUPR | 5′-TCCTGCAGTTTTCCTCAATCAGTGTACCG | PCR, upstream flanking DNA |
| GMHDNF | 5′-TCCTGCAGCCTCAATCTTCTGTGAACATT | PCR, downstream flanking DNA |
| GMHDNR | 5′-CCGCATGCGCACTAATTAAAACCGACCCG | PCR, downstream flanking DNA |
Underlined bases are restriction sites designed for cloning; sites in GMHF and GMHR ultimately were not used. Sites in other primers are as follows: GMHUPF, XbaI; GHMUPR and GMHDNF, PstI; GMHDNR, SphI.
Cloning and sequencing of Y. pseudotuberculosis gmhA.
Using the genome sequence of Y. pestis CO92 (15), PCR primers were designed to amplify chromosomal DNA of gmhA, including all 221 nucleotides intervening between gmhA and the upstream open reading frame yafH (Table 2). Two independent PCR products from YPIII template DNA were sequenced, without cloning, using the same primers. The PCR product was cloned into the vector pCR2.1-TOPO, using E. coli DH10B as host, to create pCBD32.
Construction of gmhA deletion mutants.
Approximately 900 bp of DNA on each side of gmhA was amplified by PCR using the primers shown in Table 2. The products were digested with the enzymes indicated and then ligated into the allelic replacement vector pCVD442 along with a PstI fragment containing the cat (chloramphenicol resistance) gene of pRY109. The resulting plasmid, pCBD41, was transformed into the E coli conjugation donor strain SM10λpir (20), which was then mated to Y. pseudotuberculosis YPIII and Y. pestis KIM6+. Exconjugants were selected on Yersinia selective agar (Difco) containing chloramphenicol and ampicillin. Plasmid replication requires the pir gene, which is absent in Yersinia spp., and therefore the selection recovered presumed plasmid cointegrates at the gmhA chromosomal locus. To obtain a second recombination that removed gmhA and plasmid sequences, cells were plated on L agar containing chloramphenicol and 5% sucrose but lacking NaCl; this selected against cells retaining the vector-carried sacB gene, which is lethal in the presence of sucrose. After confirmation of the loss of ampicillin resistance, deletion of gmhA and replacement with cat were confirmed by PCR assays and Southern hybridization (data not shown).
Analysis of LPS and LOS.
Bacteria were grown on NGM agar. Cells were lysed in a buffer of 0.63 M Tris-HCl, pH 6.8, 1% sodium dodecyl sulfate, 10% sucrose, 0.008 M tetrasodium-EDTA, and 0.004% bromophenol blue. After boiling for 5 min, proteinase K was added to 1.5 mg/ml and the samples incubated for 2 to 3 h at 60°C. Samples were boiled again and immediately electrophoresed in gels containing 15% acrylamide, 0.2% sodium dodecyl sulfate, and 4 M urea. Gels were fixed and silver stained as described previously (23).
Flea infections.
Xenopsylla cheopis fleas from colonies maintained at the Rocky Mountain Laboratories were infected by using an artificial feeding system (10). Bacteria were grown in brain heart infusion agar at 37°C without aeration for 16 h, quantified by direct counting in a Petroff-Hausser counting chamber, recovered by centrifugation, and resuspended in phosphate-buffered saline at ∼2.5 × 109/ml. The infectious blood meal was prepared by adding 1 ml of the bacterial suspension to 5 ml of fresh heparinized mouse blood. Fleas that took a blood meal after a 1-h feeding period were kept at 21°C and 75% relative humidity and fed twice weekly on uninfected mice. One sample of 50 male and 50 female fleas was monitored for 4 weeks for mortality and proventricular blockage as described previously (10, 12). Additional samples of 20 female fleas were placed at −80°C immediately after the infectious blood meal and after 7 and 28 days to determine the infection rate and bacterial load per flea. Thawed fleas were surface sterilized and individually triturated in 50 μl brain heart infusion and 20 μl glass sand; dilutions of the triturates were plated on Yersinia selective agar (8, 10). Y. pestis CFU were counted after 48 h of incubation at 28°C.
Growth and detection of biofilms in polystyrene dishes.
Bacteria were inoculated in 1 ml of NGM broth and incubated in 24-well polystyrene culture plates at 25°C for 4 days. Broth-only samples were the negative control. Cultures were removed, and the wells were washed three times with wheat germ agglutinin (WGA) buffer (0.01 M phosphate, 0.15 M NaCl, pH 7.3). WGA conjugated to alkaline phosphatase (EY Laboratories) was added to each well at 1 μg/ml in WGA buffer and incubated at room temperature for 90 min, after which the wells were washed three times with buffer alone. Substrate stock solution (nitroblue tetrazolium chloride [18.75 mg/ml] and 5-bromo-4-chloro-3-indolyl phosphate [9.4 mg/ml] in 67% dimethyl sulfoxide) was diluted 1:50 just before use into staining buffer (0.1 M Tris, 0.05 M MgCl2, 0.1 M NaCl, pH 9.5). One milliliter of staining solution was added to each well and incubated at room temperature for 3 to 4 h. Wells were washed three times with deionized water, 1 ml of ethanol was added, and plates were covered and wrapped with paraffin film to prevent evaporation. Plates were incubated overnight at room temperature with gentle rotation (60 rpm) to dissolve the stained material, after which absorbance was measured at 425 nm. Each strain was tested in quadruplicate.
RESULTS
Screen for biofilm-defective Y. pseudotuberculosis.
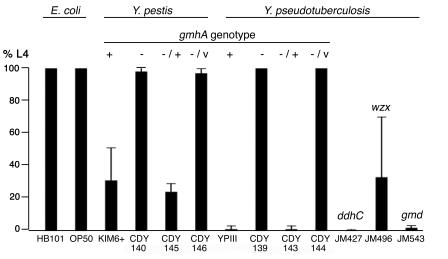
C. elegans development is highly synchronous when the nematodes are fed nonpathogenic E. coli strains. At 20 to 22°C, all animals reach the fourth larval stage of development (L4) 2 days after eggs are laid (Fig. 1). In contrast, the biofilm produced by Y. pseudotuberculosis YPIII covers the nematode mouth and inhibits feeding (Fig. 2), such that almost no animals are L4 after 2 days (Fig. 1). Under the same conditions, Y. pestis KIM6+ inhibits growth only partially, which would complicate identification of defective bacteria in a genetic screen. We therefore employed a two-step strategy to identify Y. pestis biofilm-related genes. First, we generated transposon insertions in Y. pseudotuberculosis and screened for mutants that permit normal nematode growth, indicating a biofilm defect. Next, we constructed mutations in the Y. pestis homologues of the identified genes and tested for defects in biofilm formation on C. elegans and in flea blockage.
FIG. 1.
Nematode growth on Y. pestis and Y. pseudotuberculosis strains. The percentage of L4 C. elegans was determined 2 days after eggs were laid on lawns of the indicated strains. gmhA genotypes: +, wild type; −, chromosomal deletion; −/+, deletion strain carrying complementing plasmid pCBD32; −/v, deletion strain carrying vector. Data are means and standard deviations from three independent experiments for each strain except YPIII, which was assayed six times.
FIG. 2.
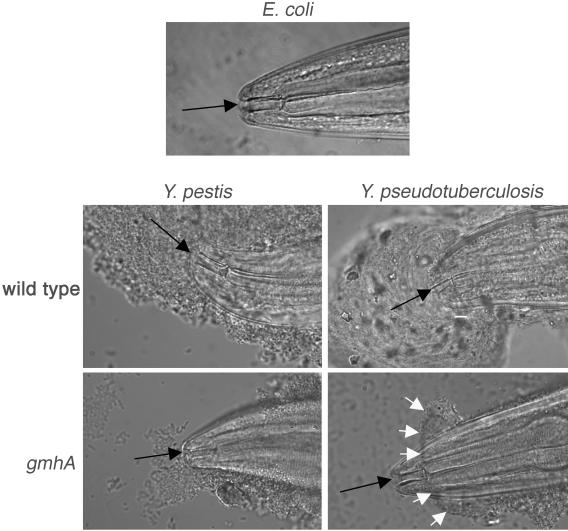
Biofilms on C. elegans made by Yersinia wild-type and gmhA mutant strains. A nematode grown on E. coli food strain OP50 is shown for comparison. Black arrows, mouth. Wild-type Y. pestis and Y. pseudotuberculosis biofilms cover the mouth completely and block feeding. Most Y. pestis gmhA biofilms fail to attach to the mouth (not shown), but a few are as shown, accounting for the 2% of C. elegans animals that fail to grow normally on this strain (Fig. 1). Y. pseudotuberculosis gmhA biofilms attach to the side of the head posterior to the mouth; white arrows mark the forward edge of the biofilm.
Y. pseudotuberculosis was mutagenized with transposon TnphoA. Of approximately 1,400 mutant colonies tested in the C. elegans growth assay, 4 were confirmed defective. One, with a mutation in hmsT, has been described previously (4), and two others have not yet been characterized. Analysis of the fourth mutant by Southern hybridization indicated that two copies of TnphoA were inserted in the chromosome (data not shown). DNA flanking one insertion was cloned and sequenced, producing a match to Y. pestis gmhA. Because a Y. pseudotuberculosis genomic sequence was not yet available, we took advantage of the high degree of sequence similarity between Y. pestis and Y. pseudotuberculosis. The genome sequence of Y. pestis was used to design PCR primers to amplify the gmhA locus, including the complete coding region and an additional 221 base pairs extending 5′ to the next open reading frame upstream. The double TnphoA mutant was transformed with the cloned PCR product from Y. pseudotuberculosis, and this restored C. elegans growth inhibition (data not shown). The second transposon insertion was not examined further.
gmhA is required for normal biofilm formation by both Y. pestis and Y. pseudotuberculosis.
Strains with complete gmhA deletions were constructed in both Y. pseudotuberculosis YPIII and Y. pestis KIM6+ backgrounds and assayed for nematode growth inhibition (Fig. 1). The Y. pestis mutant, although strongly defective, consistently inhibited growth of about 2% of nematodes. The Y. pseudotuberculosis mutant was completely defective, permitting 100% of animals to reach stage L4. Transformation with the plasmid-carried wild-type Y. pseudotuberculosis gmhA complemented the defect in both species. The growth rate of each mutant was the same as that of the corresponding wild type (data not shown).
Nematodes were incubated on bacterial lawns overnight, washed to remove loosely adhering material, and examined by differential interference contrast microscopy (Fig. 2). Wild-type bacteria of both species produced copious biofilms that covered the mouths of the animals and also adhered to their sides. gmhA mutants of both species made small biofilms, but their appearances differed. The Y. pestis mutant biofilms adhered directly to the mouths of some worms, consistent with its low level of growth inhibition. The Y. pseudotuberculosis gmhA biofilms adhered to the side of the head but never covered the mouth, so feeding was not impaired.
The failure of mutant biofilms to bind the mouth could result from a change in the amount of EPS, its composition, or both. To examine the role of EPS quantity, Yersinia lawns were made using various ratios of wild-type bacteria to hms mutants, which have defects in a polysaccharide synthesis pathway and which make no biofilms on C. elegans (4). Y. pestis KIM6+ wild type was mixed with CDY113, which contains a spontaneous deletion of the 102-kb pgm locus that includes hmsHRFS (7). Y. pseudotuberculosis YPIII wild type was mixed with CDY103, a biofilm-defective strain with a mutation in hmsT (4). For Y. pestis, a 0.17 ratio of wild-type to pgm inoculum produced small biofilms that failed to cover the mouths of the worms and allowed more than 98% of nematodes to grow normally, essentially reproducing the gmhA phenotype. The Y. pseudotuberculosis gmhA phenotypes also could be reproduced by this method, although greater dilution was required (data not shown). Although the possibility that the gmhA biofilms have an altered composition cannot be excluded, the phenotypes are explainable solely by diminished production of EPS.
The gmhA open reading frame of Y. pseudotuberculosis strain YPIII was sequenced and found to be identical to that of Y. pestis strain KIM (5), the parent strain of KIM6+. During the course of this work the genomic sequence of Y. pseudotuberculosis strain IP32953 was determined, and it contained the identical gmhA coding sequence (GenBank accession no. NC 006155). gmhA of Y. pestis CO92 differs in one nucleotide from both Y. pseudotuberculosis and Y. pestis KIM, giving CO92 an aspartic acid at residue 109 instead of glycine. The KIM and Y. pseudotuberculosis polypeptides of 193 amino acids are 91% identical to that of the E. coli protein.
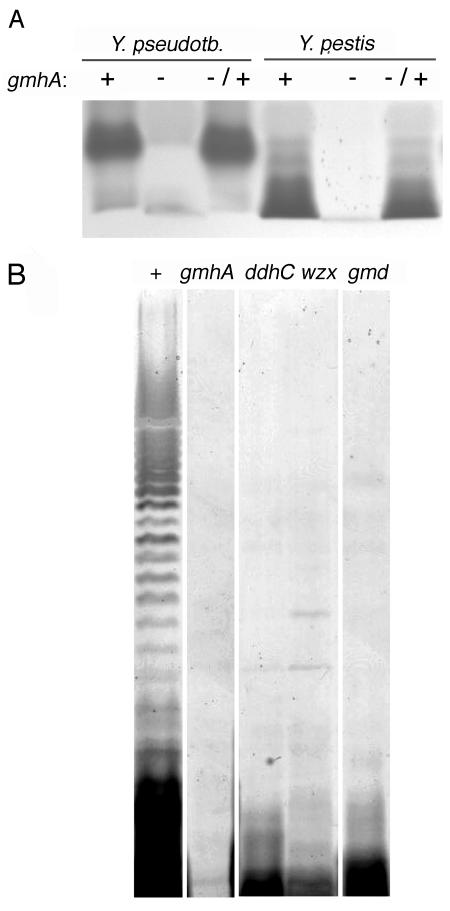
gmhA encodes phosphoheptose isomerase, an enzyme in the biosynthetic pathway of heptose (3). Mutations that abrogate heptose synthesis cause truncation of LPS or LOS (3). We examined Y. pseudotuberculosis LPS and Y. pestis LOS on silver-stained gels after electrophoresis. In both species, the core of the gmhA mutant migrated faster than that of the wild type, indicating that it was truncated (Fig. 3A). Also as expected, O antigen was absent in the Y. pseudotuberculosis gmhA mutant (Fig. 3B).
FIG. 3.
LOS and LPS of Yersinia mutants. (A) Core oligosaccharide of gmhA mutants migrates faster through gels, indicating truncation. +, wild type; −, gmhA deletion mutant; −/+, mutant complemented with pCBD32. (B) O antigen is absent in the Y. pseudotuberculosis gmhA mutant and in strains with mutations in known O-antigen genes. Wild-type YPIII (+) shows a typical ladder-like pattern produced by variable numbers of O-antigen repeat units. Faint bands in mutant samples are presumed protease-resistant proteins.
O antigen is not required for Y. pseudotuberculosis biofilms.
Wild-type Y. pestis lacks O antigen (17) yet makes biofilms on C. elegans. It therefore seemed probable that Y. pseudotuberculosis biofilms would not require O antigen. We confirmed this by examining Y. pseudotuberculosis strains with transposon insertions in O-antigen biosynthetic genes ddhC, wzx, and gmd (14). The absence of O antigen in each was confirmed by silver staining (Fig. 3B). When tested in the nematode growth assay, the ddhC and gmd mutants were indistinguishable from the wild type (Fig. 1), indicating that O antigen is not required for biofilm formation. The wzx mutant made a visible biofilm on C. elegans (data not shown) but for unknown reasons was highly variable in nematode growth assays (Fig. 1).
The Y. pestis gmhA mutant is defective for flea blockage.
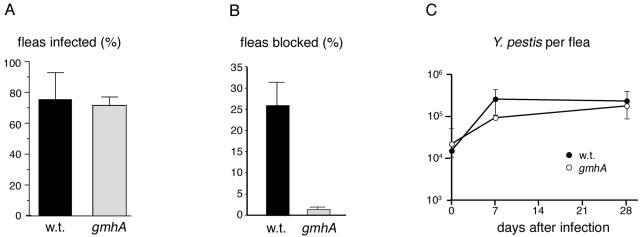
The Y. pestis gmhA mutant CDY140 was tested for flea blockage in parallel with its parent strain, KIM6+. The mutant was able to colonize fleas (Fig. 4A), but proventriculus blockage was almost entirely absent (Fig. 4B). The mutant grew in fleas to the same titer as the wild type (Fig. 4C), indicating that the failure to block fleas was not the result of a growth defect in vivo. These results indicate that gmhA plays a specific role in blockage, rather than a general role in colonization. The C. elegans results described above suggest that the defect in fleas is due to reduced EPS production.
FIG. 4.
The Y. pestis gmhA mutant is defective for flea blockage. (A) Colonization of flea digestive tract after a single blood meal containing Y. pestis. (B) Flea blockage during the 4-week period after an infected blood meal. The 25% level of blockage for the wild type is typical for strain KIM6+. (C) Y. pestis CFU taken up in the infectious blood meal (day 0) and in infected fleas 7 and 28 days postinfection. Data are means and standard deviations from two independent experiments. w.t., wild type.
Lack of correlation between biofilms in vivo and in vitro.
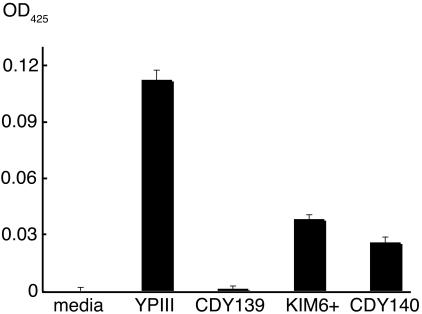
Biofilms are frequently studied in vitro by growing them on solid artificial surfaces such as glass and plastic. We examined Yersinia wild-type and gmhA mutants by growing standing broth cultures in polystyrene dishes, using the same medium (NGM) as for C. elegans biofilms except without agar (Fig. 5). Y. pseudotuberculosis YPIII made a substantial biofilm, and its isogenic gmhA mutant was severely defective (P = 0.001, two-tailed Student t test). Y. pestis KIM6+ made a smaller biofilm than YPIII, consistent with its lesser ability to inhibit C. elegans growth on NGM agar. However, the isogenic Y. pestis gmhA made only slightly less biofilm than its parent. In repeated trials the mutant always made less than the wild type, but the difference was not statistically significant (e.g., P = 0.08 for Fig. 5). This is in contrast to the almost complete failure of Y. pestis gmhA to either inhibit C. elegans growth (Fig. 1) or block fleas (Fig. 4).
FIG. 5.
In vitro biofilms. Bacteria were grown standing for 4 days in polystyrene dishes, after which broth and planktonic cells were removed and adherent biofilm matrix was detected with WGA conjugated to alkaline phosphatase (see Materials and Methods for details). Data are means and standard errors of the means for four replicates in a single experiment; results are typical of multiple experiments. OD425, optical density at 425 nm.
DISCUSSION
Recent evidence indicates that Y. pestis flea blockage is mediated by a biofilm the bacteria produce in the insect proventriculus. Microscopic examination shows bacteria embedded in an extracellular matrix that stains positively for polysaccharide (12). Blockage requires the Y. pestis hmsHFRS operon (10), which is also required to make biofilms on C. elegans (4). Although polysaccharide synthesis by hmsHFRS gene products has not been demonstrated directly, HmsR contains a conserved glycosyltransferase domain, Pfam 00535 (2). A protein containing this domain, Staphylococcus epidermidis IcaA, is required for biofilm formation and for in vitro synthesis of a homopolymer of β(1,6)-linked N-acetyl-d-glucosamine (9).
Screening for defects in C. elegans biofilms identified gmhA, and we showed that this gene is also required for Y. pestis flea blockage. Biochemical evidence from E. coli indicates that GmhA is a phosphoheptose isomerase. E. coli cells lacking this enzyme cannot synthesize heptose and have severely truncated LPS (3). The Yersinia GmhA amino acid sequence is 91% identical to that of E. coli, implying identical function. As expected from the known structures of LPS and LOS, mutations in gmhA increased the electrophoretic mobility of core oligosaccharide components for both Yersinia species and eliminated O antigen from Y. pseudotuberculosis (Fig. 3).
Although gmhA mutants make visible biofilms on C. elegans, these biofilms have scant ability to block nematode feeding (Fig. 1) and are less substantial than those made by the wild type (Fig. 2). In the case of Y. pseudotuberculosis, the aberrant biofilms were never observed to cover the mouth. With Y. pestis the biofilms attached to the mouths of some animals (Fig. 2), but only rarely was this sufficient to block feeding and inhibit growth. The presence of a visible biofilm indicates that the defect cannot be a complete loss of EPS. By titrating wild-type bacteria with hms mutants that are biofilm defective, so that the mixed cultures made less EPS, we reproduced the phenomenon of small biofilms that attach to the side of the C. elegans head but not to the mouth. This suggests that the defect in ghmA mutants is because of diminished EPS production rather than a change in the EPS composition or structure. These observations, incidentally, suggest that the surface composition of the C. elegans cuticle differs between the mouth and the side of the head nearby and that the EPS has greater affinity for the latter.
Heptose apparently is not a constituent of the normal biofilm, since none was detected by gas chromatography and mass spectrometry of Y. pestis EPS purified from biofilms on nematode (J. Cipollo, personal communication). The biofilm phenotypes of gmhA mutants therefore appear to be an indirect effect, most likely from truncation of the core oligosaccharide of LOS and LPS. An indirect role for core heptose was suggested by a study of biofilm formation in vitro by Klebsiella pneumoniae and Proteus mirabilis. Mutations in waaE, which encodes a protein involved in adding a substitution on inner-core heptose, resulted in 40% reductions in biofilm (11). A possible explanation for this defect is that a component of the biofilm synthesis and export machinery interacts with the substituted heptose in the core. Similarly, it may be that in wild-type Yersinia spp., outer membrane proteins involved in EPS synthesis, processing, or export interact with core heptose or with a distal core component.
The Y. pestis gmhA mutant is severely defective for flea blockage (Fig. 4B). This defect is not due to an inability to colonize fleas, because the mutant colonized as many fleas as did the wild type (Fig. 4A) and grew in the insect to the same extent (Fig. 4C). Rather, the defect specifically involves proventriculus blockage. Significantly, a small fraction of fleas were blocked (Fig. 4B), just as a small fraction of C. elegans animals were inhibited from growth by ghmA mutant biofilms (Fig. 1). This suggests that in the flea, as on the nematode, the gmhA mutant is able to produce a small amount of EPS. These results also indicate the C. elegans biofilm closely mimics some aspects of flea blockage, further validating the nematode model.
Screening Y. pestis mutants directly in fleas to identify genes involved in blockage is impractical due to the slow growth of the insects, the need to dissect each flea, and the fact that only a fraction of infected fleas become blocked. On the other hand, screening for biofilm-defective Yersinia spp. by using C. elegans is efficient because the nematodes grow rapidly in a simple culture system, the biofilm is visible on the exterior of intact worms, and close to 100% of animals are affected when the biofilm is produced by Y. pseudotuberculosis.
Our results with in vitro biofilms (Fig. 5) suggest that biofilms grown on polystyrene are inferior to those produced on C. elegans as a model for the in vivo flea biofilm. When grown on polystyrene, the Y. pestis gmhA mutant has only a slight defect that does not rise to the level of statistical significance. Screening in this in vitro system therefore could fail to recover informative mutants. In contrast, identification of gmhA confirms that using the C. elegans biofilm assay is a practical means to find additional Y. pestis genes with roles in flea blockage.
Acknowledgments
We thank J. W. Hsu for assistance with transposon mutagenesis, C. Jarrett for help with flea infections, J. Mecsas for bacterial strains, and E. Joyce for a splendid technical suggestion.
L.T. and S.L.A. were supported by National Institutes of Health grant AI057512 to C.D. and by institutional development funds of the University of Alabama at Birmingham. This research was initiated in the laboratory of S. Falkow at Stanford University, where it was funded by the Defense Advanced Research Projects Agency and by National Institutes of Health grant AI026195.
Editor: J. B. Bliska
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooke, J. S., and M. A. Valvano. 1996. Biosynthesis of inner core lipopolysaccharide in enteric bacteria: identification and characterization of a conserved phosphoheptose isomerase. J. Biol. Chem. 271:3608-3614. [DOI] [PubMed] [Google Scholar]
- 4.Darby, C., J. W. Hsu, N. Ghori, and S. Falkow. 2002. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417:243-244. [DOI] [PubMed] [Google Scholar]
- 5.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fetherston, J. D., P. Schuetze, and R. D. Perry. 1992. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol. Microbiol. 6:2693-2704. [DOI] [PubMed] [Google Scholar]
- 8.Gemski, P., J. R. Lazere, T. Casey, and J. A. Wohlhieter. 1980. Presence of a virulence-associated plasmid in Yersinia pseudotuberculosis. Infect. Immun. 28:1044-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerke, C., A. Kraft, R. Sussmuth, O. Schweitzer, and F. Gotz. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273:18586-18593. [DOI] [PubMed] [Google Scholar]
- 10.Hinnebusch, B. J., R. D. Perry, and T. G. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367-370. [DOI] [PubMed] [Google Scholar]
- 11.Izquierdo, L., N. Abitiu, N. Coderch, B. Hita, S. Merino, R. Gavin, J. M. Tomas, and M. Regue. 2002. The inner-core lipopolysaccharide biosynthetic waaE gene: function and genetic distribution among some Enterobacteriaceae. Microbiology 148:3485-3496. [DOI] [PubMed] [Google Scholar]
- 12.Jarrett, C. O., E. Deak, K. E. Isherwood, P. C. Oyston, E. R. Fischer, A. R. Whitney, S. D. Kobayashi, F. R. DeLeo, and B. J. Hinnebusch. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Infect. Dis. 190:783-792. [DOI] [PubMed] [Google Scholar]
- 13.Joshua, G. W., A. V. Karlyshev, M. P. Smith, K. E. Isherwood, R. W. Titball, and B. W. Wren. 2003. A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology 149:3221-3229. [DOI] [PubMed] [Google Scholar]
- 14.Mecsas, J., I. Bilis, and S. Falkow. 2001. Identification of attenuated Yersinia pseudotuberculosis strains and characterization of an orogastric infection in BALB/c mice on day 5 postinfection by signature-tagged mutagenesis. Infect. Immun. 69:2779-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 16.Perry, R. D., M. L. Pendrak, and P. Schuetze. 1990. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J. Bacteriol. 172:5929-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prior, J. L., J. Parkhill, P. G. Hitchen, K. L. Mungall, K. Stevens, H. R. Morris, A. J. Reason, P. C. Oyston, A. Dell, B. W. Wren, and R. W. Titball. 2001. The failure of different strains of Yersinia pestis to produce lipopolysaccharide O-antigen under different growth conditions is due to mutations in the O-antigen gene cluster. FEMS Microbiol. Lett. 197:229-233. [DOI] [PubMed] [Google Scholar]
- 18.Reeves, P. R. 1994. Biosynthesis and assembly of lipopolysaccharide, p. 281-317. Elsevier, Amsterdam, The Netherlands.
- 19.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Simon, R., Y. Priefer, and A. Puhler. 1983. A broad host range mobilizing system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 21.Tan, L., and C. Darby. 2004. A movable surface: formation of Yersinia sp. biofilms on motile Caenorhabditis elegans. J. Bacteriol. 186:5087-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor, R. K., C. Manoil, and J. J. Mekalanos. 1989. Broad-host-range vectors for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J. Bacteriol. 171:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 24.Vinogradov, E. V., B. Lindner, N. A. Kocharova, S. N. Senchenkova, A. S. Shashkov, Y. A. Knirel, O. Holst, T. A. Gremyakova, R. Z. Shaikhutdinova, and A. P. Anisimov. 2002. The core structure of the lipopolysaccharide from the causative agent of plague, Yersinia pestis. Carbohydr Res. 337:775-777. [DOI] [PubMed] [Google Scholar]
- 25.Wood, W. B. 1988. The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory, Plainview, N.Y.
- 26.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]